Abstract
The herpesvirus ubiquitin-specific protease (USP) family, whose founding member was discovered as a protease domain embedded in the large tegument protein of herpes simplex virus 1 (HSV-1), is conserved across all members of the Herpesviridae. Whether this conservation is indicative of an essential function of the enzyme in vivo has not yet been established. As reported here, USP activity is conserved in Marek's disease virus (MDV), a tumorigenic alphaherpesvirus. A single amino acid substitution that abolishes the USP activity of the MDV large tegument protein diminishes MDV replication in vivo, and severely limits the oncogenic potential of the virus. Expression of the USP transcripts in MDV-transformed cell lines further substantiates this hypothesis. The herpesvirus USP thus appears to be required not only to maintain a foothold in the immunocompetent host, but also to contribute to malignant outgrowths.
Keywords: chicken, deubiquitinating enzyme, herpes, Marek's disease virus
The ubiquitin-proteasome system controls cytosolic proteolysis, certain aspects of transcription, antigen presentation via major histocompatibility complex (MHC) class I products, and the trafficking of surface-exposed receptors (1–5). As for many other posttranslational modifications, both ubiquitin conjugation and its reversal by ubiquitin-specific proteases (USPs) determine the biological outcome of the reaction. The enzyme families that catalyze ubiquitin conjugation and removal are quite diverse (6–9). Consequently, bioinformatic analysis is not always adequate to identify novel USPs. To target such enzymes biochemically, we developed activity-based probes for USPs and enzymes that act on ubiquitin-like modifiers (10). These probes are equipped with an affinity handle to allow retrieval and identification of the enzymes targeted by the electrophilic warhead installed at the probe's carboxyl terminus.
Through the use of one of these probes, HA-ubiquitin vinylmethylester (HA-UbVME), we identified the large tegument protein of herpes simplex virus 1 (HSV-1), viral protein (VP) 1/2, encoded by the unique-long (UL) 36 gene, as the source of an active USP. Its sequence showed no obvious similarity to known eukaryotic USPs and failed to yield an obvious signature of residues diagnostic of known cysteine protease families. Nonetheless, sequence comparisons across the Herpesviridae show the presence of a few absolutely conserved residues (Cys, Asp, His, Glu), all of which are consistent with involvement in a potential thiol protease active site. We have, meanwhile, confirmed both the mechanism of action of such ubiquitin-based probes (11–15) and the identity of the viral cysteine protease domain as an authentic USP by crystallographic analysis of the homologous segment of the murine cytomegalovirus (MCMV) M48 protein (16). Notwithstanding the conservation of the identified USP activity in all herpesviruses, we do not know whether this activity makes a contribution to the replicative success and pathogenicity of herpesviruses in vivo.
Marek's disease virus (MDV) is a lymphomagenic alphaherpesvirus that affects chickens. It is a major source of concern for the poultry industry and necessitates vaccination of chicken flocks in ovo or shortly after hatching (17–19). The severity of the symptoms and their clockwork timing, as well as the availability of the natural host, make MDV an ideal model to assess the importance of the viral USP for alphaherpesvirus pathogenesis in vivo. The course of infection includes lytic replication, then latency, followed by reactivation and the formation of massive T cell lymphomas in multiple organs (18–20). We undertook the generation of an MDV mutant that lacks the USP activity to address its role in pathogenicity.
Results
Characterization of the MDVUSP Active Site Cysteine.
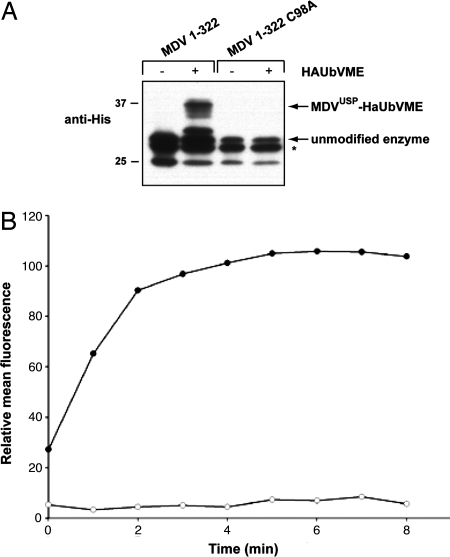
In HSV-1, the UL36-derived USP was discovered as an ≈450-aa amino-terminal fragment of the VP1/2 tegument protein, generated late during viral infection (12). So far, only HSV-1 has yielded an enzymatically active, proteolytic fragment that is distinct from the intact tegument protein in virus-infected cells. We cloned and expressed the corresponding region of the MDV UL36 gene (referred to as MDVUSP), based on homology and secondary structure predictions (13, 14). This 322-residue fragment, when expressed by in vitro transcription/translation, readily formed adducts with Ub-VME in a manner sensitive to the inclusion of N-ethylmaleimide (data not shown). This is consistent with the involvement of a cysteine residue in catalysis, similar to its Epstein–Barr virus (EBV), human cytomegalovirus (HCMV), MCMV, and HSV-1 counterparts. We confirmed this by mutagenesis of the putative active site cysteine at position 98 to alanine (MDVUSP/C98A). We further generated recombinant MDVUSP and MDVUSP/C98A in Escherichia coli, and purified these preparations by means of their carboxyl-terminal His tags. Although the purified material showed some amino-terminal heterogeneity (possibly because of the use of alternate translational start sites), covalent adduct formation was unmistakable for the wild-type version, and altogether absent for the MDVUSP/C98A mutant (Fig. 1A). We then assayed the enzyme preparations for hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin (Ub-AMC), and only the wild-type enzyme hydrolyzed Ub-AMC (Fig. 1B). These results demonstrate catalytic activity and establish C98 as the active site cysteine of the MDVUSP/UL36 protein.
Fig. 1.
Activity of recombinant UL36USP. The first 322 residues of MDV UL36 were expressed recombinantly in E. coli and purified by means of a C-terminal His tag. (A) Purified enzyme was incubated in the presence or absence of a molar excess of Ha-UbVME for 15 min at room temperature and subjected to SDS/PAGE. Anti-6-His antibody was used to detect proteins by immunoblot. Unmodified enzyme and covalent adducts are indicated with arrows. A nonspecific background protein band is indicated with an asterisk. (B) MDVUSP and MDVUSP/C98A (100 pM) were incubated with a 1,000-fold excess of Ub-AMC (100 nM). Hydrolysis was measured as an increase in fluorescence, indicating release of AMC, over time. Filled circles are representative of MDVUSP activity, and open circles are representative of MDVUSP/C98A activity.
In Vitro Growth of an MDVUSP Active Site Mutant.
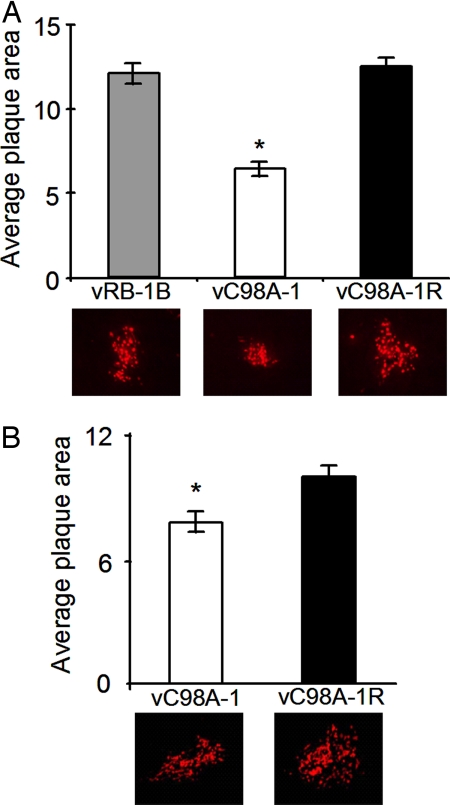
We used recombination-based mutagenesis of a modified infectious bacterial artificial chromosome (BAC) clone of MDV (pRB-1B), derived from the highly oncogenic RB-1B strain (21), to obtain two independent isolates of the C98A mutant in the USP domain of MDV (vC98A-1 and -2). In addition, we prepared revertants of each of the mutants (vC98A-1R and -2R), in which we restored the originally mutated sequences to the wild-type parental sequence. Analysis of the in vitro growth properties in primary chicken embryo cells (CEC) revealed an ≈20% reduction in plaque size [Fig. 2B (50% when plated on primary kidney epithelial cells; Fig. 2A]. The revertants, as expected, replicated in a manner indistinguishable from parental vRB-1B upon infection of chicken embryo fibroblast cells (CEC) or chicken kidney cell (CKC) cultures. When endpoint titers from infected cultures were measured, vC98A-1 grew to titers that were marginally (≈4-fold) lower when compared with the vC98A-1R revertant virus (vC98A-1, 0.6 ± 0.2 × 104; vC98A-1R, 2.2 ± 0.8 × 104). We therefore conclude that the activity of the MDVUSP is not essential for growth in tissue culture, although a slight defect in plaque formation was observed in primary chicken cells. The magnitude of such differences likely depends on the cell type on which the virus inoculum is plated.
Fig. 2.
Reduced plaque sizes in vC98A viruses. CKC (A) or CEC (B) cultures were infected with 100 plaques per well of a six-well dish with vRB-1B, vC98A-1, or vC98A-1R. Five days p.i., wells were fixed with 90% cold acetone for 10 min and air dried, and plaques were immunohistochemically stained by using anti-MDV polyclonal antibodies and anti-chicken secondary antibodies labeled with Alexa Fluor 568 (Molecular Probes). Digital images were obtained from 75–100 individual plaques by using an epifluorescence Axiovert 25 inverted microscope and an AxioCam HRc digital camera (Zeiss). Plaque areas were measured by using the NIH ImageJ software, and shown are the average plaque areas in calibrated units for each respective virus. The average area of plaques produced by the vC98A-1 was significantly smaller (≈50% for CKC and ≈25% for CEC) to the wild-type vRB-1B or revertant vC98A-1R (P < 0.01) by using Student's t tests. Shown below the histogram are representative plaques corresponding to the average plaque size of each virus.
In Vivo Replication and Pathogenicity of the MDVUSP/C98A Active Site Mutant.
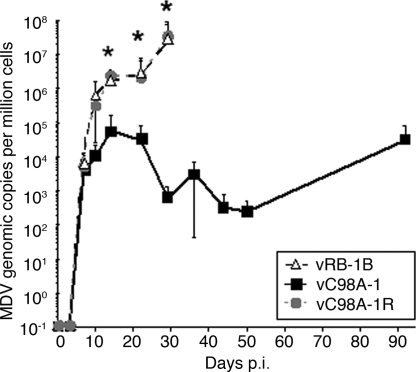
We performed two independent experiments to assess the role of the virus-encoded USP in vivo. Chickens (n = 13–15 per group) were inoculated with virus reconstituted from the parental pRB-1B (vRB-1B), the USP-defective mutants (vC98A-1 and -2) or their revertants (vC98A-1R and 2R). We measured viral load in peripheral blood mononuclear cells (PBMC) by qPCR. Early replication [7 days after infection (p.i.)] was similar between groups. The C98A mutation thus does not impair the ability of the virus to replicate in vivo. However, after 7 days p.i., viremia levels were reduced 1,000- to 10,000-fold for the USP-defective mutants when compared with parental vRB-1B or their respective revertant viruses (Fig. 3).
Fig. 3.
qPCR assays of chicken blood. DNA was obtained from blood of chickens infected with parental vRB-1B, the C98A-1 mutant, or its revertant C98A-1R. Shown are MDV genome copies per million cells normalized to the chicken iNOS gene. Considerable numbers of chickens were terminated in the vRB-1B and revertant groups by day 36 p.i. because of development of multifocal lymphoma, and blood samples could be used for comparison with the vC98A-1 mutant only up to 29 days p.i. Levels of viral DNA were significantly different between the mutant (vC98A-1) and both the vRB-1B and vC98A-1R groups at 14, 22, and 29 days p.i. by using Student's t test (P < 0.01) as indicated with an asterisk. In the repeat experiment, a similar deficiency in the vC98A-1 group was seen (data not shown).
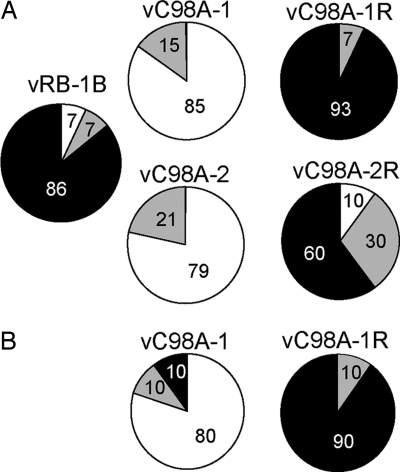
We examined chickens for signs of clinical Marek's disease, mainly characterized by wasting and gross T cell tumors, from 3 to 7 weeks p.i. In >90% of animals infected with vRB-1B or the revertant viruses, we observed clinical signs of Marek's disease. Also, vRB-1B and the revertant viruses regularly caused tumors at more than two distinct organ sites, evidence for a high level of metastasis (Fig. 4A). In stark contrast, none of the chickens infected with either of the two vC98A USP mutants presented with clinical signs of Marek's disease, even at 13 weeks p.i. At the termination of the experiment at 13 weeks p.i., ample time for Marek's disease to develop even with low-virulence strains (21), only five (three in the C98A-1 group and two in the C98A-2 group) of 27 chickens infected with the mutant virus contained tumors. However, these tumors were very small and isolated, with no signs of metastasis. We observed similar results in the repeat experiment (Fig. 4B).
Fig. 4.
Tumorigenesis in viruses lacking USP activity is severely impaired. Marek's disease incidence was evaluated over the course of 92 days in two experiments (A and B) and is shown as a percentage of Marek's disease incidence for each group separated by lymphoma incidence. Lymphoma incidence show the percentage of chickens of each group that failed to develop lymphomas (0, white) or chickens in which less than (≤2, gray) or more than (>2, black) two sites of lymphoma manifested.
Natural Reversion of USP Mutant in Vivo.
Using DNA obtained from surviving chickens at the termination of the experiment, we amplified a 444-bp PCR product, corresponding to the amino terminus of UL36, from DNA preparations of PBMC isolated from 10 chickens infected with the mutant virus. One of the viral samples contained wild-type virus sequences encoding a cysteine at position 98, indicating that natural reversion to the virulent, parental genotype had occurred in this chicken. We sequenced a PCR product from two of the three chickens with small tumors in the C98A-1 mutant group. One chicken maintained the mutant virus, at least in peripheral circulation, whereas another chicken contained virus with a revertant genotype. Although the frequency of reversion to the parental genotype is low (≈10%), the phenomenon was observed in a fairly short time frame. We do not know when reversion occurred, but because the chicken showed no overt signs of disease at termination of the experiment except a small lesion, we surmise that reversion must have taken place shortly before termination of the experiment.
Analysis of Horizontal Spread.
Finally, we examined whether animals housed in direct contact with either the C98A mutant or the revertant viruses, but not themselves inoculated directly with virus, contracted Marek's disease. This type of experiment, involving contact animals, is used to assay horizontal transmission of MDV. MDV starts to spread horizontally from chicken to chicken after ≈14 days p.i. Infection of naïve birds occurs by inhalation of chicken dander shed from the feather follicles of infected chickens (18, 20). As expected from the reduced viremia levels and the nearly complete absence of macroscopic tumors in the C98A mutant groups, contact animals remained free of Marek's disease lesions; however, four of eight contact chickens did contain viral DNA in PBMC as shown by qPCR assays performed for that group. In contrast, 78% (seven of nine) of animals in contact with parental or revertant virus succumbed to Marek's disease and all animals were virus positive, establishing that USP activity is dispensable for horizontal transmission of the virus. Similar results were seen in the second experiment in which 78% (seven of nine) in the vC98A-1R revertant group developed Marek's disease, whereas no chickens housed with the vC98A-1-infected group developed Marek's disease, but six6 of eight did contain MDV DNA, and one chicken contained naturally reverted sequences. MDV with deficiencies in lytic replication fails to spread horizontally, because the levels of production of infectious virus are insufficient for efficient delivery to the site of virus exit—the skin—and consequently transfer to contact animals is impossible (22, 23). Thus, although replication of MDVUSP mutant in vitro was mildly impaired, depending on the cell type examined, efficient lytic replication—at least at early stages of infection and sufficient for interindividual transfer—was achieved in vivo and accounts for horizontal transmission of even the mutant virus.
MDVUSP mRNA Expression in Transformed Cells.
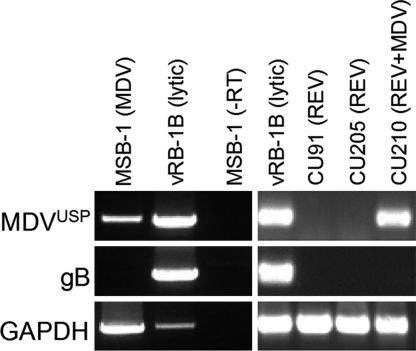
The discrepancy between efficient lytic virus replication in chickens at early times p.i. and the spread to sentinel animals, on the one hand, and the severely reduced incidence of lymphomas, on the other hand, prompted us to examine expression of MDVUSP in the MDV-transformed lymphoblastoid cell line (LCL) MSB-1 (24). These experiments were designed to examine whether MDVUSP would be expressed in the latent/tumor phase of infection. We performed RT-PCR analyses on RNA obtained from MSB-1 or LCL derived from retrovirus [reticuloendotheliosis virus (REV)]-infected birds (CU91 and CU205) used as negative controls. CKC cultures lytically infected with vRB-1B served as positive controls. We also examined glycoprotein B (gB) mRNA expression, which is observed only during lytic replication of MDV. MDV-infected CKC cultures expressed both the UL36/MDVUSP and gB transcripts but, as expected, the tumor MSB-1 cell line lacked expression of gB (Fig. 5). In contrast, we detected expression of UL36/MDVUSP mRNA in the LCL (Fig. 5). To confirm these findings, we examined another cell line transformed with the REV and superinfected with MDV. Similar to MSB-1, expression of latency/transformation-related MDV genes is tightly regulated in the CU210 cell line, and lytic transcripts were never detected (25). In the CU210 LCL, we observed the same pattern of expression of UL36/MDVUSP transcripts in the absence of gB. Although RNA samples were DNase-treated to exclude possible DNA contamination, the primers designed for amplification of gB and GAPDH span introns and exons to discriminate between cDNA and DNA, and we found no evidence for DNA contamination. When the MSB-1 LCL RNA was amplified in the absence of reverse transcriptase (−RT), we observed no amplification of the target genes, consistent with the absence of DNA contamination. Amplification of the UL36/MDVUSP sequence thus requires the presence of its mRNA. We conclude that MDVUSP mRNA is expressed in the latent and tumor stage of MDV infection, consistent with its proposed role for tumorigenesis in vivo.
Fig. 5.
MDVUSP mRNA expression in MDV-transformed LCL. RT-PCR assays were performed on RNA obtained from the MDV-transformed cell line MSB-1 by using primers specific for the MDVUSP and gB genes as described in Materials and Methods. As negative controls, two cell lines transformed with REV (CU91 and CU205) were used, in addition to positive controls of MDV-infected CKC cultures. The CU210 cell line that was initially transformed with REV and then was superinfected and “supertransformed” with MDV was also used because tight transcriptional control of MDV genes was reported in this cell line (25). As an internal RNA control, GAPDH mRNA expression was also examined. One sample was used without addition of reverse transcriptase (−RT) to show the PCR products produced are specific for RNA and not DNA contamination.
Discussion
In this report, we show that formation of T cell lymphomas by an oncogenic herpesvirus, MDV, is drastically reduced if the virus-encoded USP (MDVUSP) is absent. These findings implicate DUB activity in the establishment and/or maintenance of T cell transformation. Although MDV mutants defective in USP activity show moderate impairment of replication in vitro, this impairment appears minimal. First, the importance of the MDVUSP for viral replication may well be cell-type dependent, as evidenced by the fact that in vitro growth in primary CEC cultures were affected considerably less than growth in CKC cultures, with a mean reduction in plaque size of only ≈20% (Fig. 2). Second, after infection of a high dose of MDV in vivo, there was no detectable difference in early lytic replication (<7 days p.i.) between the USP-negative and parental or repaired viruses. Just before this time point after infection, initial replication in B cells by MDV switches to establishing latent infection in CD4+ T cells, which occurs at 7–10 days p.i (20). Note that the primary targets of MDV infection and replication in vivo cannot be used to assess plaque size or growth in vitro. Third, the fact that the USP-defective virus spreads to uninfected naïve chickens indicates that viral replication is sufficient to reach and replicate in the skin, the port of exit of infectious virus, and to establish an infection in contact chickens. Usually, attenuation and severe reduction of MDV lytic replication results in the absence of replication in the skin, and in such cases horizontal transmission does not occur (23). The reduced replication observed in this study is mild compared with many mutant MDV studied (22, 26, 27), and the dramatic reduction in oncogenic transformation of CD4+ T cells and tumor metastasis suggest that MDVUSP plays a more direct role in transformation or maintenance of the transformed state. Further evidence to support this rationale includes the detection of MDVUSP mRNA transcripts in MDV-transformed T cells (Fig. 5), which suggests MDVUSP activity in latently infected and transformed cells.
Although the identification of the exact role of MDVUSP in MDV tumorigenesis would require the molecular identification of its substrates and/or interacting partners, there is precedent in the literature that links deubiquitinating enzymes and oncogenesis. For example, USP7 deubiquitinates the tumor suppressor protein, p53, thereby rescuing it from degradation, and reduced expression of this protease is linked to increased cancer incidence (28). In line with this finding, USP28 is involved in stabilizing the myc protooncogene (29). A virally encoded DUB, such as MDVUSP, may well deubiquitinate proteins involved in transformation or maintenance of the transformed state. A possible role of MDVUSP might involve deubiquitination of the MDV oncoprotein, Meq, thus blocking its degradation via the proteasomal pathway and thereby allowing transformation and/or maintenance of transformation. Removal of ubiquitin from MDV's oncoprotein may also offer an explanation for the conundrum that activity of cytotoxic T lymphocytes toward transformed and Meq-expressing CD4+ T cells is not observed. As such, multiple roles for MDVUSP in transformation and lymphomagenesis are conceivable. Clearly, the absence of the herpesviral USP activity encoded by UL36 has profound effects in vivo and results in severely reduced tumor formation and metastasis. The observed natural reversion to the wild-type coding sequence further suggests that USP activity is beneficial to MDV survival in the host. Interestingly, horizontal transmission continues, albeit at reduced efficiency, indicating that transport of virus to the skin and assembly of infectious virus in feather follicle epithelia is still functional.
A number of herpesviruses express gene products that interact with USP7. Everett and colleagues (30) discovered that the HSV-1 viral immediate-early ICP0 binds to USP7, giving USP7 the nickname of herpesvirus-associated USP or HAUSP. This interaction contributes to the role of ICP0 as an activator of viral gene expression (31). In addition, the EBV EBNA-1 protein also interacts with USP7 and has been implicated to play a role in establishment or maintenance of EBV latency in B cells (32).
USP7 is not the only deubiquitinating enzyme associated with tumorigenesis. Mutations in CYLD, a USP implicated in modulation of NF-κB signaling, lead to cylindromatosis, a malignant condition of the skin attributed to a failure to deubiquitinate TRAF2. In the case of MDV, we observe the opposite phenomenon, the requirement of USP activity for tumor formation and metastasis. We know of no other situation where the expression of an active viral USP is involved in tumorigenesis, making the example of the MDVUSP unique thus far. Our results raise the possibility that inappropriate expression or increased activity of cellular USPs, likewise, could contribute to malignant transformation.
All eukaryotes use ubiquitin, and it is, therefore, no surprise that herpesviruses, dependent on eukaryotic hosts for replication, also exploit the ubiquitin machinery. The enzymatic cascade involved in the attachment and removal of ubiquitin is complex and involves numerous proteins capable of ubiquitination and its reversal. Genetic analysis in yeast demonstrates that none of the 17 USPs (except for the metalloprotease RPN11) is essential for survival (33), however, and even compound mutants deficient in several USPs survive, presumably due in part to the redundant nature of many of these enzymes. When viewed from that perspective, herpesviruses present a unique situation, and potentially a very beneficial one from a therapeutic point of view. The essential nature of the herpesvirus USP, demonstrated here, combined with the unique structure of its active site (12) suggests that this enzyme may be a promising candidate for antiviral drug therapy. Viruses that lack this activity might be appropriate for the construction of attenuated herpesviruses.
Materials and Methods
Cells and Viruses.
CEC and CKC cultures were prepared from specific-pathogen-free (SPF) embryos or chickens, respectively, following methods described (34) and were used to reconstitute BAC DNA viruses.
Lymphoblastoid T cell lines (LCL) transformed with either MDV (21) or REV (25) were grown in RPMI medium 1640 plus 10% FBS at 41°C in a humidified atmosphere of 5% CO2. The MSB-1 cell line was kindly provided by Mark S. Parcells (University of Delaware, Newark, DE) whereas the CU91, CU205, and CU210 cell lines were kindly provided by Karel A. Schat (Cornell University, Ithaca, NY). Before RNA extraction, dead cells were removed by centrifugation over Ficoll-Hypaque Plus (Amersham Pharmacia).
Cloning and Recombinant Expression.
The nucleotides encoding the amino-terminal 322 aa of MDV UL36 were cloned from the pRB-1B BAC by standard procedure and ligated into the pBAD24 vector in-frame with a carboxyl-terminal His Tag. The C98A mutant was constructed by using QuikChange site-directed mutagenesis kit (Stratagene) according to manufacturer instructions. Proteins were expressed in E. coli DH5alpha cells and purified by using Ni-NTA resin (Qiagen). Purified protein was dialyzed overnight in storage buffer [50 mM Tris, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, 10% (vol/vol) glycerol, (pH 7.5)] and snap frozen for storage at −80°C.
HA-UbVME Labeling.
Labeling reactions were performed with 1 μM enzyme in the absence or presence of 1 μM HA-UbVME in reaction buffer [50 mM Tris, 100 mM NaCl, 1 mM DTT (pH 7.5)] for 30 min at room temperature. After incubation, samples were resuspended in reducing sample buffer without boiling and subjected to SDS/PAGE analysis, followed by immunoblot with Penta-His antibody (Qiagen).
Ub-AMC Assays.
Ub-AMC was purchased from Boston Biochem. Ub-AMC hydrolysis assays were performed in assay buffer [50 mM Tris·HCl, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, (pH 7.5)], supplemented with 1 mg/ml BSA (Roche) at 25°C. Enzyme concentrations were determined with the Bradford reagent (Bio-Rad) and BSA (Pierce) as a standard. Measurements were performed in triplicate with 100 pmol of the respective enzyme and 100 nmol of substrate in a total volume of 30 μl per well in a 384-well plate. Measurements were taken by using a Spectramax M2 plate reader (Molecular Devices) with a 368-nm/467-nm filter pair and a 455-nm cutoff.
Construction of Mutant MDV.
Two-step red-mediated recombination was performed exactly as described (21, 35). See supporting information (SI) Fig. 1 for more detailed description and SI Table 1 for a list of primers used.
PCR and DNA Sequencing.
PCR assays were performed by using Reddy Mix (ABgene), 100 ng of template DNA, and 0.2 mM concentrations of the forward and reverse primers (SI Table 1). PCR products were gel purified by using the QIAquick Gel Extraction kit (Qiagen), and sequencing was performed by the DNA sequencing facility at Cornell University.
RNA Extraction and RT-PCR Analysis.
Total RNA was extracted from 1 × 107 cells by using RNA STAT-60 (Tel-Test) according to the manufacturer's instructions. RNA samples were DNase-treated with the DNA-free system from Ambion by using the manufacturer's instructions.
Reverse transcription was performed by using 1 μg of total RNA with the GeneAmp Gold RNA PCR Kit (Applied Biosystems) as described (36). The primers specific for GAPDH were described (36), whereas the gB- and UL36/MDVUSP-specific primers are shown in SI Table 1.
Animal Studies.
SPF P2a (MHC: B19B19) chickens were obtained from Cornell University's departmental flocks and housed in isolation units. Water and food were provided ad libitum. All experimental procedures were conducted in compliance with approved Institutional Animal Care and Use Committee (IACUC) protocols (2002-0085). Chickens were inoculated intraabdominally with 2,000 plaque-forming units (PFU) of the various viruses at 1 day of age, whereas control chickens were inoculated with uninfected CKC cultures. MDV spreads horizontally between animals after 14 days p.i.; therefore, groups of chickens were housed in separate rooms. Chickens were assigned to inoculation groups by using a randomization table.
Chicken Blood DNA Extraction and qPCR Assays.
To measure viral DNA copies in chicken blood by using qPCR assays, DNA was extracted from chicken blood as follows: 40 μl of blood was obtained from the wing vein and mixed with 20 μl of 0.1 M EDTA and then frozen at −80°C until all samples were collected. Ten microliters of blood/EDTA mix was used to collect DNA by using the DNeasy 96 Tissue kit from Qiagen following the manufacturer's instructions as described (21). Quantification of MDV genomic copies by using qPCR was performed as described (21, 26). Briefly, primers and probe specific for the MDV-ICP4 gene were used to detect MDV genomes, whereas DNA loading for each sample was normalized by using primers and probe specific for the chicken inducible nitric oxide synthase (iNOS) gene. All qPCR assays were performed in an ABI Prism 7500 Fast Real-time PCR System (Applied Biosystems), and the number of copies for ICP4 and iNOS DNA were determined and expressed as the number of MDV copies per 1 × 106 cells.
Supplementary Material
Acknowledgments
We thank Neil Margulis for excellent technical assistance and the Animal Care Staff in the Poultry Virus Isolation Unit of Cornell University for their help during the animal studies. This work was supported in part by grants from the National Institutes of Health (to N.O. and H.L.P.) and the U.S. Department of Agriculture (to N.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706295104/DC1.
References
- 1.Glickman MH, Ciechanover A. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Raiborg C, Rusten TE, Stenmark H. Curr Opin Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 3.Sigismund S, Polo S, Di Fiore PP. Curr Top Microbiol Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- 4.Sun LJ, Chen ZJ. Curr Opin Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Weissman AM. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 6.Kim JH, Park KC, Chung SS, Bang O, Chung CH. J Biochem. 2003;134:9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- 7.Soboleva TA, Baker RT. Curr Protein Pept Sci. 2004;5:191–200. doi: 10.2174/1389203043379765. [DOI] [PubMed] [Google Scholar]
- 8.Wing SS. Int J Biochem Cell Biol. 2003;35:590–605. doi: 10.1016/s1357-2725(02)00392-8. [DOI] [PubMed] [Google Scholar]
- 9.Love KR, Catic A, Schlieker C, Ploegh HL. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 10.Hemelaar J, Galardy PJ, Borodovsky A, Kessler BA, Ploegh HL, Ovaa H. J Proteome Res. 2004;3:268–276. doi: 10.1021/pr0341080. [DOI] [PubMed] [Google Scholar]
- 11.Wang JL, Loveland AN, Kattenhorn LM, Ploegh HL, Gibson W. J Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. Mol Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Misaghi S, Galardy PJ, Meester WJN, Ovaa H, Ploegh HL, Gaudet R. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 14.Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. J Virol. 2005;79:15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gredmark S, Schlieker C, Quesada V, Spooner E, Ploegh HL. J Virol. 2007;81:10300–10309. doi: 10.1128/JVI.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gauger L, Ploegh HL. Mol Cell. 2007;25:677–687. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baigent SJ, Smith LP, Nair VK, Currie RJW. Vet Immunol Immunopathol. 2006;112:78–86. doi: 10.1016/j.vetimm.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Jarosinski KW, Tischer BK, Trapp S, Osterrieder N. Exp Rev Vaccines. 2006;5:761–772. doi: 10.1586/14760584.5.6.761. [DOI] [PubMed] [Google Scholar]
- 19.Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. Nat Rev Microbiol. 2006;4:283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- 20.Calnek BW. In: Marek's Disease. Hirai K, editor. Berlin: Springer; 2001. pp. 25–55. [Google Scholar]
- 21.Jarosinski KW, Margulis NG, Kamil JP, Spatz SJ, Nair VK, Osterrieder N. J Virol. 2007;81:10575–10587. doi: 10.1128/JVI.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parcells MS, Anderson AS, Morgan TW. J Virol. 1995;69:7888–7898. doi: 10.1128/jvi.69.12.7888-7898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schat KA, Calnek BW, Fabricant J, Graham DL. Avian Pathol. 1985;14:127–146. doi: 10.1080/03079458508436213. [DOI] [PubMed] [Google Scholar]
- 24.Akiyama Y, Kato S, Iwa N. J Biken. 1973;16:177–179. [PubMed] [Google Scholar]
- 25.Pratt WD, Morgan RW, Schat KA. J Virol. 1992;66:7239–7244. doi: 10.1128/jvi.66.12.7239-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarosinski KW, Osterrieder N, Nair VK, Schat KA. J Virol. 2005;79:11647–11659. doi: 10.1128/JVI.79.18.11647-11659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamil JP, Tischer BK, Trapp S, Nair VK, Osterrieder N, Kung HJ. J Virol. 2005;79:6984–6996. doi: 10.1128/JVI.79.11.6984-6996.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuya D, Huang C, Liu D, Nakashima T, Yokomise H, Ueno M, Nakashima N, Sumitomo S. J Pathol. 2006;208:724–732. doi: 10.1002/path.1931. [DOI] [PubMed] [Google Scholar]
- 29.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. Nat Cell Biol. 2007;9:765–771. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 30.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everett RD, Meredith M, Orr A. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holowaty MN, Frappier L. Biochem Soc Trans. 2004;32:731–732. doi: 10.1042/BST0320731. [DOI] [PubMed] [Google Scholar]
- 33.Amerik AY, Li SJ, Hochstrasser M. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- 34.Schat KA, Purchase HG. In: A Laboratory Manual for the Isolation and Identification of Avian Pathogens. Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM, editors. Kennett Square, PA: Am Assoc Avian Pathol; 1998. pp. 223–234. [Google Scholar]
- 35.Tischer BK, von Einem J, Kaufer B, Osterrieder N. BioTechniques. 2006;40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 36.Jarosinski KW, O'Connell PH, Schat KA. Virus Genes. 2003;26:255–269. doi: 10.1023/a:1024447230464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.