Abstract
Alloreactive memory T cells may be refractory to many of the tolerance-inducing strategies that are effective against naive T cells and thus present a significant barrier to long-term allograft survival. Because CD4+CD25+ regulatory T cells (Tregs) are critical elements of many approaches to successful induction/maintenance of transplantation tolerance, we used MHC class I and II alloreactive TCR-transgenic models to explore the ability of antigen-specific Tregs to control antigen-specific memory T cell responses. Upon coadoptive transfer into RAG-1−/− mice, we found that Tregs effectively suppressed the ability of naive T cells to reject skin grafts, but neither antigen-unprimed nor antigen-primed Tregs suppressed rejection by memory T cells. Interestingly, different mechanisms appeared to be active in the ability of Tregs to control naive T cell-mediated graft rejection in the class II versus class I alloreactive models. In the former case, we observed decreased early expansion of effector cells in lymphoid tissue. In contrast, in the class I model, an effect of Tregs on early proliferation and expansion was not observed. However, at a late time point, significant differences in cell numbers were seen, suggesting effects on responding T cell survival. Overall, these data indicate that the relative resistance of both CD4+ and CD8+ alloreactive memory T cells to regulation may mediate resistance to tolerance induction seen in hosts with preexisting alloantigen-specific immunity and further indicate the multiplicity of mechanisms by which Tregs may control alloimmune responses in vivo.
Keywords: suppression, tolerance, transplantation
Donor-specific tolerance, which is readily achieved by many means in inbred rodent models, has been difficult to reproduce in outbred rodents and primates. Memory T cells, derived from prior exposure to alloantigen or generated by heterologous immunity or lymphopenia-induced proliferation, are believed to be an important part of this barrier (1–3). Compared with naive T cells, memory T cells respond more rapidly after TCR activation, respond to lower concentrations of antigen, and are less dependent on CD28 costimulation (4, 5). Additionally, they are relatively long-lived in the periphery and can mount immune responses within parenchymal tissues directly without a need for prior priming in lymphoid organs (4–9).
Deletion and regulation play key roles in the induction and maintenance of tolerance (10). Although both CD4+ and CD8+ memory T cells are relatively resistant to apoptosis and antibody-mediated deletional therapy (7, 8, 11, 12), our knowledge about the susceptibility of memory T cells to regulation in the context of transplantation is incomplete. Naturally occurring CD4+CD25+ regulatory T cells (Tregs) are an extensively characterized cell type within the Treg pool and are required for transplantation tolerance in many model systems (13, 14). Such Tregs have been demonstrated to have multiple effects on T cell responses to alloantigen, including suppression of proliferation, cytokine production, survival, and alloreactive T cell migration to the graft (15–18). However, the behavior of Tregs in the context of transplantation has primarily been investigated in models using inbred nonsensitized mice. In the single study that examined Tregs and memory cells, it was reported that polyclonal CD4+CD25+ Tregs from donor splenocyte transfusion (DST)-treated animals, but not from unmanipulated mice, were capable of controlling graft rejection mediated by memory T cells in splenectomized alymphopenic mice (19).
Here we used alloreactive CD4 and CD8 TCR-transgenic (Tg) mice to investigate the issue of regulation of memory T cell responses in transplantation by using systems where alloantigen-reactive T cells can be identified and the number, specificity, and affinity for alloantigen are defined and controlled. We show that, unlike naive T cells, alloreactive memory T cells are comparatively resistant to regulation because neither CD4+ nor CD8+ memory T cells could be prevented from rejecting skin allografts by CD4+CD25+ Tregs. Consistent with this, memory CD4+ and CD8+ T cells are less susceptible in vitro and in vivo to inhibition of proliferation, cytokine production, and survival after alloantigen challenge. These data suggest that part of the barrier to tolerance induction presented by memory cells may be mediated by resistance to regulation.
Results and Discussion
Experimental Models.
To study the ability of Tregs to control alloreactive memory T cell responses, we used two lines of well defined alloreactive TCR-Tg mice: ABM and BM3 mice. ABM mice are on a B6 background and carry TCR transgenes, which encode specificity for the MHC class II molecule I-Abm12 (20). Antigen-reactive cells are CD4+Vα2+Vβ8+, comprise ≈90–95% of the peripheral repertoire of adult mice, and are sufficient to induce bm12 skin graft rejection upon adoptive transfer into immunodeficient mice (20). Moreover, we have shown recently that ABM CD4+Vα2+Vβ8+CD25+ cells express foxp3 and exhibit regulatory function in vitro and in vivo, which is specific for I-Abm12 (13), thereby enabling us to investigate the activity of a clonal population of Tregs with known antigen-specificity.
BM3.RAG-1−/− (H2k) carry TCR transgenes that encode reactivity for the MHC class I molecule H2Kb (21). BM3 CD8+ T cells can be identified by using the anticlonotypic mAb, Ti98 (22), and acutely reject H2Kb+ skin allografts when adoptively transferred into immunodeficient mice (23). BM3 T cells do not express foxp3 and, therefore, do not possess intrinsic regulatory activity (data not shown).
Together this body of work indicates the suitability of each of these models for our studies to compare the ability of clonal and polyclonal populations of CD4+CD25+foxp3+ Tregs to control rejection initiated by naive and memory direct pathway CD4+ and CD8+ T cells, where the specificity and affinity of naive and memory T cells were defined.
Skin Graft Rejection Mediated by Memory T Cells Is Relatively Resistant to Regulation.
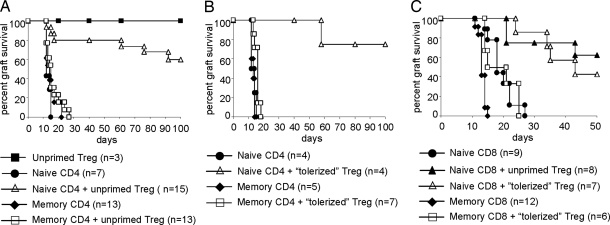
We used skin transplantation models with the previously mentioned alloreactive, TCR-Tg CD4+ and CD8+ T cells to test the susceptibility of memory T cells to regulation in vivo. Naive or memory CD4+ or CD8+ TCR-Tg T cells (defined respectively as CD44− cells from unmanipulated mice and CD44+ T cells from mice that rejected the antigen-specific allograft) (see Materials and Methods), together with CD4+CD25+ Tregs from unmanipulated mice, were coadoptively transferred into congenic B6 RAG-1−/− or CBA RAG-1−/− animals, which then received skin allografts 1 day later (Fig. 1). Consistent with previous studies (23), naive ABM and BM3 T cells were able to effect prompt graft rejection (Fig. 1). The cotransfer of ABM Tregs, a clonal population, along with naive ABM T cells significantly delayed rejection induced by the naive T cells. In striking contrast, ABM Tregs had no effect on memory T cell-induced graft rejection (Fig. 1A). Similar findings were observed in the BM3 system, where Tregs isolated from naive CBA mice were able to prevent the rejection of skin allografts mediated by naive BM3 T cells (Fig. 1C), but not by memory BM3 T cells (data not shown).
Fig. 1.
Skin allograft rejection mediated by memory T cells is resistant to regulation. (A and B) TCR-Tg ABM CD4+ naive or memory T cells (3.3 × 104) were adoptively transferred to B6 RAG-1−/− mice, either alone or with 1 × 105TCR-Tg ABM CD25+CD4+ Tregs. Tregs were either unprimed (A) or alloantigen-primed (B) as described in the text. As a control group, Tregs were injected with CD25−CD4+ T cells. Bm12 skin transplants were performed 1 day after adoptive transfer. (C) TCR-Tg BM3 CD8+ naive or memory T cells (1 × 105) were adoptively transferred to CBA RAG-1−/− mice, either alone or together with 3 × 105 polyclonal Tregs. Tregs were either unprimed or isolated from mice that had received YTS177/DST pretreatment (primed) as described in the text. B10 skin transplants were performed 1 day after adoptive transfer. P values comparing naive versus naive plus Tregs are as follows: A, P = 0.0025; B, P = 0.0062; C, P = 0.0011 (unprimed) and P = 0.0002 (tolerized). P values comparing naive T cells plus Tregs versus memory T cells plus Tregs are as follows: A, P < 0.0001; B, P = 0.0028; C, P = 0.0018.
We considered the possibility that Tregs derived from animals tolerized to the relevant alloantigen (i.e., displayed long-term allograft acceptance) might be better able to control memory T cell responses than Tregs from naive mice. Thus, in the next set of experiments, we used Tregs from ABM mice tolerized to bm12 skin grafts by treatment with DST plus CTLA4Ig plus anti-CD154 plus rapamycin or Treg from CBA mice tolerized by pretreatment with a nondepleting anti-CD4 (YTS177) and a DST 28 days before Treg isolation (Fig. 1 B and C). As was the case with Tregs isolated from unmanipulated mice, Tregs from tolerized mice prevented skin allograft rejection elicited by naive ABM or BM3 T cells, but remained unable to block memory ABM or BM3 T cell-mediated skin graft rejection (Fig. 1 B and C).
Graft Acceptance Is Associated with an Increase in Antigen-Specific Tregs.
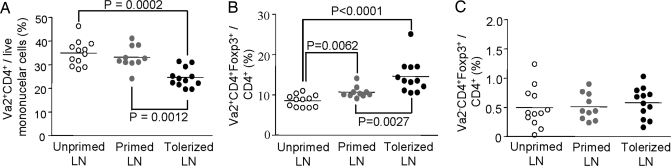
We next asked whether induction of long-term graft acceptance was associated with changes in the numbers of antigen-specific effector or Tregs in relevant secondary lymphoid tissues. The ABM system was most suitable for this analysis because these mice contain transgene-expressing Tregs. To this end, we analyzed cells from the spleen and draining lymph nodes of three groups of animals: (i) untransplanted (naive) ABM mice (24); (ii) primed ABM mice, which received a bm12 skin allograft without treatment (these grafts were rejected after ≈2 weeks, and the animals were killed 10 weeks after transplantation (3); and (iii) tolerized ABM mice, which received a bm12 skin allograft plus treatment with DST plus CTLA4Ig plus anti-CD154 plus rapamycin (as per Fig. 1B). This regimen was sufficient to enable long-term graft survival, and animals were killed at 10 weeks.
As shown in Fig. 2, animals that accepted their grafts had a statistically significant decrease in the percentage of total ABM (Vα2+CD4+) T cells in draining lymph nodes, compared with naive ABM mice or ABM mice that rejected their grafts (Fig. 2A). This finding was accompanied by a reciprocal increase in the percentage of ABM Tregs (Vα2+CD4+foxp3+) (Fig. 2B). The percentage of non-ABM Tregs (Vα2−CD4+foxp3+) did not change among the three groups (Fig. 2C). No alterations were seen in the relative frequencies of these cells in the spleen (data not shown).
Fig. 2.
Deletion of antigen-specific effector T cells and accumulation of antigen-specific regulatory T cells in long-term engrafted mice. (A–C) ABM mice were either untransplanted or received bm12 skin grafts without (primed) or with (tolerized) treatment by DST plus CTLA4Ig plus anti-CD154 plus rapamycin (see Materials and Methods). Ten weeks after transplantation, animals were killed, and lymph nodes and spleen were analyzed by flow cytometry for Vα2, CD4, and foxp3.
Taken together, these data show that, although Tregs are able to prevent skin allograft rejection mediated by naive T cells, both alloreactive CD4+ and CD8+ memory T cells are resistant to regulation by alloantigen-reactive Tregs. Moreover, graft acceptance by ABM mice is associated with an increase in the frequency of antigen-specific (but not other) Tregs. This latter point, although important by itself, also serves to further validate the use of the BM3 system, where Tregs do not express the transgene, for our studies of Tregs and memory.
Resistance of Memory T Cells to Suppression by CD4+CD25+ Tregs in Vitro.
The ability to isolate naive, memory, and Tregs with defined specificities in the ABM system enabled us to use in vitro studies to begin to dissect the mechanisms by which alloreactive T cells were able to control naive but not memory T cell alloresponses.
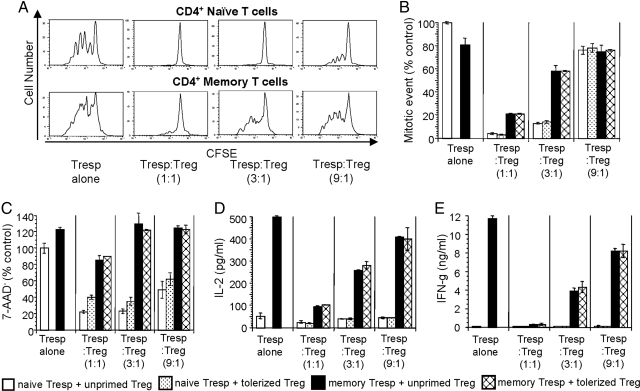
We found that, although the in vitro proliferation of memory ABM T cells in response to bm12 APCs could be inhibited in a dose-dependent manner by Tregs, the degree of suppression was far lower than that observed in naive ABM T cells (Fig. 3 A and B). Similar trends were seen on cytokine production, where the high levels of IL-2 and IFN-γ made by memory T cells could only be effectively inhibited at 1:1 ratios of memory cells to Tregs (Fig. 3 D and E). Last, we observed that Tregs strongly inhibited the survival of naive T cells, with comparatively little effect on memory T cell viability (Fig. 3C). Perhaps surprisingly, despite the ability of Tregs to suppress skin allograft rejection mediated by naive, albeit not by memory, BM3 T cells (Fig. 1) in vitro, the addition of Tregs failed to inhibit either proliferation or cytokine production by either population (data not shown). Taken together, these data suggest that Tregs may inhibit naive but not memory CD4+ T cell responses at the level of T cell priming, but that the suppression mechanism of alloreactive CD8+ T cell responses may be distinct and not accurately modeled in vitro.
Fig. 3.
Resistance of memory T cells to in vitro regulation. (A) CFSE histograms of naive and memory ABM CD4+ T cells in suppression assays (see Materials and Methods) with variable numbers of ABM CD4+CD25+ Tregs isolated from naive mice. (B and C) Mitotic events calculated from CFSE dilution curves and cell viability measured by 7-AAD staining of naive and memory ABM T cells at 72 h were normalized to the value of the naive ABM T cell group without Tregs. (D and E) IL-2 (24 h) and IFN-γ (72 h) secretion by naive and memory ABM T cells in suppression assays. These data (mean ± SD) are representative of three to nine independent experiments.
In Vivo Suppressive Effect of CD25+CD4+ Tregs on Memory T Cells.
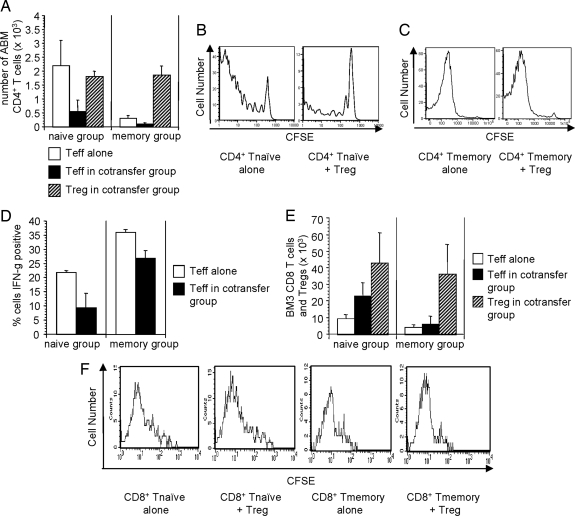
We next analyzed the in vivo effects of Tregs on memory T cell proliferation and cytokine production. For these studies, B6 RAG-1−/− mice received bm12 skin grafts on day 0. Five days later, such mice were adoptively transferred with carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled Thy1.1+ naive or memory ABM T cells either alone or together with Thy1.2+ ABM Tregs. Animals were killed for analysis on days 10–11, and draining lymph nodes, spleen, and skin were harvested. We observed that Tregs significantly inhibited the accumulation of transferred naive ABM T cells in draining lymph nodes (P < 0.05) (Fig. 4A Left). In contrast, although far fewer numbers of memory T cells could be detected in draining lymph nodes (or spleen) (data not shown), they could nonetheless be quantified, and, importantly, their accumulation was not significantly affected by Tregs (P > 0.05) (Fig. 4A Right). Consistent with these results, Tregs were able to attenuate the division of transferred naive T cells, but had little to no effect on transferred memory T cells (Fig. 4 B and C). In addition to their effects on cell numbers, Tregs reduced the frequency of naive ABM T cells producing IFN-γ by >50% (P < 0.05) (Fig. 4D Left), but were relatively less effective at reducing IFN-γ producing memory ABM T cells (P > 0.05) (Fig. 4D Right). These results are distinct from those seen in another system, where Tregs prevention of graft rejection was associated with isolated inhibition of cytokine production by antigen-reactive T cells without an effect on their proliferation or accumulation (25).
Fig. 4.
Resistance of memory T cells to in vivo regulation. (A–D) B6 RAG-1−/− mice received bm12 skin grafts and 5 days later were adoptively transferred with CFSE-labeled Thy1.1+ naive or memory ABM T cells plus Thy1.2+ ABM Tregs. Animals were killed on days 10 and 11 for analysis. (A) Total number of live, alloantigen-reactive CD4+ naive T cells, memory T cells, and Tregs (Thy1.1−CD3+) in draining lymph nodes of skin graft recipients. (Left) Animals receiving naive T cells plus Tregs. (Right) Animals receiving memory T cells plus Tregs. Open bars are effector T cells (naive or memory Thy1.1+ cells) from animals that did not receive cotransfer of Tregs. Filled bars are effector T cells from animals that also received cotransfer of Tregs. Hatched bars are Tregs in animals that received cotransfer of Tregs. (B and C) CFSE histograms of transferred naive and memory T cells. (D) Transferred naive and memory ABM T cells were restimulated in vitro with PMA plus ionomycin for 5 h and then stained for intracellular IFN-γ. (E and F) Naive or memory BM3 T cells were transferred to CBA RAG-1−/− mice with or without tolerized Tregs. All mice received a B10 skin graft 1 day after cell transfer. (E) Total number of live, BM3 CD8+ naive and memory T cells and polyclonal Tregs (CD4+) in draining lymph nodes 10 days after B10 skin transplantation. (F) CFSE histograms of live naive and memory CD8+ BM3 T cells in the draining lymph nodes of mice 10 days after B10 skin transplantation. These data show the mean number of BM3 T cells with one standard deviation (n = 2–3 mice per group). The data are representative of one to three independent experiments.
The ability of Tregs to control naive and memory BM3 T cell responses to skin allografts also was investigated. In contrast to results obtained by using ABM T cells, Tregs from BM3 CBA animals treated with a tolerizing protocol were unable to prevent initial proliferation (day 10) of either naive or memory BM3 T cells (Fig. 4F) in response to the skin allograft despite being able to prevent rejection mediated by naive but not memory BM3 T cells (Fig. 1C). Furthermore, to our surprise, the absolute number of BM3 T cells was significantly increased by day 10 in draining lymph nodes in mice that had received cotransfer of Tregs plus naive but not memory BM3 T cells (Fig. 4E). This finding suggests a distinct mechanism for the failure of BM3 versus ABM Tregs to control memory T cell-mediated rejection. Therefore, we next investigated the possibility that the presence of memory T cells might alter the homing of Tregs to the graft.
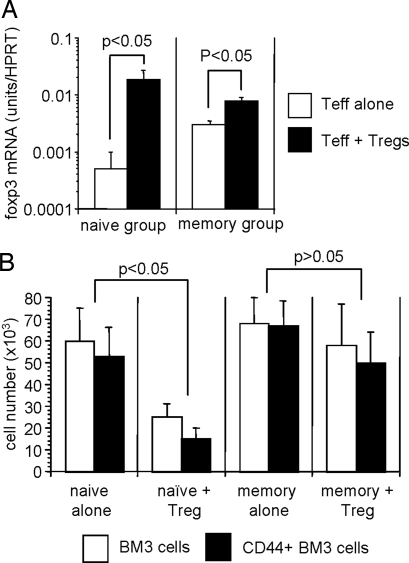
We found that, although foxp3 expression (used as a surrogate marker of Treg infiltration) within skin allografts was increased in mice that received Tregs, there was no difference in foxp3 expression (and therefore graft infiltration) between mice that also received either naive or memory BM3 T cells (Fig. 5A). Therefore, altered homing of Tregs to the graft did not account for the inability of such cells to regulate memory BM3 T cell-mediated rejection. In addition, the infiltration of allografts by BM3 T cells was found to be the same regardless of whether mice had received naive or memory BM3 T cells with and without Tregs (data not shown).
Fig. 5.
Tregs infiltrate skin allografts and block accumulation of naive but not memory T cells. (A) Real-time PCR analysis was performed on skin allografts harvested from CBA RAG−/− mice that had (Tnaive or Tmemory plus Treg) or had not (Tnaive alone or Tmemory alone) received cotransfer of 3 × 105 CD25+CD4+ Tregs, along with a transfer of 1 × 105 BM3 T cells. Skin allografts were harvested 10 days after transplantation and analyzed for the expression of Foxp3. Results are expressed as mRNA (units per HPRT) ± SD. Two to three grafts were analyzed per time point per experiment. (B) Mice generated as discussed previously were killed 50 days after transplantation, and pooled peripheral lymphoid tissues were analyzed for the number of BM3 T cells and CD44+ BM3 T cells (two to three mice analyzed per group). Data are presented as the mean ± SD.
We recently showed that alloreactive Tregs maintain BM3 T cell tolerance to skin allografts by the late deletion of intragraft effector T cells, which ultimately results in a diminished memory T cell pool (M.C.-G. and N.D.J., unpublished data). Consistent with these data, we found that mice that had received both naive BM3 T cells and Tregs had far lower numbers of total or CD44+ BM3 T cells 50 days after transplantation than mice that received BM3 T cells alone (P < 0.05) (Fig. 5B). In clear contrast, in mice where Tregs failed to control rejection mediated by memory BM3 T cells, the number of total or CD44+ BM3 T cells was undiminished and was not significantly different, compared with mice that had received memory BM3 T cell alone (P > 0.05) (Fig. 5B). Therefore, these data suggest that memory T cells may be more resistant to deletion by Tregs than naive T cells, resulting in graft rejection proceeding unabated.
Concluding Remarks.
In conclusion, we have shown that one of the ways in which alloreactive memory T cells may form a barrier to tolerance induction is through a reduced susceptibility to suppression by alloreactive Tregs. The fact that high doses of ABM Tregs had inhibitory effects of memory ABM T cell proliferation and cytokine production raises the possibility that differences seen between naive and memory T cells are quantitative and not qualitative. Thus, extremely high numbers of Tregs might be able to modulate memory T cell-mediated graft rejection, but such numbers are not easily achievable either experimentally or physiologically.
A number of previous studies have investigated mechanisms by which Tregs control allograft rejection. These studies have shown effects within both regional lymphoid tissue (13, 14) and grafts (26). Our results are consistent with this finding, which indicates that, in the ABM CD4-Tg model, Tregs have marked effects on graft priming as manifest by a reduction of the early expansion of effector cells in lymphoid tissue. In contrast, in the BM3 model, a Tregs effect on early proliferation and expansion is not observed. However, at a later time point, significant differences in cell numbers are seen, suggesting effects on BM3 survival. This difference may reflect the fact that ABM Tregs are monoclonal, whereas CBA Tregs are polyclonal. Thus, the latter population may vary in specificity and potency and/or different mechanisms of suppression of CD4 versus CD8 T cell responses.
Although the TCR-Tg approach used here cannot fully replicate the complexity of a polyclonal allogeneic response, it allows for precise identification of graft-reactive effector T cells and (in the case of the ABM model) Tregs, which greatly enables the differentiation between qualitative and quantitative effects. ABM Tregs also have been shown to modulate polyclonal T cell-rejection responses against the bm12 antigen (13), emphasizing the relevance of this system. Our findings contrast with a previous study, which concluded that CD8+ memory T cell-mediated rejection was susceptible to tolerance (19). This may be caused by a variety of differences in the models used and emphasizes the need to test principles in multiple settings.
Materials and Methods
Mice.
ABM mice (20), which are TCR-Tg mice (H2b) reactive to I-Abm12, were backcrossed >15 generations on a B6 background. BM3 TCR-Tg mice (BM3; H2k) were kindly provided by A. L. Mellor (Medical College of Georgia, Augusta) (21) and were crossed to a CBA RAG-1−/− background. Bm12, C57BL/6 (B6) RAG-1−/−, and C57BL/10 (B10; H2b) mice were purchased from The Jackson Laboratory. CBA RAG-1−/− mice, a generous gift of D. Kioussis (Mill Hill, London), CBA.Ca (CBA; H2k), and B10 mice were bred in the Biomedical Services Unit (Oxford). All experimental protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee or performed in accordance with the 1986 Animals (Scientific Procedure) Act of the United Kingdom.
Flow Cytometry and Antibodies.
Sorting was performed on a FACSVantage or FACSAria, and flow-cytometric analyses were performed on FACSCalibur, FACSort, or LSR II instruments (BD Biosciences). All antibodies used in flow cytometry and immunohistochemistry were purchased from BD PharMingen or eBioscience or were produced and labeled in house (Ti98) (22).
Preparation of Naive and Memory T Cells.
ABM naive T cells (Vα2+CD25−CD44−) were isolated by FACS from lymph nodes and spleens of unmanipulated ABM mice. Memory ABM T cells (Vα2+CD25−CD44+) were FACS-purified from ABM mice 60–100 days after bm12 skin transplantation, at which time all animals had rejected their grafts (median graft survival was ≈11 days). Naive BM3 T cells (CD8+CD44−) were FACS-purified from BM3 RAG−/− mice. To obtain memory BM3 T cells, CBA RAG−/− mice received both naive BM3 T cells and a B10 skin graft, and CD8+CD44+ cells were purified from such mice 100 days after transplantation, at which time all animals had rejected their grafts (median graft survival was 26 days).
Preparation of CD4+CD25+ Tregs.
For ABM experiments, unprimed alloantigen-specific CD25+CD4+ T cells (Vα2+CD4+CD25+) were sorted from unmanipulated ABM mice, and primed or tolerized alloantigen-specific CD4+CD25+ T cells were sorted from ABM mice 60–100 days after rejection or tolerance induction to a bm12 skin graft, respectively. Long-term engraftment was accomplished as follows: DST (D−3), 0.5 mg of human CTLA4Ig/MR-1 (D−3, D0, D3, and D7) (BioExpress), and 3 mg/kg rapamycin (D0, D1, D2, and every other day until D14) (LC Laboratories). For BM3 T cell experiments, unprimed CD4+CD25+ T cells were sorted from unmanipulated CBA mice. Primed CD4+CD25+ T cells were purified either from CBA mice tolerized to a B10 cardiac graft (0.5 mg of MR-1, D0, D2, and D4) or DST/YTS177-pretreated CBA mice (250 μl of B10 blood, D−27; 200 μg of YTS177, D−28 and D−27) (a kind gift from Herman Waldmann, University of Oxford, Oxford).
In Vitro Suppression Assays.
CFSE-labeled naive or memory ABM CD4+ T cells (1 × 105) were cultured with 3 × 105-irradiated, T cell-depleted bm12 splenocytes with or without unprimed or tolerized ABM CD4+CD25+ Tregs. Mitotic events were calculated at 72 h as described (27). Survival rates were measured by 7-amino actinomycin D (7-AAD). IL-2 and IFN-γ were assayed by BD OptEIA ELISA kit after 24 and 72 h, respectively.
Adoptive Transfer and Skin Graft Survival Analysis.
Naive or memory T cells were injected i.v. into RAG−/− mice alone or together with Tregs. One day after adoptive transfer, full-thickness skin transplantation was conducted. Graft rejection was defined as >80% necrosis.
In Vivo Suppression Analysis on Proliferation, Survival, and Cytokine Production.
B6 RAG-1−/− animals received bm12 skin grafts. Five days later, they were adoptively transferred with 5 × 105 CFSE-labeled Thy1.1+ naive or memory ABM CD4+ T cells alone or together with Thy1.2+ ABM CD25+CD4+ Tregs at a ratio of 1:2. Draining lymph nodes were harvested 10–11 days after transplantation. After in vitro restimulation by 10 ng/ml phorbol 12-myristate 13-acetate (PMA) (Sigma–Aldrich) and 1 μM ionomycin (Sigma–Aldrich) for 5 h, harvested cells were stained for intracellular IFN-γ by using GolgiStop kit (BD Bioscience). BM3 CD8+ naive or memory T cells (1 × 105) were adoptively transferred to CBA RAG−/− mice with or without 3 × 105 DST/YTS177-treated CD4+CD25+ Tregs 1 day before B10 skin transplantation. Draining lymph nodes and skin grafts were harvested 10 days after transplantation.
Real-Time PCR.
Skin grafts were harvested at different time points after transplantation and snap frozen. DNase I-treated total RNA was later isolated by using the Absolutely RNA Miniprep kit (Stratagene) and reverse-transcribed by the M-MLV reverse transcriptase (Invitrogen) as previously described (28). Real-time quantification was performed by using the PRISM 7700 sequence detection system (PE Applied Biosystem) using a fluorogenic probe as described previously (28). Specific primers and probe for hypoxanthine phosphoribosyltransferase (HPRT) and foxp3 have been previously described (23, 29). Samples were standardized for HPRT, and quantification of the gene of interest was given by 2δ−Ct, where δ−Ct is obtained by calculating the difference between Ct of the gene of interest and HPRT (30).
Statistical Analysis.
Statistical analysis was performed by using the two-tailed Mann–Whitney U test. Graft survival data were analyzed by the log-rank test. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Tynua Gray for expert assistance with animal screening, the University of Pennsylvania Flow Cytometry Core for outstanding technical assistance, the staff of Biomedical Services for excellent technical support, and Curie Ahn and laboratory members for helpful discussions. This work was supported by National Institutes of Health Grants AI-41521 (to M.H.S. and L.A.T.), AI-62789 (to H.E.R. and L.A.T.), and AI-37691 (to L.A.T.); the Biotechnology and Biological Sciences Research Council; National R&D Program Grant M10417060002-05N1706-00210 from the Ministry of Science and Technology of Korea (to J.Y.); a Wellcome Trust Prize Studentship (to M.O.B.); a Kidney Research U.K. Senior Research Fellowship (to N.D.J.); and a Royal Society Wolfson Research Merit Award (to K.J.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Adams AB, Williams MA, Jones TR, Shirasugi N, Durham MM, Kaech SM, Wherry EJ, Onami T, Lanier JG, Kokko KE, et al. J Clin Invest. 2003;111:1887–1895. doi: 10.1172/JCI17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantenburg B, Heinzel F, Das L, Heeger PS, Valujskikh A. J Immunol. 2002;169:3686–3693. doi: 10.4049/jimmunol.169.7.3686. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, Bensinger SJ, Zhang J, Chen C, Yuan X, Huang X, Markmann JF, Kassaee A, Rosengard BR, Hancock WW, et al. Nat Med. 2004;10:87–92. doi: 10.1038/nm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprent J, Surh CD. Annu Rev Immunol. 2002;20:551–579. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- 5.Valujskikh A, Lakkis FG. Immunol Rev. 2003;196:65–74. doi: 10.1046/j.1600-065x.2003.00087.x. [DOI] [PubMed] [Google Scholar]
- 6.Chalasani G, Dai Z, Konieczny BT, Baddoura FK, Lakkis FG. Proc Natl Acad Sci USA. 2002;99:6175–6180. doi: 10.1073/pnas.092596999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grayson JM, Murali-Krishna K, Altman JD, Ahmed R. J Immunol. 2001;166:795–799. doi: 10.4049/jimmunol.166.2.795. [DOI] [PubMed] [Google Scholar]
- 8.Inaba M, Kurasawa K, Mamura M, Kumano K, Saito Y, Iwamoto I. J Immunol. 1999;163:1315–1320. [PubMed] [Google Scholar]
- 9.Dai Z, Nasr IW, Reel M, Deng S, Diggs L, Larsen CP, Rothstein DM, Lakkis FG. J Immunol. 2005;174:1165–1170. doi: 10.4049/jimmunol.174.3.1165. [DOI] [PubMed] [Google Scholar]
- 10.Wood KJ, Sakaguchi S. Nat Rev Immunol. 2003;3:199–210. doi: 10.1038/nri1027. [DOI] [PubMed] [Google Scholar]
- 11.Neujahr DC, Chen C, Huang X, Markmann JF, Cobbold S, Waldmann H, Sayegh MH, Hancock WW, Turka LA. J Immunol. 2006;176:4632–4639. doi: 10.4049/jimmunol.176.8.4632. [DOI] [PubMed] [Google Scholar]
- 12.Pearl JP, Parris J, Hale DA, Hoffmann SC, Bernstein WB, McCoy KL, Swanson SJ, Mannon RB, Roederer M, Kirk AD. Am J Transplant. 2005;5:465–474. doi: 10.1111/j.1600-6143.2005.00759.x. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Fueyo A, Sandner S, Habicht A, Mariat C, Kenny J, Degauque N, Zheng XX, Strom TB, Turka LA, Sayegh MH. J Immunol. 2006;176:329–334. doi: 10.4049/jimmunol.176.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MKT, Moore DJ, Jarrett BP, Lian MM, Deng S, Huang X, Markmann JW, Chiaccio M, Barker CF, Caton AJ, Markmann JF. J Immunol. 2004;172:6539–6544. doi: 10.4049/jimmunol.172.11.6539. [DOI] [PubMed] [Google Scholar]
- 15.Billiard F, Litvinova E, Saadoun D, Djelti F, Klatzmann D, Cohen JL, Marodon G, Salomon BL. J Immunol. 2006;177:2167–2174. doi: 10.4049/jimmunol.177.4.2167. [DOI] [PubMed] [Google Scholar]
- 16.Grossman WJ, Verbsky JW, Barchet W, Colonna M, Atkinson JP, Ley TJ. Immunity. 2004;21:589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. J Immunol. 2005;174:1783–1786. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 18.Sarween N, Chodos A, Raykundalia C, Khan M, Abbas AK, Walker LS. J Immunol. 2004;173:2942–2951. doi: 10.4049/jimmunol.173.5.2942. [DOI] [PubMed] [Google Scholar]
- 19.Dai Z, Li Q, Wang Y, Gao G, Diggs LS, Tellides G, Lakkis FG. J Clin Invest. 2004;113:310–317. doi: 10.1172/JCI19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandner SE, Salama AD, Houser SL, Palmer E, Turka LA, Sayegh MH. Am J Transplant. 2003;3:1242–1250. doi: 10.1046/j.1600-6143.2003.00220.x. [DOI] [PubMed] [Google Scholar]
- 21.Sponaas AM, Tomlinson PD, Antoniou J, Auphan N, Langlet C, Malissen B, Schmitt-Verhulst AM, Mellor AL. Int Immunol. 1994;6:277–287. doi: 10.1093/intimm/6.2.277. [DOI] [PubMed] [Google Scholar]
- 22.Hua C, Boyer C, Buferne M, Schmitt-Verhulst AM. J Immunol. 1986;136:1937–1944. [PubMed] [Google Scholar]
- 23.Jones ND, Carvalho-Gaspar M, Luo S, Brook MO, Martin L, Wood KJ. J Immunol. 2006;176:2316–2323. doi: 10.4049/jimmunol.176.4.2316. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin JD, Dorn IG., II Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 25.Lin CY, Graca L, Cobbold SP, Waldmann H. Nat Immunol. 2002;3:1208–1213. doi: 10.1038/ni853. [DOI] [PubMed] [Google Scholar]
- 26.Graca L, Cobbold SP, Waldmann H. J Exp Med. 2002;195:1641–1646. doi: 10.1084/jem.20012097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells AD, Gudmundsdottir H, Turka LA. J Clin Invest. 1997;100:3173–3183. doi: 10.1172/JCI119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho-Gaspar M, Billing JS, Spriewald BM, Wood KJ. J Immunol Methods. 2005;301:41–52. doi: 10.1016/j.jim.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Hori S, Nomura T, Sakaguchi S. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]