Abstract
To characterize proteins associated with active transcription complexes, we purified RNA polymerase II (pol II) from Saccharomyces cerevisiae after fixing live cells with formaldehyde. The approach mimics ChIP and requires solubilizing cross-linked complexes with sonication. Pol II was affinity-purified, and associated proteins were identified by MS. Several classes of proteins depended on cross-linking, including Mediator, general transcription factors, elongation factors, ribonucleoprotein particle (RNP) proteins, and histones. A tagged RNP protein reciprocally purified pol II under identical cross-linking conditions, and the association between RNP proteins and pol II was largely RNase-sensitive. The data indicate that the cross-linked Pol II purification contains elongating pol II with associated nascent RNP. Consistent with this view, some elongation factors no longer associate with pol II after inactivation of transcription in the temperature-sensitive pol II mutant, rpb1-1. Taken together, our data suggest that the cross-linked pol II purification contains a mixed population of pol II, including initiating pol II and elongating pol II.
Keywords: mass spectrometry, nascent RNP, RNA processing, transcription, affinity purification
Purification and mass spectrometric analysis of protein complexes is a ubiquitous approach in modern molecular biology (1). The identification of interacting proteins and their subsequent characterization provides valuable insight into the complexes that carry out essential cellular functions. Purifications can use multiple biochemical fractionations (e.g., ion exchange, gradients, gel filtration, and affinity purification) or a streamlined tandem affinity purification (TAP) approach (2). Conventional purification methods, however, often preclude the accurate identification of in vivo-relevant complexes. Major complications include dissociation of protein complexes, as a function of off-rates and extract dilution. Moreover, strong affinities can promote in vitro associations that do not exist in vivo. These can occur as a consequence of protein exchange and the absence of subcellular localization. Finally, major complexes may be insoluble, or harsh methods required to effect solubility can perturb complex composition. Ribonucleoprotein particles (RNPs) and especially transcription complexes are particularly subject to these caveats.
The synthesis of protein-encoding mRNAs is directed by RNA polymerase II (pol II), an extensively studied macromolecular machine. The composition of pol II is dynamic and changes with the stage of transcription, namely, initiation, elongation, and termination (3, 4). Difficulties with complex purification underlie controversies, for example, to what extent initiation occurs by stepwise assembly of a preinitiation complex on DNA. Moreover, elongating pol II is tightly associated with DNA, which makes its characterization challenging (5).
ChIP is a major tool for characterizing DNA-associated proteins and complexes, including transcription and cotranscriptional RNA processing (6, 7). Because ChIP uses in vivo formaldehyde cross-linking, it avoids many of the drawbacks of extract formation with more normal protocols and covalently links many proteins to their position along the DNA. Moreover, cross-linking of multisubunit complexes may be very efficient because of the large number of intracomplex contacts. Efficient cross-linking is suggested by studies of transcriptional activators and the proteosome (8, 9) and the fact that numerous members of a given complex generally provide comparable ChIP signals.
Nonetheless, ChIP has some drawbacks. It is a single-candidate protein assay that is then labor-intensive to assay large numbers of factors. A subtle orientation or flexibility problem with a key lysine epsilon amino group may give rise to a false negative result. For example, the addition of protein–protein cross-linkers with longer spacer arms can enhance ChIP signal from proteins more distal from DNA (10). Moreover, cross-linking can reduce binding to affinity resins. It also causes high background binding, which necessitates washing under high stringency conditions that further lower signals. Therefore, most tags suffer poor yields under these conditions, which is why quantitative PCR is a key feature of ChIP. However, this difficulty can be circumvented by using a modified tandem affinity tag system based on a biotinylated peptide flanked by two hexahistidine tags [hexahistidine-biotin-hexahistidine (HBH) tag]. This tag provides sufficiently strong affinities to accommodate the high stringency conditions that normally reduce signal and has been used to purify in vivo cross-linked complexes for MS, including the proteasome (9, 11, 12). The 75-aa peptide in the HBH tag is recognized and efficiently biotinylated in vivo by endogenous biotin ligases in yeast and mammalian systems.
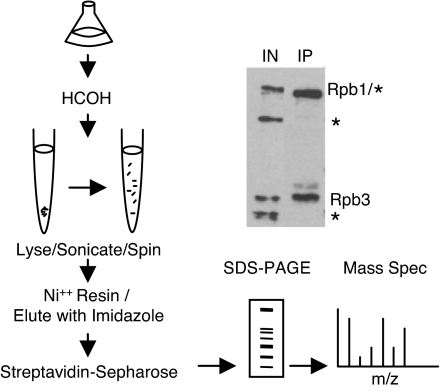
To investigate the distribution of in vivo transcription complexes, we have purified pol II by using a modified version of the published HBH-based purification. This scheme included binding sonicated lysates to nickel resin, eluting complexes with imidazole, and then binding them to streptavidin-Sepharose under high stringency conditions [Fig. 1 and supporting information (SI) Fig. 5]. Purified complexes can then be characterized by MS. Importantly and consistent with work by others (9, 11, 13), the reversed formaldehyde cross-links appear not to interfere with standard MS peptide identification.
Fig. 1.
Schematic of cross-linked pol II purification. Cells expressing HBH-tagged pol II were cross-linked, lysed, sonicated, and affinity-purified over consecutive nickel and streptavidin resins. SDS/PAGE-resolved proteins were then identified by LC-MS/MS. A Western blot for biotinylated proteins shows the specific selection of pol II over endogenous biotinylated proteins (*). IN, input.
Following this strategy, we characterized proteins associated with purified HBH-tagged pol II from Saccharomyces cerevisiae. Several categories of proteins depended on cross-linking, and the data indicate that the cross-linked pol II purification contains elongating pol II containing nascent messenger RNP (mRNP). Consistent with this view, a copurifying RNP protein was tagged and shown to purify pol II only under the same cross-linking conditions; moreover, these associations were RNase-dependent. The temperature-sensitive RNA polymerase mutant rpb1-1 showed that selected elongation factors associate with pol II in a transcription-dependent manner. Taken together, our results suggest that the cross-linked pol II purification contains initiating pol II and an elongating pol II fraction.
Results
Cross-Linked Pol II Contains Active Transcription Complexes.
To characterize the protein composition of pol II complexes with cross-linking, we introduced the HBH tag into two pol II subunits (Rpb1p and Rpb3p) at the chromosomal loci in the same strain. We considered that tagging two proteins in the same complex might improve recovery and mitigate epitope masking by the large size of pol II complexes and cross-linking. Indeed, this strategy modestly increased yields, especially of the first purification step on a histidine affinity matrix, and was therefore used for all subsequent purifications (data not shown). We cross-linked and purified double-tagged pol II complexes with a modified two-step method (histidine affinity matrix followed by streptavidin Sepharose) and achieved ≈20% recovery of pol II by Western blotting. This assay also indicated that the first purification step significantly reduced copurifying the major yeast biotinylated proteins on streptavidin-Sepharose (Fig. 1).
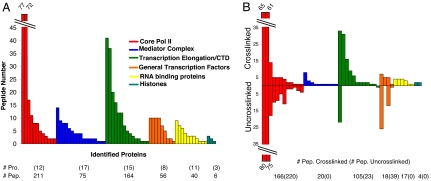
Pol II-associated proteins were identified by tandem MS. Proteins that were identified in a side-by-side purification with untagged pol II and common contaminants were removed from the list of candidates. Approximately 70 proteins (545 peptides) specifically copurified with pol II under these conditions (Fig. 2A and SI Table 1). We provisionally defined “specific” as those proteins previously identified as associated with active transcription complexes or with an assigned role in transcription or RNA processing. We also required two or more identified peptides or the presence of other members of a well characterized protein complex.
Fig. 2.
Cross-linking enriches the composition of purified pol II. (A) Cross-linked purification of pol II. Copurifying proteins are grouped based on subcomplexes or function. The y axis is the number of peptides identified for each protein (x axis). The number of proteins (Pro.) and peptides (Pep.) are indicated below each color-coded category. (B) Comparison of pol II purified with (Upper) and without (Lower) cross-linking. The number of identified peptides is located below each group.
The first group of proteins includes all core pol II subunits as expected (Fig. 2A and SI Table 1, red). The number of peptides identified from each pol II protein, and from the subunits of other protein complexes, is roughly proportional to molecular weight, i.e., the larger subunits have more identified peptides. The second identified pol II-associated complex is Mediator (Fig. 2A and SI Table 1, blue), with most subunits identified. The third, large category of proteins functions in transcriptional elongation and/or is associated with the pol II C-terminal domain (CTD) (Fig. 2A and SI Table 1, green). Many of these proteins are individual members of characterized protein complexes, e.g., the PAF and FACT complexes. Two enzymes involved in 5′ capping of pol II transcripts, Ceg1p and Cet1p, are also present. The fourth group is general transcription factors (GTFs; Fig. 2A and SI Table 1, orange), including TFIIB, TFIIF (Tfg1, Tfg-2, Tfg-3), TFIIS (Dst1p), TFIIE, and TFIIA. The fifth group contains RNP or 3′ end formation factors, presumably associated with nascent RNA (Fig. 2A, yellow), and the final group contains histones (Fig. 2A, teal).
Because the proteins associated with cross-linked pol II are significantly different from those in the literature obtained by more traditional purification strategies, we verified that this difference is caused by in vivo formaldehyde cross-linking rather than some other idiosyncrasy of our doubly tagged yeast strain or purification. To this end, we repeated the purification but also purified pol II in parallel with a standard extract protocol without formaldehyde. Similar quantities of pol II were subject to MS from the two extracts.
Although somewhat fewer peptides were identified from this second cross-linked preparation (compare the number of core pol II and Mediator peptides in the cross-linked column of SI Table 2 with the number in SI Table 1), most of the same proteins and multiprotein factors were identified. This finding indicates that the in vivo association of many of these complexes and factors with pol II is reproducible. This experiment also identified many fewer proteins and complexes associated with pol II without cross-linking (Fig. 2B and SI Table 2), which resembled more closely several previously published results (14–17).
In the category of Elongation/CTD proteins, for example, only the elongation protein Spt5p was identified in the absence of cross-linking (Fig. 2B and SI Table 2). Spt6p was not identified, despite comparable numbers of peptides in the cross-linked samples (SI Tables 1 and 2). The standard purification failed to identify a single Mediator peptide despite containing all of the core pol II subunits (Fig. 2B and SI Table 2). A different mix of GTFs was identified without cross-linking, with some factors and subunits missing (TFIIA, TFIIB) and others overrepresented (TFIIF) relative to the cross-linked sample (Fig. 2B and SI Table 2). Finally, no RNP proteins or histones were identified. Although few histone peptides were identified in the parallel, cross-linked samples, the negative result is consistent with the literature as these proteins have not been identified in previous pol II purifications. We conclude from this side-by-side comparison that cross-linking can maintain the complex integrity or in vivo associations that are otherwise invisible by conventional purification approaches.
Purification of Nascent RNP Complexes.
Well studied RNP proteins such as Yra1p, Npl3p, and Sub2p (Fig. 2 and SI Tables 1 and 2) have been shown to associate with active genes by ChIP (18–20). Their presence in the set of cross-linked proteins (SI Tables 1 and 2) supports the notion that some fraction of the purified pol II is actively engaged in transcription. Because the RNP protein Yra2p is in this set and had not been previously to be characterized with respect to transcription, we verified that it, too, is associated with an active gene by ChIP (SI Fig. 6).
We then assayed whether the association between pol II and RNP proteins could be validated by the opposite strategy, namely, would purification of an RNA binding protein identify the transcription machinery? To this end, we HBH-tagged and purified Npl3p from cross-linked cells; purity and yield were similar to the pol II purifications.
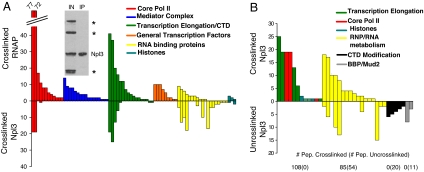
As predicted, many peptides from the two largest core pol II subunits, Rpb1p and Rpb2p, copurified with Npl3p-HBH. Also present were several of the elongation proteins identified in the pol II purification, e.g., Spt5p and Spt6p, and FACT and PAF complex proteins and histones (SI Table 3). This composition appears to reflect a subset of the transcription complexes purified with tagged pol II, which is evident from Fig. 3, where the lack of blue and orange proteins reflects the absence of mediator and GTFs. Moreover, the number of pol II proteins and peptides is much lower in the Npl3p purification than in the pol II purification, suggesting that the former recovers only a small fraction of pol II-containing complexes. Conversely, the Npl3p-HBH is much more enriched in RNA-binding proteins than the pol II purification (compare SI Table 3 with SI Table 1; note the larger number of yellow proteins in Fig. 3A). This finding is consistent with an association of Npl3p with non-nascent and nascent RNA.
Fig. 3.
Cross-linking increases association of Npl3p with transcription machinery. (A) Comparison of cross-linked Pol II (Upper) and cross-linked Npl3p (Lower) purifications. Protein categories are color-coded as in Fig. 2. (Inset) Western blot shows specific purification of Npl3p-HBH with endogenous biotinylated proteins marked by *. (B) Comparison of cross-linked (Upper) and uncross-linked (Lower) Npl3p-HBH. The number of identified peptides is located below the corresponding group.
Because copurification of transcription-related proteins with Npl3p has not been reported (21), we purified Npl3p-HBH without cross-linking (SI Table 3). These results were much more similar to those of Hurt et al. (21) with no detectable purification of transcription proteins (Fig. 3B and SI Table 3; note the absence of red and blue proteins in the bottom uncross-linked sample). In addition to the many RNP proteins, there were CTD kinases in the uncross-linked purification, which is consistent with a previous report (17). There was also a unique association with BBP and Mud2, which bind the branch point region of intron-containing genes (22–24). Interestingly, these two splicing proteins, Air2p, and the CTD kinases were not detected in the cross-linked purification. This difference may reflect inefficient protein–protein cross-linking for some proteins and complexes (see Discussion).
Verification of Transcription-Dependent Interactions.
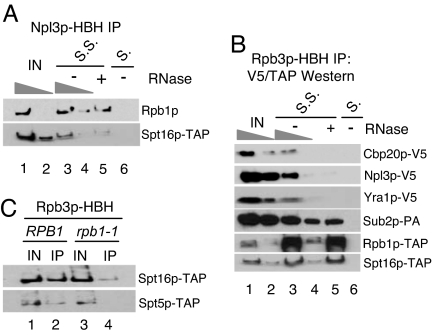
We presume that the association of transcription complexes with Npl3p and RNP proteins more generally is through their association with nascent RNA. To test this possibility, we immunoprecipitated Npl3p-HBH, treated beads with RNase or buffer alone, and Western-blotted for either Rpb1p or Spt16p-TAP. Both proteins are ≈40–60% RNase-sensitive (Fig. 4A, compare lanes 3 and 5). The lack of greater RNase sensitivity may be caused by direct cross-linking between these transcription proteins and Npl3p or poor RNase digestion of nascent RNA after cross-linking. Another possibility is that there are multiple Npl3p proteins associated with each nascent RNA. In this case, a substantial fraction of RNase-resistant Rpb1p and Spt16p might obscure a much smaller fraction of RNase-resistant Npl3p. This situation predicts that a larger percentage of RNP protein associated with transcription complexes may be sensitive to RNase in the reciprocal immunoprecipitation (IP). To this end, we immunoprecipitated pol II via Rpb3p-HBH with and without RNase treatment and blotted for epitope-tagged RNP proteins (Fig. 4B, compare lanes 3 and 5). Indeed, all RNP proteins tested (Cbp20p, Npl3p, Yra1p, and Sub2p) were ≈80% RNase-sensitive. In contrast, the association of Rpb1p-TAP and Spt16p-TAP were RNase-resistant under identical conditions, indicating that other pol II subunits and some transcription factors are cross-linked to Rpb3p-HBH via direct or indirect protein–protein and/or protein–DNA contacts.
Fig. 4.
Co-IPs confirm transcription-dependent associations. (A) Rpb1p and Spt16p are partially RNase-sensitive when immunoprecipitated with Npl3p-HBH. Five-fold dilutions of the −RNase are shown. Extracts were applied to Streptavidin-Sepharose (S.S.) and empty Sepharose (S.) as a negative control. (B) RNP proteins are highly RNase-sensitive. Rpb3p-HBH was immunoprecipitated and blotted for RNP or transcription proteins. (C) Spt5p and Spt16p associate with pol II in a transcription-dependent manner. RPB1 or rpb1-1 cells were shifted to 37°C for 1 h, and cross-linked complexes were immunopreciptated with Rpb3-HBH and blotted for Spt5-TAP and Spt16-TAP. IN, input.
The ability to purify dynamic transcription complexes suggests that copurification of some elongation factors with pol II should be transcription-dependent. To test this idea directly, we monitored the association of Spt5p and Spt16p with pol II by co-IP and Western blotting in a strain harboring an RPB1 ts allele (rpb1-1). The rpb1-1 allele confers temperature sensitivity and causes pol II to dissociate from active genes after a shift to the nonpermissive temperature (25, 26). Importantly, there is not an appreciable decrease in Rpb1p levels after this temperature shift (data not shown).
There is a substantial decrease in the association of both Spt5p-TAP and Spt16p-TAP with pol II after a 1-h shift to 37°C in the rpb1-1 strain compared with a 1-h shift in a WT strain (Fig. 3C). This finding suggests that both proteins associate with active pol II, but only the association of Spt5p is sufficiently strong to survive conventional purification or can occur after extract formation (see Discussion).
Discussion
To begin a characterization of proteins associated with active transcription, we purified tagged pol II from S. cerevisiae after fixing cells with formaldehyde. Several categories of transcription-relevant proteins were copurified only after cross-linking. Based on this data set, we suggest that a substantial fraction of the cross-linked pol II preparation contains elongating pol II complexes. This notion is consistent with estimates that ≈60–70% of pol II is engaged in transcription (27, 28). The dependence on cross-linking is based on the restricted set of elongation factors and the lack of pol II-associated RNP proteins and histones without cross-linking (Fig. 2B and SI Table 2), consistent with other reports (14–17). Moreover, copurification of RNP proteins is RNase-sensitive, indicating it is associated with nascent RNA. Finally, the reciprocal purification of the RNP protein Npl3p after cross-linking captures proteins engaged in active transcriptional elongation, including pol II.
The copurification of many Mediator and GTF proteins indicates that cross-linked pol II contains initiating pol II complexes. Some of these proteins are undetectable in the Npl3p purification, suggesting that they represent a specific fraction of pol II distinct from the elongating pol II with nascent mRNP. Moreover, there is no evidence of Mediator and specific GTFs associated with noncross-linked pol II (Fig. 2B and SI Table 2), which indicates that initiating and/or holoenzyme complexes are stabilized by cross-linking. Moreover, the presence of TFIIA and TFIIB, which are not holoenzyme components yet associate with initiation complexes (4), further indicates the presence of initiating pol II in our purification. The presence of the 5′ capping machinery is also consistent with the presence of initiation or early elongation complexes; Ceg1p and Cet1p localize to promoter proximal regions and their association depends on phosphorylation of Ser-5 of the CTD of Rpb1p (29).
The sonication step of this protocol may also solubilize a more abundant pool of initiating pol II and elongating pol II complexes. Their remarkable stability (5) suggests that they may be largely insoluble and poorly recovered with a more conventional protocol. Moreover, the absence of DNase treatment suggests that some proteins may interact only indirectly with pol II, e.g., with DNA adjacent to a pol II molecule; GTFs and histones are likely candidates for this type of indirect interaction.
Spt5p and the more limited complement of GTFs associated with pol II in the absence of cross-linking could reflect specific pol II subcomplexes that remain intact without cross-linking. Indeed, the reported copurification of Spt5, TFIIF, and TFIIS, but not Mediator, with pol II supports a bona fide complex (30). Also consistent with our data is the observation in this same study that Spt16p and Pob3p (FACT complex components) only immunoprecipitated with hyperphosphorylated pol II, indicating a specific association with elongating pol II (20). However, some of these reported associations with pol II could form after extract formation. A virtue of in vivo cross-linking is that it may prevent protein–protein associations from taking place in vitro and freezing associations that occur in vivo.
Nonetheless, the lack of further fractionation of the fixed pol II complexes reported here makes it impossible to distinguish between different interpretations, for example, the presence of Mediator and GTFs within a pol II holoenzyme rather than the presence of an initiating pol II–Mediator fraction plus a promoter-associated GTF–pol II fraction (31–35). Moreover, genomewide profiling indicates that Mediator can associate with gene-coding regions, suggesting that a portion of copurifying Mediator may even be in elongating pol II complexes (36, 37). Purification of a tagged Mediator subunit under cross-linking conditions might distinguish between some of these possibilities.
It is tempting to correlate the pol II protein complexes with ChIP results, which is one of the reasons for considering an initiating pol II fraction, because most studies report that Mediator and GTFs primarily coimmunoprecipitate to upstream DNA and promoters, respectively (34, 35). However, Mediator is reported not to interact by ChIP with some actively transcribed yeast genes, which may reflect some gene-specific dependence on Mediator (38). Another possibility is that ChIP capture after in vivo formaldehyde incubation may miss interactions on some genes that are not oriented properly for covalent cross-linking of a protein to DNA or RNA. Indeed it is possible inefficient cross-linking also explains the absence of splicing proteins associated with pol II. We had expected to find them associated with elongating pol II based on their association with DNA by ChIP (39–41), and Reed and colleagues (42) purified U1–pol II complexes from mammalian systems. This difference more likely reflects the low frequency of intron-containing transcription units in S. cerevisiae and the inefficiency of MS relative to the PCR-based ChIP assay.
It is possible that protein–protein interactions within some multiprotein transcription complexes are more efficiently formaldehyde cross-linked than protein–DNA complexes (8), e.g., most Mediator–pol II complexes may not be DNA-associated. Nonetheless, some protein–protein cross-linking may be inefficient for the same orientation or proximity reasons that can affect protein–DNA cross-linking, which might explain the absence of expected pol II-associated proteins, for example, Elongator, TFIIH, and TFIID. The stringent washing conditions required by the cross-linking protocol may make this problem more severe and further reduce the levels of associated but noncovalently linked proteins, which could explain why Npl3p associates with the CTD kinases, Air2p, and the early splicing proteins BBPp and Mud2p only in the absence of cross-linking. The association of these splicing proteins with Npl3p may influence the robust cotranscriptional association of U1 small nuclear RNP and associated factors with intron-containing pre-mRNAs (39–41). Alternatively, the association may occur only in vitro after extract formation.
Cross-linking generally increases the number of proteins and protein complexes associated with Npl3p. For example, our uncross-linked purification fails to reveal an association between Npl3p and some TREX (transcription/export) complex components, consistent with the literature (43). There is, however, a cross-linking-dependent association of Npl3p with four TREX components (Yra1p, Sub2p, Tho2p, and Hrb1p), suggesting that Npl3p interacts directly or indirectly with TREX on nascent transcripts (Fig. 3B and SI Table 4). Nascent mRNP is presumably too insoluble or the association too labile without sonication or in vivo cross-linking, respectively. Cross-linking may also provide a more accurate picture of in vivo non-nascent mRNA populations associated with specific RNP proteins (44, 45).
Cross-linking also increases the detection of many proteins and protein complexes with pol II. For example, RNP proteins, histones, and multiple elongation factors are not detectable without cross-linking (Fig. 2B and SI Table 2), suggesting that in vivo cross-linking maintains protein associations with DNA and nascent RNA and pol II. The increase is despite very straightforward purification and MS protocols; indeed, we typically used 4–8 liters of midlog cells with a modified TAP and a standard liquid chromatography (LC)-MS/MS approach from the Taplin MS Facility, Harvard Medical School, Boston (Steve Gygi). Our results suggest that improvements could easily be made to increase sensitivity and identify more peptides and proteins. Taken together with a greater use of mutants or physiological manipulations that affect transcription or RNA processing, and with further fractionation of pol II complexes, this cross-linking–MS approach should be able to provide more insights into active transcription and its regulation. With a large number of highly active genes per cell (high copy plasmids and/or a large number of gene copies in mammalian cells), a variation on this tagging and cross-linking theme might even be able to characterize the protein composition of a specific gene.
Materials and Methods
Strain Construction.
HBH tags were integrated at chromosomal loci in haploid W303 yeast by standard PCR-based homologous recombination (11, 46). A strain list is in SI Table 4. Tag integration and expression was confirmed by PCR and Western blotting with streptavidin-HRP (Amersham). Strains for co-IP studies were generated by crossing an RPB3-HBH strain with either TAP-tagged or V5-tagged strains of the opposite mating type. The resulting meiotic segregants were genotyped, and tags were confirmed by Western blotting.
Purification of Cross-Linked Complexes.
Purifications were performed with between 4 and 16 liters of cells. For large-scale purifications, multiple 2-liter chromatin preparations were performed. For a single 2-liter culture, cells were grown to an OD600 of ≈1.0–1.5 in yeast extract/peptone/dextrose. Growth media were supplemented with 6 μM biotin (Sigma) to ensure biotin was not limiting for biotinylation of the HBH tag. Two-liter cultures were cross-linked with 200 ml of 11% formaldehyde for 20 min and quenched with 300 ml of Tris-glycine (10 mM–2.5 M) for 5 min. Cultures were centrifuged for 5 min at 3,000 rpm in a Sorvall G-3 rotor. Cell pellets were resuspended and washed twice with ice-cold 1× TBS and once with cold FA lysis buffer [50 mM Hepes-KOH (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 1% Triton, 0.1% NaDoc, 0.1% SDS]. Cell pellets were frozen and stored at −80°C. Pellets were resuspended in 8 ml of FA lysis buffer and lysed by bead beating (MiniBead Beater 8; BioSpec Products) with glass beads for 2 min. Glass beads were removed, and lysates were centrifuged in 1-ml aliquots at 14,000 rpm. The supernatant was removed, the cell pellet was washed with 1 ml of FA lysis buffer, and the lysate was centrifuged. The cell pellet was then resuspended in 8 ml of FA lysis buffer and split into two 15-ml Falcon tubes. Each lysate was sonicated with a 550 Sonic Dismembranator (Fisher Scientific) for 2 min, 40 s at setting 3 (20 s on, 20 s off). Sonicated lysates were transferred to Eppendorf tubes and centrifuged at 3,000 rpm for 5 min and then at 10,000 rpm for an additional 5 min. The chromatin-containing supernatant was removed and stored at −80°C.
Lysates were adjusted to 10 mM imidazole, added to 200 μl of washed Ni++ resin (Amersham), and rotated overnight at 4°C. Resin was washed three times for 5 min with 10 ml of FA lysis buffer supplemented with 10 mM imidazole. Complexes were eluted for 5 min with 1 ml of FA lysis buffer supplemented with 150 mM imidazole. Eluates from multiple IPs were combined and added to washed streptavidin-Sepharose (100 μl of slurry; Amersham) for 6 h at 4°C. Beads were washed three times for 5 min with 10 ml of FA lysis buffer with 750 mM NaCl and increasing SDS concentration (0.1%, 0.2%, and 0.5%). Washed beads were then transferred to a 1.5-ml Eppendorf tube and boiled for 10 min with SDS loading buffer (≈100 μl) containing 50 mM Tris·Cl, pH 7.5.
For MS, decross-linked complexes were loaded onto 4–20% or 4–12% SDS/PAGE gels and stained with Coomassie G-250. Gel lanes were excised in between 2 and 10 slices of ≈1 cm by 1 cm. Gel slices were sent to the Taplin Biological MS Facility for MS analysis.
MS.
MS was performed by the Taplin Biological Mass Spectrometry Facility with an LTQ linear ion-trap mass spectrometer (ThermoFinnigan). Excised gel bands were subjected to a modified in-gel trypsin digestion procedure (47). Peptides were subjected to electrospray ionization before entering an LTQ linear ion-trap mass spectrometer (ThermoFinnigan). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences and protein identity were determined by matching protein or translated nucleotide databases with the acquired fragmentation pattern by the software program Sequest (ThermoFinnigan) (48).
Co-IP Experiments.
Co-IP experiments were performed with RPB3-HBH or NPL3-HBH strains with additional TAP- or V5-tagged proteins (SI Table 4). Cultures were grown to an OD600 of ≈0.6–1.0 and cross-linked according to a previously modified protocol (20). Sonicated lysates were applied to either streptavidin-Sepharose (Amersham) or Sepharose CL-4B (Sigma) and incubated overnight at 4°C. For RNase experiments, beads were washed in 1× FA lysis buffer (275 mM NaCl), 2× high-salt FA lysis buffer (500 mM NaCl), and 1× low-salt FA lysis buffer (150 mM NaCl). One milliliter of FA lysis buffer (150 mM NaCl) supplemented with 7.5 units of RNase A and 300 units of RNase T1 (RNase A/T1 Mixture; Ambion) or an equivalent volume of RNase storage buffer [10 mM Hepes (pH 7.2), 20 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, and 50% glycerol (vol/vol)] was added to the beads and incubated with rotation at room temperature for 30 min. RNase-treated beads were then washed in 1× FA (275 mM NaCl) and 1× FA (500 mM NaCl).
Input and immunoprecipitated samples were resolved by SDS/PAGE and proteins were detected by standard Western blot analysis. TAP- or V5-tagged proteins were detected with peroxidase-antiperoxidase (1:5,000; Sigma) or anti-V5 antibody (1:2,000; Invitrogen), respectively, using Tris-buffered saline Tween-20 and 5% milk.
Supplementary Material
Acknowledgments
We thank P. Kaiser (University of California, Irvine) for the HBH plasmids; the laboratories of F. Stutz (University of Geneva, Geneva) and D. Bentley (University of Colorado, Boulder) for yeast strains; S. Gygi for the use of the Taplin Biological Mass Spectrometry Facility; and J. Agar, D. Rio, M. Marr, K. Struhl, R. Kornberg, and M. Green for helpful discussions and comments on the manuscript. This work was supported in part by National Institutes of Healtha Grant R01 GM23549 (to M.R.) and National Institutes of Health Genetics Training Grant GM07122 (to Brandeis University).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710179104/DC1.
References
- 1.Gingras AC, Gstaiger M, Raught B, Aebersold R. Nat Rev. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 2.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 3.Sims RJ, III, Belotserkovskaya R, Reinberg D. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 4.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 5.Wuarin J, Schibler U. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlando V. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 7.Keogh MC, Buratowski S. Methods Mol Biol. 2004;257:1–16. doi: 10.1385/1-59259-750-5:001. [DOI] [PubMed] [Google Scholar]
- 8.Hall DB, Struhl K. J Biol Chem. 2002;277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 9.Guerrero C, Tagwerker C, Kaiser P, Huang L. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Kurdistani SK, Grunstein M. Methods. 2003;31:90–95. doi: 10.1016/s1046-2023(03)00092-6. [DOI] [PubMed] [Google Scholar]
- 11.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Tagwerker C, Zhang H, Wang X, Larsen LS, Lathrop RH, Hatfield GW, Auer B, Huang L, Kaiser P. Yeast. 2006;23:623–632. doi: 10.1002/yea.1380. [DOI] [PubMed] [Google Scholar]
- 13.Vasilescu J, Guo X, Kast J. Proteomics. 2004;4:3845–3854. doi: 10.1002/pmic.200400856. [DOI] [PubMed] [Google Scholar]
- 14.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 15.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 16.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 18.Lei EP, Krebber H, Silver PA. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei EP, Silver PA. Genes Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abruzzi KC, Lacadie S, Rosbash M. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Proc Natl Acad Sci USA. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abovich N, Liao XC, Rosbash M. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 23.Abovich N, Rosbash M. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 24.Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 25.Schroeder SC, Schwer B, Shuman S, Bentley D. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonet M, Scafe C, Sexton J, Young R. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Struhl K. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 28.Svejstrup JQ, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg RD. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komarnitsky P, Cho EJ, Buratowski S. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, III, Hartzog GA. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 32.Koleske AJ, Young RA. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 33.Thompson CM, Koleske AJ, Chao DM, Young RA. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 34.Kuras L, Borggrefe T, Kornberg RD. Proc Natl Acad Sci USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pokholok DK, Hannett NM, Young RA. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 36.Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 38.Fan X, Chou DM, Struhl K. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 39.Tardiff DF, Lacadie SA, Rosbash M. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lacadie SA, Rosbash M. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 43.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 44.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 45.Vasudevan S, Steitz JA. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 48.Eng JK, McCormick A, Yates JR., III J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.