Abstract
Comprehensive knowledge of the genomic alterations that underlie cancer is a critical foundation for diagnostics, prognostics, and targeted therapeutics. Systematic efforts to analyze cancer genomes are underway, but the analysis is hampered by the lack of a statistical framework to distinguish meaningful events from random background aberrations. Here we describe a systematic method, called Genomic Identification of Significant Targets in Cancer (GISTIC), designed for analyzing chromosomal aberrations in cancer. We use it to study chromosomal aberrations in 141 gliomas and compare the results with two prior studies. Traditional methods highlight hundreds of altered regions with little concordance between studies. The new approach reveals a highly concordant picture involving ≈35 significant events, including 16–18 broad events near chromosome-arm size and 16–21 focal events. Approximately half of these events correspond to known cancer-related genes, only some of which have been previously tied to glioma. We also show that superimposed broad and focal events may have different biological consequences. Specifically, gliomas with broad amplification of chromosome 7 have properties different from those with overlapping focalEGFR amplification: the broad events act in part through effects on MET and its ligand HGF and correlate with MET dependence in vitro. Our results support the feasibility and utility of systematic characterization of the cancer genome.
Keywords: bioinformatics, comparative genomic hybridization, glioblastoma, copy-number alterations, single-nucleotide polymorphism arrays
Comprehensive knowledge of the mutational events responsible for cancer is a critical foundation for future diagnostics, prognostics, and targeted therapeutics. Various efforts are now underway aimed at systematically obtaining this information. The first challenge in such a program is to study large collections of tumors to characterize the alterations that have occurred in their genomes. With recent advances in genomic technology, this is becoming increasingly feasible. For example, DNA arrays containing probes for hundreds of thousands of genetic loci have made it possible to detect regional amplifications and deletions with high resolution. Once the genomic alterations have been detected, the second challenge is to distinguish between “driver” mutations that are functionally important changes (that is, that confer a biological property that allows the tumor to initiate, grow, or persist) and “passenger” mutations that represent random somatic events (that is, changes that occurred before a clonal expansion and are simply carried along despite conferring no selective advantage).
The importance of this second challenge is evident from recent studies of chromosomal aberrations in cancer. Strikingly, different studies of the same tumor type often report “regions of interest” that are highly discordant. For example, two recent studies of lung cancer, with similar sample sizes and analytic methods, reported 48 and 93 regions of interest, respectively (1, 2); the overlap between the lists was <5%.
Although perfect agreement should not be expected (in part because of differences in analytic methods), such a high level of discordance is disconcerting. There are two potential explanations. One possibility is that the true number of cancer-related regions is extremely large, with each tumor containing only a small and variable subset of the alterations and each study detecting only a small subset of the regions. An alternative possibility is that many of the regions of interest reported in current studies are random events of no biologic significance, such as random passenger mutations. Current analysis methods do not explicitly account for the background rate of random chromosomal aberrations. Similar issues arise in interpreting studies of point mutations in cancer resequencing projects (3–6).
In this article we describe a statistical approach, called Genomic Identification of Significant Targets in Cancer (GISTIC), for identifying regions of aberration that are more likely to drive cancer pathogenesis. The method identifies those regions of the genome that are aberrant more often than would be expected by chance, with greater weight given to high-amplitude events (high-level copy-number gains or homozygous deletions) that are less likely to represent random aberrations.
We then apply GISTIC to a newly generated, high-resolution data set of chromosomal aberrations in 141 gliomas. Glioma is an excellent model in which to test the approach because the functional roles of a substantial number of copy number alterations have already been validated in preclinical models (7, 8). We find 32 statistically significant events of genomic amplification or loss. Using standard analytic methods, the regions of interest found in this study and those reported in two other recent studies of glioma are highly discordant. Strikingly, we find that the discordance largely vanishes when the GISTIC methodology is applied to the underlying data from all three studies. Moreover, the regions we find contain nearly all cancer genes previously known to be involved in glioma.
The significant aberrations in the glioma genome fall into two types: focal and broad (near the size of a chromosome arm). By studying the biological properties of tumors, we find evidence that overlapping focal and broad events can have very different consequences. Focusing on chromosome 7 (chr7), we show that focal high-level amplification at the EGFR gene is associated with activation of EGFR itself whereas broad lower-level amplification of the whole chromosome often activates the MET axis by increasing the dosage of both MET and its ligand HGF, suggesting that a subset of glioblastoma (GBM) patients with polysomy 7 might benefit from MET inhibition.
Results
GISTIC Methodology.
GISTIC identifies significant aberrations through two key steps [Fig. 1 and supporting information (SI) Text]. First, the method calculates a statistic (G score) that involves both the frequency of occurrence and the amplitude of the aberration. Second, it assesses the statistical significance of each aberration by comparing the observed statistic to the results that would be expected by chance, using a permutation test that is based on the overall pattern of aberrations seen across the genome. The method accounts for multiple-hypothesis testing using the false-discovery rate (FDR) framework (9) and assigns a q value to each result, reflecting the probability that the event is due to chance fluctuation. For each significant region, the method defines a “peak region” with the greatest frequency and amplitude of aberration. Each peak is tested to determine whether the signal is due primarily to broad events, focal events, or overlapping events of both types.
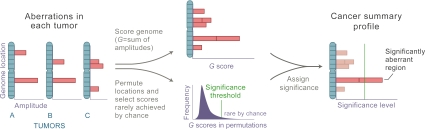
Fig. 1.
Overview of the GISTIC method. After identifying the locations and, in the case of copy-number alterations, magnitudes (as log2 signal intensity ratios) of chromosomal aberrations in multiple tumors (Left), GISTIC scores each genomic marker with a G score that is proportional to the total magnitude of aberrations at each location (Upper Center). In addition, by permuting the locations in each tumor, GISTIC determines the frequency with which a given score would be attained if the events were due to chance and therefore randomly distributed (Lower Center). A significance threshold (green line) is determined such that significant scores are unlikely to occur by chance alone. Alterations are deemed significant if they occur in regions that surpass this threshold (Right). For more details see SI Text.
Application to Glioma.
We applied the method to a collection of 141 gliomas, including 107 primary GBMs, 15 secondary GBMs, and 19 lower-grade gliomas (SI Table 2). We hybridized genomic DNA to microarrays containing probes for ≈100,000 SNPs to identify copy-number changes and loss of heterozygosity (LOH). A genome-wide view of the copy-number alterations is shown in Fig. 2a (LOH results are described in SI Note 1 in SI Text). The overall pattern is complex, with almost every region of the genome being altered in at least one tumor. Nonetheless, only 16 broad and 16 focal events are significant. Focal events are superimposed on four broad events (including two focal events on chromosome 7 and single events on chromosomes 9, 10, and 13), resulting in a total of 28 peak regions of amplification, deletion, and LOH.
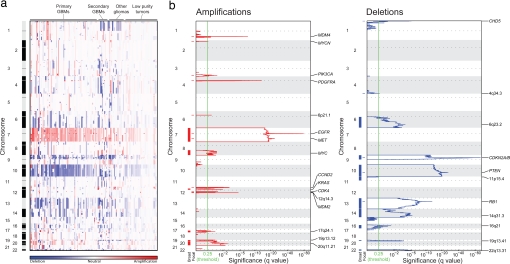
Fig. 2.
Significant broad and focal copy-number alterations in the glioma genome. (a) Amplifications (red) and deletions (blue), determined by segmentation analysis of normalized signal intensities from 100K SNP arrays (see SI Text), are displayed across the genome (chromosome positions, indicated along the y axis, are proportional to marker density) for 141 gliomas (x axis; diagnosis is displayed on top, and gliomas with low purity are segregated to the right). Broad events near the size of a chromosome arm are the most prominent, including amplifications of chr7 and deletions of chr10 observed among >80% of GBMs. (b) GISTIC analysis of copy-number changes in glioma. The statistical significance of the aberrations identified in a are displayed as FDR q values (9) to account for multiple-hypothesis testing. Chromosome positions are indicated along the y axis with centromere positions indicated by dotted lines. Fifteen broad events (indicated by red bars for amplifications and blue bars for deletions) and 16 focal events (indicated by dashes) surpass the significance threshold (green line). The locations of the peak regions and the known cancer-related genes within those peaks are indicated to the right of each panel. Several broad regions, including chr7 and chr10, contain superimposed focal events, leading to needle-shaped peaks superimposed on highly significant plateaus.
The 16 broad events include six amplifications (chromosomes 7, 8q, 12p, 17q, 19p, and 20), nine deletions (6q, 9p, 10, 11p, 13, 14, 16q, 19q, and 22), and one region of copy-neutral LOH (17p) (Fig. 2b, SI Table 3, and SI Note 1 in SI Text). These events occur at high frequency (range 10–70%, median 27%). In particular, amplification of chr7 and deletion of chr10 each affect >60% of our samples, including >80% of our primary GBMs. For broad regions without superimposed focal events, the peak regions are large (median of 110 genes) (SI Table 3).
The 16 focal events tend to occur at lower frequencies than the broad aberrations (range 6–49%, median 14%) (SI Table 3). Among these, amplifications of 4q12 and 7p11.2 (18–26% of samples) and deletions of 1p36.31 and 9p21.3 (35–49%) are the most frequent. In some cases, a high degree of amplification renders amplifications highly significant even though they occur in only 6–7% of samples (for example, the regions containing CDK4 and MDM2 on chr12). Because the background rate of deletions across the genome is higher, deletions usually must occur at higher frequencies than amplifications to attain similar levels of significance (SI Table 3). The peak regions for the focal events can be localized to small regions (median of four genes).
Analysis Confirms Known Genes and Identifies New Loci.
We compared the 28 peak regions to the locations of oncogenes and tumor-suppressor genes previously implicated in the pathogenesis of glioma. A recent review (10) lists 12 such genes reported to be altered in multiple studies of glioma (TP53, RB1, CDKN2A/B, PTEN, EGFR, PDGFRA, MET, CDK4, CDK6, MDM2, MDM4, and MYC). We found that 11 of these 12 genes each correspond with one of the 28 peak regions (with one of these, MYC, lying just beyond the boundaries defined by GISTIC; as described below, this slight discrepancy is resolved with additional data) (SI Table 3). The 12th gene (CDK6) lies within the broad region of significant amplification on chr7, although it does not correspond to a peak. Interestingly, TP53 is within the single peak region of LOH that is not reflected in a peak of copy-number change, suggesting that it is primarily inactivated through copy-neutral LOH (SI Note 1 in SI Text).
An additional five peak regions contain genes that are known to play a role in other cancers (MYCN, PIK3CA, CCND2, KRAS, and CHD5) (11, 12). Our analysis suggests that chromosomal aberrations involving these genes are also relevant for glioma pathogenesis. These genes should therefore be carefully characterized in glioma.
The remaining 12 regions (43% of the total) are not associated with known cancer-related genes. These events occur at substantial frequencies (6–37%), but nine are due to broad events and two others rarely reach high amplitude. The final region (12q14.3) undergoes high-level amplifications, but always in concert with amplifications of either of two neighboring regions containing CDK4 and MDM2 (data not shown), suggesting that it may be due to structural features required to amplify these genes [as is the case in dedifferentiated liposarcoma (13, 14)]. The fact that nearly half the regions (including regions affected by highly prevalent aberrations) are not yet associated with known cancer-related genes underscores the importance of systematic analysis of the cancer genome.
Previous studies of copy-number alterations in glioma have shown distinct patterns for certain subtypes such as primary vs. secondary GBMs (15) or astrocytic vs. oligodendroglial tumors (16). To explore whether our combined analysis of these glioma subtypes prevented the detection of alterations specific to primary GBMs, we performed a separate analysis on only the 107 primary GBMs in our sample set. No additional statistically significant alterations were identified (SI Fig. 4).
Consistency Across Independent Data Sets.
We then sought to compare our results with two previous studies of copy-number alterations in glioma (178 samples on 100K SNP arrays; 37 samples on a 16K CGH arrays) (15, 17). At first glance, there appear to be striking differences. The previous studies reported many more regions of interest (208 and 97) (Table 1), but the regions included fewer of the known glioma-associated genes and show low concordance with one another. The differences are attributable to the methodology used in these studies, in which minimal common regions of copy-number change are reported, without explicitly taking into account the degree of background noise (see SI Note 2 in SI Text). Applying a similar analysis to our own data identifies a similarly large number of regions (144) (see SI Note 2 in SI Text), but these include fewer known glioma-associated genes and show low concordance with the other studies. By contrast, applying GISTIC to the raw data from the other two studies identifies 24 and 26 significant regions each. Importantly, these regions agree closely with the 27 regions (excluding copy-neutral LOH of 17p, because it is not observed in the copy-number analyses) identified above (SI Fig. 5), and they include essentially the same glioma-associated genes. Moreover, these results are specific to glioma, as seen in a comparison to lung cancer (SI Fig. 6) (2). The strong concordance across three independent data sets and two different platforms supports the validity of both the GISTIC methodology and our results for glioma.
Table 1.
Comparison of results between copy-number analyses of the glioma genome
| Data set | Platform | No. of tumors | MCR analysis* |
GISTIC analysis |
|||
|---|---|---|---|---|---|---|---|
| No. of MCRs | No. of glioma genes in MCRs† | No. of peaks | No. of glioma genes in peaks† | ||||
| Initial | 100K SNP | 141 | 144 | 5 | 27 | 9 | |
| Kotliarov et al. (17) | 100K SNP | 178 | 208 | 3 | 26 | 9 | |
| Maher et al. (15) | 16K aCGH | 37 | 97 | 8 | 24 | 9 | |
*Minimal common regions (MCR) analysis, as presented in cited publications.
†Eleven glioma genes affected by copy-number aberrations are considered ″known″: PTEN, RB1, CDKN2A/B, EGFR, PDGFRA, MET, CDK4, CDK6, MDM2, MDM4, and MYC (10).
Given the close agreement across these data sets, we combined our initial data set with the data from the prior study performed on the same platform to obtain a pooled data set with 319 glioma samples. This analysis identified 34 significant regions, including 27 of the 28 identified in the initial data set (SI Table 3). For the additional regions, the prevalence of aberrations is similar in the initial and combined data sets (11.0% vs. 11.3%), but they now exceed the significance threshold due to the larger sample size. Increasing the sample size also leads to narrower peak regions, with the median number of genes decreasing from 12 to five per region. Inclusion of additional data sets may define these regions with even greater precision and facilitate identification of the gene targets.
Overlapping Broad and Focal Events May Have Different Consequences.
Having identified various instances of overlapping broad and focal events, we explored whether they have distinct functional consequences, using chr7 as an example (SI Fig. 7). We first compared copy-number profiles to gene expression among a group of 43 primary GBMs for which we had sufficient material for a combined analysis. EGFR was overexpressed in most GBMs with focal EGFR amplification (7gainEGFRamp) but in none of the GBMs with broad amplification of chromosome 7 in the absence of focal EGFR amplification (7gain) (Fig. 3a). We found three additional sources of evidence supporting the biologic distinction between these two classes of tumors. First, recognizing that EGFR-amplified tumors are known to carry a low rate of mutations in the TP53 gene (18), we sequenced TP53 and found these mutations more frequently associated with 7gain compared with 7gainEGFRamp (two-sided Fisher's exact test, P = 0.03). Second, we found that 7gain is less frequently associated with EGFR point mutations or expression of the EGFRvIII deletion mutant [determined for many of our tumors in a prior study (19)] (P = 0.001). Third, we found that 7gain but not 7gainEGFRamp occurs frequently in secondary GBMs (P = 0.16). These three findings are consistent with earlier observations regarding 7gainEGFRamp (20, 21) and further suggest that 7gain has distinct functional consequences.
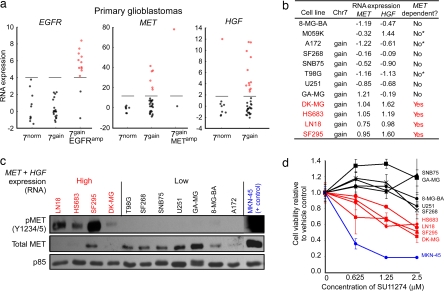
Fig. 3.
Broad gains of chromosome 7 often activate the MET pathway but not EGFR. (a) Expression levels of EGFR, MET, and its ligand HGF (all located on chr7) in primary GBMs. These data are log2-transformed signal intensities from all concordant probe sets for each gene from Affymetrix U133 arrays, centered and normalized according to the median and median absolute deviation of samples with 7norm. Samples with 7gainEGFRamp but not 7gain overexpress EGFR (highlighted in red) relative to 7norm. Conversely, a subset of tumors with 7gain overexpress MET or its ligand HGF, even in the absence of focal amplification. (b) A subset of glioma cell lines with 7gain also overexpress MET and HGF (MET/HGF+ lines, highlighted in red). We characterized lines as having 7gain if SNP array analysis showed them to be amplified across most of chr7. Cell lines are classified as being MET-dependent based on the results shown in d or previously published results (asterisks) (35). (c) Constitutive phosphorylation of MET in MET/HGF+ lines. Immunoblots to the indicated epitopes were performed on whole-cell lysates prepared after 24-h serum starvation. Decreased MET protein levels in activated lines are a result of HGF-induced degradation (36). MET-dependent gastric cancer cells (MKN-45) were included as positive controls (35). (d) Decreased viability of MET/HGF+ cell lines (red) compared with non-MET/HGF+ lines (black) when treated with the MET inhibitor SU11274. Viability was measured by using Trypan blue exclusion after exposure to inhibitor at the indicated concentrations for 96 h. MKN-45 cells (blue) were included as positive controls.
To explore the function of 7gain, we identified genes on the chromosome that show extreme outliers in expression in at least 10% of tumors with 7gain, compared with 7normal (SI Note 3 in SI Text). The notion behind this “comparative outlier analysis” is that broad events such as 7gain may have heterogeneous effects across various tumors. We are interested in identifying genes that are strongly up-regulated in even a subset of the samples.
Strikingly, two of the top four genes in this analysis are those encoding the receptor MET and its ligand HGF (SI Table 4). Approximately one-third of 7gain events are associated with either MET or HGF overexpression (Fig. 3a), and tumors that overexpress one tend to overexpress both (P = 0.06) (data not shown). The overexpression of MET and HGF appears to be functionally relevant: we studied glioma cell lines with 7gain and increased expression of MET and HGF (Fig. 3b) and found phosphorylation and activation of the MET receptor even under serum-starved conditions (Fig. 3c). These cell lines showed enhanced responsiveness to the MET kinase inhibitor SU11274 (22) (Fig. 3d) at drug concentrations that inhibit the MET signaling pathway (SI Fig. 8 A and B). None of the GBM cell lines with 7gain showed constitutive activation of EGFR or were responsive to the EGFR kinase inhibitor erlotinib (SI Fig. 8 C and D). Compared with the relatively rare (5–7%) focal amplification of the MET gene locus (SI Table 3), 7gain with overexpression of MET and HGF may provide a more common mechanism for cell-autonomous activation of the MET signaling pathway in glioma. This finding may be relevant for the clinical deployment of inhibitors targeting this network in glioma and other cancers (23, 24).
Discussion
Statistical approaches such as GISTIC identify those recurrent changes that are concordant across data sets and less likely to represent random passenger events. Indeed, we have now successfully used this approach to identify biologically significant aberrations in lymphoma (25), melanoma (W. M. Lin, A. C. Baker, R.B., W. Winckler, W. Feng, et al., unpublished work), and lung cancer (26). Although we believe it is likely that most events identified by GISTIC are drivers that recur because of positive selection during tumor evolution, some events may recur because of biases in the DNA mutation or repair machinery. In addition, some driver aberrations may occur at low frequency and therefore may be missed. The design of future experiments should consider the number of tumors needed to power detection of such rare events.
Ultimately, the utility of systematic efforts to characterize the cancer genome is an empirical question. There are at least two potential concerns: on one hand, that the vast majority of cancer-related genes are already known with little left to learn; on the other hand, that cancer is hopelessly complicated, with a large number of cancer genes, each altered in a small fraction of tumors. The results here suggest a more favorable situation, at least for copy-number alterations. With appropriate statistical methodology, three studies reveal a concordant picture of the glioma genome. There appears to be a tractable number of recurrent events, in the range of 40. Larger tumor collections may identify some additional low-prevalence events, but it seems likely that the majority of significant recurrent copy-number alterations at this scale have been found. Approximately half are likely to involve known cancer-related genes, with some not having previously been established to be involved in glioma; all of these genes should be systematically characterized in glioma. The remaining events likely point to cancer-related genes and other functional elements that remain to be discovered; the identification of the genes associated with the broad events is particularly important and will likely require the application of orthogonal approaches, such as expression profiling, mutational analysis, and RNA interference. Finally, copy-number aberrations are only one form of the genomic changes in glioma. Identification of other cancer-associated events, including mutations, rearrangements, and epigenetic alterations, will require similar statistical approaches and large data sets, as presented here.
Methods
Clinical Samples and Cell Lines.
Genomic DNA was extracted from fresh-frozen tumors samples using DNeasy (Qiagen). Non-tumor tissue, including paired normal brain corresponding to 10 gliomas, was used for germ-line control DNA. Collection and analysis of all clinical samples were approved by the University of California Institutional Review Board. RNA and DNA were also obtained from the GBM cell lines 8-MG-BA, A172, DK-MG, GAMG, HS683, LN-18, SF-268, SF-295, SNB-75, T98G, and U251. Gastric and lung cancer cell lines MKN-45 (high level MET amplified) and H3255 (L858R EGFR mutant) were included as positive controls in experiments with the MET kinase inhibitor SU11274 and the EGFR kinase inhibitor erlotinib, respectively.
SNP Arrays.
Genomic DNA was applied according to the manufacturer's instructions to oligonucleotide arrays (Affymetrix) interrogating 116,204 SNP loci on all chromosomes except Y. Arrays were scanned by using the GeneChip Scanner 3000, and genotyping was performed by using Affymetrix Genotyping Tools Version 2.0. Probe-level signal intensities were normalized to a baseline array with median intensity using invariant set normalization (27). SNP-level signal intensities were obtained by using a model-based (PM/MM) method (28). Further analytic steps are described in SI Text. SNP, gene, and cytogenetic band locations are based on the hg16 (July 2003) genome build (http://genome.ucsc.edu). Data from SF-268, SF-295, SNB-75, and six gliomas (along with paired normals) were previously published (29, 30). Data are available at www.broad.mit.edu/cancer/pub/GISTIC and through the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/projects/geo, accession no. GSE9635).
Mutation Detection.
PTEN and TP53 were sequenced in 134 of the 141 samples undergoing SNP analysis as previously described (19). All exons were covered in >70% of samples except exons 1, 8, and 9 in PTEN and exons 7 and 10 in TP53. EGFR point mutations and vIII expression were determined for 133 and 58 samples, respectively, in a prior study (19).
Expression Arrays.
Expression data were obtained by using Affymetrix U133A/B and plus 2 arrays from 43 primary GBMs undergoing SNP array analysis. CEL files from U133A and plus 2 arrays were preprocessed separately by using RMA (31). Probe sets common to both arrays were used after equalizing the mean and standard deviation of the U133A and plus 2 arrays. Expression data from GBM cell lines were generated by Affymetrix cartridge arrays, except SF-268, SF-295, and SNB-75, where available U133A data were used (http://wombat.gnf.org/index.html).
EGFR and MET Inhibition.
Cell proliferation assays.
Cell lines were maintained in RPMI medium 1640 containing 10% serum and 1% penicillin/streptomycin. Stock solutions of erlotinib (10 mM; WuXi Pharmatech) and SU11274 (1 mM; Calbiochem) were prepared in DMSO and maintained at −20°C or 4°C according to the manufacturer's instructions. Drugs were diluted in fresh medium before each experiment. Cells were cultured in the presence of drug or vehicle for 4 days, and viability was determined by using the WST assay (Roche) or Trypan blue exclusion assay as previously described (32, 33).
Western blot analysis.
To examine basal MET and EGFR phosphorylation, cells were grown in serum-free DMEM with 1% l-glutamine 200 mM and 1% penicillin/streptomycin for 24 h. Cells were harvested and lysed by using cell lysis buffer (Cell Signaling Technology) with 1% protease and 1% phosphatase inhibitors (Calbiochem). Total protein concentration was determined by using Bio-Rad Protein Assay Standard I (Bio-Rad). Equal protein amounts were resolved by SDS/PAGE and electrotransferred to nitrocellulose membrane blots (34). Blots were probed with antibodies against MET (25H2), P-MET (P-Tyr 1234/5), P-EGFR (P-Tyr 1173) (Cell Signaling Technology), and EGFR (sc-03; Santa Cruz Biotechnology) and exposed to standard x-ray film after application of peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) and ECL Western blotting detection reagents (GE Healthcare).
Supplementary Material
Acknowledgments
Nadav Kupiec and Bang Wong provided help with illustrations. Howard Fine, Elizabeth Maher, Cameron Brennan, Ron Depinho, and Lynda Chin provided data. This work was supported by funds from the Broad Institute of Harvard University and Massachusetts Institute of Technology, the Dana–Farber/Harvard Cancer Center Prostate Specialized Program of Research Excellence (R.B.), Department of Defense Grant PC040638 (to R.B.), National Cancer Institute Grants CA109038 and CA126546 (to M.M.), the Henry E. Singleton Brain Cancer Program at the University of California, Accelerate Brain Cancer Cure (I.K.M.), and the Brain Tumor Funders' Collaborative. J.B. is a Beatriu de Pinos Fellow of the Departament d'Educació i Universitats de la Generalitat de Catalunya. J.D. and R.K.T. are supported by fellowships of the Leukemia and Lymphoma Society and the International Association for the Study of Lung Cancer, and R.K.T. is a Mildred Scheel Fellow of the Deutsche Krebshilfe. I.K.M. is a Sontag Foundation Distinguished Scientist.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/projects/geo (accession no. GSE9635).
This article contains supporting information online at www.pnas.org/cgi/content/full/0710052104/DC1.
References
- 1.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry D, Protopopov A, You MJ, Aguirre AJ, et al. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, et al. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 3.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, et al. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 4.Greenman C, Stephens P, Smith R, Dalgliesh G, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 6.Getz G, Höfling H, Mesirov JP, Golub TR, Meyerson ML, Tibshirani R, Lander ES. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 7.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 8.Fomchenko EI, Holland EC. Clin Cancer Res. 2006;12:5288–5297. doi: 10.1158/1078-0432.CCR-06-0438. [DOI] [PubMed] [Google Scholar]
- 9.Benjamini Y, Hochberg Y. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 10.Reifenberger G, Collins VP. J Mol Med. 2004;82:656–670. doi: 10.1007/s00109-004-0564-x. [DOI] [PubMed] [Google Scholar]
- 11.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 13.Mandahl N, Heim S, Johansson B, Bennet K, Mertens F, Olsson G, Rooser B, Rydholm A, Willen H, Mitelman F. Int J Cancer. 1987;39:685–688. doi: 10.1002/ijc.2910390605. [DOI] [PubMed] [Google Scholar]
- 14.Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, Michiels JF, Terrier P, Ranchere-Vince D, Collin F, Myklebost O, Turc-Carel C. Genes Chromosomes Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 15.Maher EA, Brennan C, Wen PY, Durso L, Ligon KL, Richardson A, Khatry D, Feng B, Sinha R, Louis DN, et al. Cancer Res. 2006;66:11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 16.Kraus JA, Koopmann J, Kaskel P, Maintz D, Brandner S, Schramm J, Louis DN, Wiestler OD, von Deimling A. J Neuropathol Exp Neurol. 1995;54:91–95. doi: 10.1097/00005072-199501000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Kotliarov Y, Steed ME, Christopher N, Walling J, Su Q, Center A, Heiss J, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA. Cancer Res. 2006;66:9428–9436. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe K, Tachibana O, Sata K, Yonekawa Y, Kleihues P, Ohgaki H. Brain Pathol. 1996;6:217–223. doi: 10.1111/j.1750-3639.1996.tb00848.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, Debiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, et al. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galanis E, Buckner J, Kimmel D, Jenkins R, Alderete B, O'Fallon J, Wang CH, Scheithauer BW, James CD. Int J Oncol. 1998;13:717–724. [PubMed] [Google Scholar]
- 21.Ekstrand AJ, Sugawa N, James CD, Collins VP. Proc Natl Acad Sci USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sattler M, Pride YB, Ma P, Gramlich JL, Chu SC, Quinnan LA, Shirazian S, Liang C, Podar K, Christensen JG, Salgia R. Cancer Res. 2003;63:5462–5469. [PubMed] [Google Scholar]
- 23.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 24.Peruzzi B, Bottaro DP. Clin Cancer Res. 2006;12:3657–3660. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- 25.Takeyama K, Monti S, Manis JP, Cin PD, Getz G, Beroukhim R, Dutt S, Aster JC, Alt FW, Golub TR, Shipp MA. Oncogene. 2007 Jul 16; doi: 10.1038/sj.onc.1210650. [DOI] [PubMed] [Google Scholar]
- 26.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Nature. 2007 Nov 4; doi: 10.1038/nature 06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Wong WH. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Wong WH. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. research0032.1–research0032.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beroukhim R, Lin M, Park Y, Hao K, Zhao X, Garraway LA, Fox EA, Hochberg EP, Mellinghoff IK, Hofer MD, et al. PLoS Comput Biol. 2006;2:e41. doi: 10.1371/journal.pcbi.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 31.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 32.Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR, Meyerson M. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 34.Mellinghoff IK, Tran C, Sawyers CL. Cancer Res. 2002;62:5254–5259. [PubMed] [Google Scholar]
- 35.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC, et al. Proc Natl Acad Sci USA. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeffers M, Taylor GA, Weidner KM, Omura S, Vande Woude GF. Mol Cell Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.