Abstract
An essential step in the release of an extracellular enveloped virus particle is a budding event that ultimately separates virion and host cell membranes. For many enveloped viruses, membrane fission requires the recruitment of the class E vacuolar protein sorting (VPS) machinery by short, virally encoded peptide sequences termed “late-budding” or “L” domains. Some L-domain peptide sequences (e.g., PSAP) bind directly to components of class E VPS machinery, whereas others (e.g., PPxY) access it indirectly by recruiting ubiquitin ligases. Additionally, ubiquitin itself is known to be generally important for the fission of virion from cellular membranes, and because ubiquitination of cellular transmembrane proteins can signal the recruitment of class E machinery, a popular model is that deposition of ubiquitin on viral structural proteins mediates class E machinery recruitment. To test this model, we took advantage of a retroviral Gag protein from the prototypic foamy virus (PFV) that is almost devoid of ubiquitin acceptors, and we engineered it to generate extracellular virus-like particles in the complete absence of other viral proteins. Notably, we found that particle budding, induced by a class E VPS machinery-binding L domain (PSAP), proceeded efficiently in the absence of ubiquitin acceptors in PFV Gag. Moreover, when particle release was engineered to be dependent on a viral PPXY motif, the requirement for a catalytically active ubiquitin ligase was maintained, irrespective of the presence or absence of ubiquitin acceptor sites in PFV Gag. Thus, in this model system, ubiquitin conjugation to transacting factors, not viral proteins, appears critical for ubiquitin-dependent enveloped viral particle release.
An essential step in the formation of an extracellular enveloped virus particle is the separation of virion and host cell membranes. For many enveloped viruses, this requires the recruitment of an array of cellular proteins, including components of the class E vacuolar protein sorting (VPS) machinery that normally induce topologically analogous membrane fission events in cells (reviewed in refs. 1–4). Recruitment of membrane fission machinery is achieved via peptide motifs (PT/SAP, LYPxL, PPxY, or FPIV), encoded within viral structural proteins, termed late-budding or “L” domains (3, 4). These motifs can bind directly to components of the class E VPS machinery; for example, PT/SAP and LYPxL motifs bind endosomal sorting complex required for transport (ESCRT)-I and AIP-1/ALIX, respectively (5–10). Alternatively, PPxY and FPIV motifs require class E VPS machinery to induce virus particle release but do not bind directly to known components of it (3, 4, 7, 11).
In addition to the class E VPS machinery, ubiquitin ligases and ubiquitin are important for the fission of virion and cellular membranes, but their precise role is enigmatic (12, 13). This has been studied most intensively by using retroviruses, and several pieces of evidence suggest a general role for ubiquitin. For example, free ubiquitin is enriched in retrovirus particles (14) and a variable fraction of the major retroviral structural protein (Gag) can be mono- or oligoubiquitinated (15–18). Furthermore, proteasome inhibitors block the release of retroviruses that encode PT/SAP or PPxY L domains, most likely via ubiquitin depletion (17, 19–21). Moreover, known docking sites for ubiquitin ligases can induce virus or virus-like particle (VLP) release (17). Indeed, we and others have shown that particular HECT ubiquitin ligases (e.g., WWP1/AIP4) are recruited by viral PPxY motifs that bind to ligase-encoded “WW” domains and that a catalytically active ubiquitin ligase is essential for the efficient budding of viruses that encode PPxY L domains (22–30). Nevertheless, how ubiquitin ligases and ubiquitin functionally interface with class E factors, which are also required for PPxY motif-dependent viral budding, is not entirely clear (3, 12). Because ubiquitination can serve as a signal for recruitment of class E factors (7, 31–34), a popular assumption is that deposition of ubiquitin monomers or oligomers on viral structural proteins leads to class E machinery recruitment. Consistent with this notion, linkage of ubiquitin to the PTAP-encoding portion of HIV-1 Gag markedly increases affinity for Tsg101, an ESCRT-I subunit to which both PTAP and ubiquitin bind (7). Notably, two studies have shown that mutation of L-domain proximal ubiquitin acceptor sites in Gag proteins results in a strong block in retroviral particle release (35, 36), whereas a third study has noted weaker effects (30).
Although viral protein–ubiquitin conjugates might recruit machinery required for viral budding, there are examples of ubiquitin-dependent cellular processes, including certain class E VPS-dependent sorting events in which the ubiquitination of the cargo protein appears irrelevant (37–40). Additionally, the ubiquitin ligases involved in viral budding might interact directly with the class E VPS machinery (22, 41, 42). Therefore, to determine whether ubiquitination of viral proteins is generally required for enveloped viral particle release, we took advantage of an unusual retroviral Gag protein that is almost devoid of ubiquitin acceptor sites. Notably, we found that VLP release was efficient in the absence of ubiquitin acceptors in the viral protein, when dependent on a motif (PSAP) that directly recruits the class E VPS machinery. Strikingly, when particle release was engineered to be PPxY motif and ubiquitin ligase dependent, viral protein ubiquitin acceptors remained dispensable. Thus, ubiquitin conjugation to transacting factors rather than viral proteins appears critical for ubiquitin-dependent enveloped viral particle release.
Results
We noticed that the Gag proteins of the foamy viruses are remarkably lysine poor. This is an extremely unusual property and provided an opportunity to test whether ubiquitination of viral proteins is essential for viral particle budding. The prototypic foamy virus (PFV) encodes PT/SAP as its primary L domain (Fig. 1A), which binds to the Tsg101 component of ESCRT-I to induce budding (43, 44) and only a single lysine (residue 396; Fig. 1A). Nonetheless, there are potential ways in which the near absence of lysine acceptors in foamy virus (FV) Gag proteins could be circumvented while maintaining a requirement for viral protein ubiquitination in particle release. For example, unlike many retroviral Gag proteins, the N terminus of PFV Gag is not myristoylated and could provide a ubiquitin acceptor. Additionally, FV Pol proteins encode numerous lysines that could provide numerous opportunities for virion protein ubiquitination. Finally, FVs are unusual among retroviruses in that they assemble complete capsids in the cell cytoplasm, and the cognate Env protein is required to direct FV capsids to membranes for envelopment (45, 46). Notably, the cytoplasmic portion of PFV Env is extensively ubiquitinated (47). Thus, because virion proteins other than Gag could substitute for the presumed role of Gag ubiquitination in virion release, we first derived an experimental system in which extracellular PFV Gag VLPs could be generated in the absence of other viral proteins.
Fig. 1.
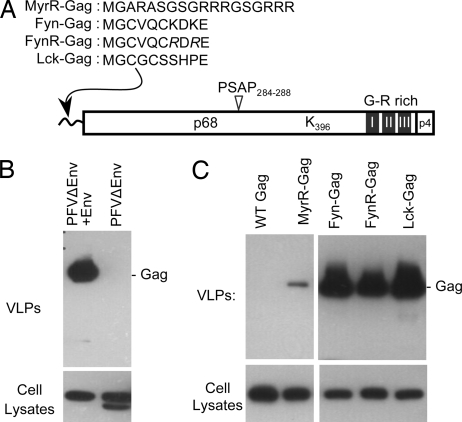
Envelope-independent release of VLPs generated by membrane-targeted PFV Gag proteins. (A) Schematic representation of the PFV Gag protein, indicating relevant landmarks [the PSAP L domain, a single lysine residue, glycine/arginine (G-R)-rich domains, and a proteolytic cleavage site near the C terminus of the ≈72-kDa protein]. Also shown are the various membrane-targeting peptides that were appended to the PFV Gag N terminus. (B) Virion production by 293T cells transfected with the PFVΔEnv proviral plasmid, either alone or along with a PFV Env expression vector. Virion and cell lysates were analyzed by Western blotting with αPFV human serum. (C) VLP production by 293T cells transfected with plasmids expressing PFV Gag appended with the indicated membrane-targeting signals.
Generation of PFV Gag VLPs in the Absence of Other Virion Proteins.
Previous work, using the Src oncoprotein myristoylation signal, has shown that appending the N terminus of PFV Gag with a membrane-targeting signal can bypass the requirement for Env coexpression in particle release (48). However, like the Src signal, most peripheral membrane-targeting signals contain lysine residues. Because this would complicate the interpretation of experiments that test the functional role of Gag ubiquitin acceptors, we designed PFV Gag proteins appended with candidate lysine-free plasma membrane-targeting peptides (Fig. 1A). These consisted of the following: (i) an artificial membrane-targeting peptide (MyrR), comprising the minimal six-residue myristoylation signal of HIV-1 Gag and an arginine-rich linker; (ii) a 10-residue myristoylated and palmitoylated peptide derived from the Fyn oncoprotein in which two lysines therein were mutated to arginine (FynR); and (iii) a naturally lysine-free 10-residue myristoylated and palmitoylated peptide from the Lck oncoprotein (Fig. 1A). Control experiments confirmed that the authentic PFV Gag protein, expressed from a PFV proviral plasmid, was released as particles only when Env was coexpressed (Fig. 1B). Concordantly, the PFV Gag protein, expressed in the absence of any other viral protein, did not generate extracellular VLPs (Fig. 1C). However, the membrane-targeted Gag proteins formed extracellular particles in the absence of Env. In the case of MyrR–Gag, VLP formation was inefficient, but the myristoylated/palmitoylated proteins (Fyn–Gag, FynR–Gag, and Lck–Gag) efficiently generated VLPs (Fig. 1C). Because Lck–Gag generated VLPs with marginally greater efficiency than FynR–Gag, without introducing lysine residues, it was selected for further experiments.
Lck–Gag Targets the Plasma Membrane and Requires Its PSAP Motif to Form Extracellular VLPs.
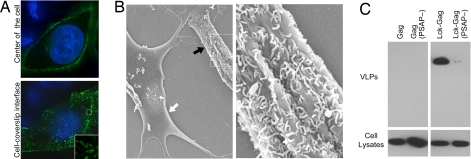
We next verified that our targeting strategy had the intended consequences. An Lck–Gag–CFP fusion protein efficiently targeted the plasma membrane in HeLa cells (Fig. 2A). Indeed, like most retroviral Gag–GFP fusion proteins, Lck–Gag–CFP formed fluorescent puncta at the cell surface, particularly when the cell surface–coverslip interface was imaged. To confirm that Lck–Gag assembled into VLPs, it was expressed in HeLa and DF1 cells, which have unusually smooth surfaces, facilitating the detection of surface VLPs by using scanning electron microscopy. Although we sometimes observed short filamentous particles on the surface of Lck–Gag-expressing HeLa or DF1 cells, it was often difficult to observe retroviral VLPs by this technique, unless their release was inhibited by coexpressing a dominant negative version of VPS4 [VPS4(E228Q)] (M.C.J, data not shown). When VPS4(E228Q) was coexpressed, Lck–Gag in DF1 cells we observed numerous filamentous particles extending from the cell's surface (Fig. 2B). Although these differed from the spherical cytoplasmic particles that were assembled by non-membrane-targeted PFV Gag (data not shown), they were indistinguishable from those assembled by murine leukemia virus (MLV) Gag, which has been shown to be filamentous in the absence of coexpressed glycosylated Gag (49) or when L-domain function is ablated (22, 50) [supporting information (SI) Fig. 7B].
Fig. 2.
Lck–Gag forms VLPs at the plasma membrane that are released in an L-domain-dependent manner. (A) HeLa cells expressing Lck–Gag–CFP (green) were fixed, and nuclei were stained with Hoechst (blue). Single deconvolved optical sections acquired at the center of the vertical dimension of the cell (Upper) or at the cell–coverslip interface (Lower) are shown. (B) Scanning electron micrograph of DF1 cells transfected with Lck–Gag and GFP–VPS4(E228Q). Representative transfected (black arrow) and untransfected (white arrow) cells, identified by GFP expression, are shown (Left). A higher-magnification view of area indicated by the dashed box is also shown (Right). (C) VLP production by 293T cells expressing wild-type PFV Gag or Lck–Gag proteins encoding either wild-type or mutant (PSAP284–287 mutated to AAAT) L domains.
As additional verification of the authenticity of Lck–Gag assembly, we determined whether VLP release required an intact L domain. Indeed, both the membrane-targeting signal and the PSAP motif in Lck–Gag were required (Fig. 2C). Thus, Lck–Gag mimics a conventional retroviral Gag protein in that it assembles, and is released, at the cell surface via a plasma-membrane-targeting signal and engagement of the class E VPS machinery.
Class E VPS Machinery and Ubiquitin-Ligase-Dependent Budding of Lysine-Free Gag.
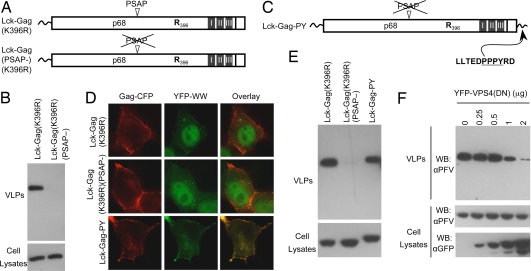
Next, we constructed Gag proteins devoid of ubiquitin acceptors by replacing the single lysine therein with arginine (Fig. 3A). Because the amino terminus of Lck–Gag is occupied by myristoylation, this manipulation should remove all ubiquitin acceptor sites. Remarkably, the Lck–Gag(K396R) protein efficiently generated extracellular VLPs (Fig. 3B), and this depended on engagement of the class E VPS machinery, because Lck–Gag(K396R)(PSAP-) failed to generated VLPs (Fig. 3B). Notably the efficiency with which Lck–Gag and Lck–Gag(K396R) formed VLPs was indistinguishable, strongly suggesting that viral protein ubiquitination is dispensable for the function of PT/SAP L domains.
Fig. 3.
PT/SAP and PPxY-type L domains induce budding of lysine-free Gag proteins. (A) Schematic representation of lysine-free Lck–Gag proteins. (B) Western blot (αPFV) analysis of Lck–Gag expression in and VLP release from 293T cells transfected with Lck–Gag(K396R) expression plasmids encoding either the wild-type or mutant L domains. (C) Schematic representation of the Lck–Gag–PY protein, which is lysine free and encodes an MLV-derived L domain, appended at its C terminus. (D) YFP–WW fusion protein localization in HeLa cells coexpressing lysine-free Lck–Gag–CFP proteins encoding a PSAP motif, no L domain, or the MLV L domain, as indicated. (E) Western blot analysis (αPFV) of VLP production by 293T cells expressing lysine-free Lck–Gag proteins encoding the indicated late domains. (F) Western blot analysis (αPFV) (Top and Middle) of VLP production by 293T cells expressing Lck–Gag–PY. Cells were cotransfected with the indicated amounts of a YFP–VPS4(E228Q) plasmid, the expression of which was monitored by using αGFP Western blotting (Bottom).
Because P(T/S)AP-type L domains bind directly to Tsg101 to recruit the class E VPS machinery, they could conceivably bypass the requirement for ubiquitination and thereby supplant the perceived role of ubiquitin in viral budding. Therefore, we next determined whether Gag ubiquitination is required for the function of an L domain that acts via ubiquitin ligase recruitment. To this end, we appended a MLV-derived L domain to the C terminus of Lck–Gag(K369R)(PSAP-), generating Lck–Gag–PY (Fig. 3C). PPxY motifs recruit HECT ubiquitin ligases by binding their WW domains; therefore, to verify that the transplanted MLV-derived L domain could indeed recruit a ubiquitin ligase (WWP1) that induces MLV budding (22), Lck–Gag–PY–CFP was coexpressed with YFP fused to the WW domains of WWP1 (YFP–WW). The YFP–WW fusion protein constitutively localizes to the nucleus but is relocalized, in a PPxY-dependent manner, when coexpressed with MLV Gag or Ebola virus matrix (22). Notably, YFP–WW was efficiently recruited to the plasma membrane in cells coexpressing Lck–Gag–PY–CFP but remained in the nucleus of cells expressing Lck–Gag proteins lacking the MLV L domain (Fig. 3D). Thus, the MLV PPxY motif was capable of recruiting WWP1 in the context of Lck–Gag–PY.
Remarkably, The PPxY L domain stimulated the budding of lysine-free Lck–Gag (Fig. 3E). Indeed, lysine-free Lck–Gag–PY generated VLPs only marginally less efficiently than Lck–Gag encoding the natural PSAP L domain (Fig. 3E). Moreover, Lck–Gag–PY particle release was inhibited by VPS4(E228Q), indicating that a requirement for a functioning class E VPS pathway in Lck–Gag–PY budding was maintained (Fig. 3F). Thus, an L domain that functions by recruiting a ubiquitin ligase was fully capable of inducing the release of VLPs via class E VPS factors, despite the absence of ubiquitin acceptor sites in the viral protein.
A Catalytically Active Ubiquitin Ligase Can Stimulate Budding of a Viral Protein Lacking Ubiquitin Acceptors.
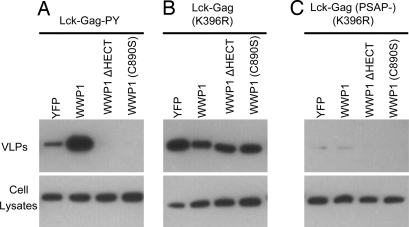
PPxY-dependent virus budding can sometimes be stimulated by overexpression of a HECT ubiquitin ligase. This effect is particularly evident with a “leaky” MLV PPxY mutant with a budding defect that can be reversed by WWP1 overexpression (22). Notably, overexpression of YFP–WWP1 markedly enhanced the release of particles generated by Lck–Gag–PY (Fig. 4A). This effect required the PPxY motif in Lck–Gag–PY because PSAP-dependent Lck–Gag VLP release was not enhanced (Fig. 4B), and release of an Lck–Gag lacking an L domain was not induced by WWP1 (Fig. 4C). Despite the fact that Lck–Gag–PY lacked ubiquitin acceptor sites, stimulation of VLP release required WWP1 catalytic activity. Indeed, expression of a truncated WWP1 lacking the catalytic domain (WWP1–ΔHECT) or an enzymatically inactive point mutant [WWP1(C890S)] inhibited rather than stimulated Lck–Gag–PY release (Fig. 4A). Again, these effects were specific to the PPxY L domain, because PSAP-dependent Lck–Gag VLP release was not affected (Fig. 4B). Thus, Lck–Gag–PY can recruit WWP1, and effect of this recruitment on particle budding was critically dependent on the catalytic activity of WWP1, even in the absence of ubiquitin acceptor sites in the viral protein.
Fig. 4.
Ubiquitin–ligase-stimulated, PPxY-dependent budding of lysine-free Gag. Western blot (αPFV) analysis of Gag expression and VLP production by 293T cells expressing lysine-free Lck–Gag proteins: Lck–Gag–PY (A); Lck–Gag(K396R) (B); or Lck–Gag(PSAP-)(K396R) (C). Each Lck–Gag protein was coexpressed with unfused YFP as a control or YFP fused to WWP1, WWP1 lacking the HECT domain (WWP1–ΔHECT), or a catalytically inactive mutant of WWP1 [WWP1(C890S)], as indicated.
Effects of Addition of Ubiquitin Acceptor Sites on Gag Ubiquitination.
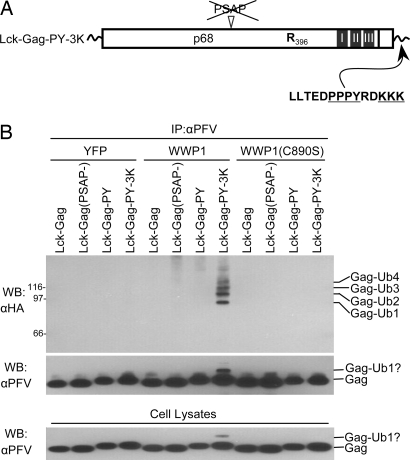
To exclude the possibility that our PFV Gag proteins became ubiquitinated by an alternative mechanism, independent of lysine or amino terminal acceptors, we determined whether ubiquitinated Gag proteins were generated in our assays. As a control, we prepared an Lck–Gag expression plasmid containing L-domain-proximal ubiquitin acceptors by appending three lysine residues to the C terminus of Lck–Gag–PY (generating Lck–Gag–PY–3K, Fig. 5A). We immunoprecipitated Lck–Gag proteins from 293T cell lysates, prepared 24 h after cotransfection with plasmids expressing (i) Lck–Gag variants, (ii) HA-tagged ubiquitin, and (iii) wild-type or inactive mutant WWP1. Thereafter, ubiquitinated Lck–Gag species were detected by immunoblotting. Lck–Gag proteins were efficiently immunoprecipitated and detected by using αPFV antibody (Fig. 5B), but ubiquitinated Lck–Gag was undetectable in cells expressing lysine-free proteins, regardless of which L domain was present and even when WWP1 was overexpressed (Fig. 5B). However, immunoprecipitates from cells expressing Lck–Gag–PY–3K contained several HA-reactive species corresponding to mono- and oligoubiquitinated Gag (Fig. 5B, Top). A secondary band of slightly higher molecular weight than Lck–Gag–PY–3K, likely corresponding to a monoubiquitinated species, was also detected by using αPFV in cells expressing Lck–Gag–PY–3K, but not Lck–Gag–PY (Fig. 5B, Middle and Lower). These ubiquitinated forms reflect WWP1-mediated ubiquitination, because they were observed in only cells overexpressing catalytically active, but not catalytically inactive, WWP1. Thus, despite the ability of Lck–Gag–PY to recruit a WWP1, its ubiquitination was detected only as a consequence of insertion of lysine residues. Conversely, lysine-free Lck–Gag proteins appear to be ubiquitination resistant.
Fig. 5.
Lysine-dependent, WWP1-induced ubiquitination of Lck–Gag. (A) Schematic representation of the Lck–Gag protein appended with the MLV L domain and three lysine residues at its C terminus (Lck–Gag–PY–3K). (B) Western blot (WB)/immunoprecipitation (IP) analysis of Lck–Gag proteins encoding the indicated late domains from 293T cells cotransfected with plasmids expressing Lck–Gag proteins, HA–ubiquitin, and unfused YFP, YFP–WWP1, or YFP–WWP1(C890S). All Lck–Gag proteins were lysine free except Lck–Gag–PY–3K. (Top and Middle) The αPFV immunoprecipitates were analyzed by Western blotting with an αHA monoclonal antibody (Top) or αPFV serum (Middle). (Bottom) Alternatively, unfractionated cell lysates were probed with αPFV serum. For the αHA blot, the migration of molecular weight markers is indicated to facilitate assignment of ubiquitinated forms.
Lck–Gag Ubiquitination Does Not Enhance Particle Release.
We next determined whether the presence of ubiquitin acceptors, and ubiquitin conjugation, could enhance PPxY-dependent particle release. First, we compared Lck–Gag–PY with Lck–Gag–PY–3K, the release of which depends on HECT ubiquitin ligase(s) and differ only in the presence or absence of L-domain proximal ubiquitin acceptors. Notably, Lck–Gag–PY–3K generated extracellular VLPs with no greater efficiency than lysine-free Lck–Gag–PY (Fig. 6A, Left). Moreover, ubiquitinated Gag species were detectable in VLPs assembled by Lck–Gag–PY–3K but not lysine-free Gag proteins (Fig. 6A, Right). Whereas WWP1 overexpression was required to detect Lck–Gag–PY–3K ubiquitination in cell lysates (Fig. 5), this manipulation was not required to observe ubiquitinated Gag in VLPs (Fig. 6A), perhaps because ubiquitinated forms are enriched in VLPs. Nevertheless, under conditions in which ubiquitinated Gag was, or was not, generated, overall VLP release was equivalent. Additionally, WWP1 stimulated the release of particles generated by using Lck–Gag–PY–3K (Fig. 6B). Importantly, however, the magnitude of this effect was no greater than that observed with lysine-free Lck–Gag–PY (Fig. 6B). Thus, WWP1-stimulated, PPxY-dependent particle release occurred equivalently whether or not it was accompanied by Gag ubiquitination.
Fig. 6.
Presence of ubiquitin acceptors or Gag ubiquitination does not affect ubiquitin-ligase-induced Lck–Gag budding. (A) Western blot analysis of VLP production by 293T cells coexpressing HA–ubiquitin and the indicated PFV Gag proteins. VLP lysates were analyzed by Western blotting with αPFV serum (Left) or monoclonal αHA antibody (Right). Migration of molecular weight markers is indicated to facilitate assignment of ubiquitinated forms. (B) Western blot (αPFV) analysis of VLP production by Lck–Gag–PY proteins that were lysine-free or that bore three appended lysine residues at the C terminus, as indicated. The Lck–Gag–PY proteins were expressed either with unfused YFP, as a control, or with YFP-fused WWP1, as indicated.
Discussion
Although the major structural proteins of enveloped viruses are often ubiquitinated as a consequence of L-domain function, it was not clear whether this is critical for the release of viral particles or a bystander reactions (3, 12, 13). Whereas mutation of a single lysine in HTLV-I Gag substantially reduces Gag ubiquitination but has rather modest effects on virion budding (30), two other studies have shown that mutation of multiple lysine residues within the Gag proteins of HIV-1 and Rous sarcoma virus particles can strongly inhibit particle release (35, 36). However, it is possible that the assembly defects observed in these studies were the result of structural perturbations, rather than attributable to loss of ubiquitin acceptors. Here, we used a remarkable, naturally occurring, lysine-poor retroviral Gag protein to demonstrate that L domains can function in the complete absence of viral protein ubiquitination. Notably, this is true of an L domain that depends on the catalytic activity of a ubiquitin ligase (22). Although it is possible that the role, if any, played by viral protein ubiquitination could vary according to cell and virus type, the mechanisms by which membrane domains are selected for assembly or with particle morphology, our findings indicate that viral protein ubiquitination is not universally required for L domain function. Rather, these findings strongly suggest that ubiquitination of unidentified transacting cellular proteins rather than viral proteins can be responsible, at least in some cases and perhaps as a general rule, for ubiquitin-dependent enveloped virus particle release.
Conceptually related findings have recently been made during analyses of the sorting of certain cellular proteins. Specifically, SnaIII sorting into the interior of the yeast vacuole (and by inference engagement of the class E VPS machinery) depends on a PPxY motif and recruitment of the yeast ortholog of WWP1, Rsp5. However, Sna3p sorting appears to be at least partly independent of lysine residues in Sna3p itself and perhaps Rsp5 catalytic activity (38–40). Additionally, Rsp5 stimulates Ste2p endocytosis, even when lysines are removed from Ste2p (37). Thus, in both viral and cellular systems, ubiquitin-dependent or ubiquitin-ligase-dependent processes do not always require substrate protein ubiquitination.
In principle, there are several ways, other than viral protein ubiquitination, in which HECT ubiquitin ligases could function to stimulate viral particle release via the class E VPS machinery. First, we previously found that the isolated HECT domain of WWP1 behaves like a soluble class E factor in that it is recruited to mutant VPS4(E228Q)-induced compartments (22) (on which all know class E factors accumulate). Moreover, there may be physical interactions between HECT ubiquitin ligases and class E VPS machinery (41, 42). Thus, HECT ubiquitin ligases could function as adaptor proteins to physically bridge PPxY motifs and the class E machinery. Second, ubiquitination could regulate the activity and stability of components of the class E VPS machinery. Indeed, certain class E VPS factors, or proteins that bind to class E VPS factors, can be ubiquitinated by HECT ubiquitin ligases (42, 51), and this could facilitate or disrupt their interactions with each other. Third, the ubiquitin-conjugated HECT domain that forms as an intermediate before ubiquitin transfer to the presumed substrate, or alternatively, autoubiquitinated forms of the ligase, could serve to recruit class E VPS machinery. If the ubiquitin conjugated ligase were simultaneously bound to a PPxY-encoding viral protein, then it could, in principle, nucleate the assembly of the class E VPS machinery at the site of viral budding. Clearly, the role of ubiquitin ligases in cellular and viral protein sorting and membrane fission processes is more complex than simply tagging cargos with ubiquitin, and further work will be required to determine precisely how they determine the fate of the proteins to which they bind.
Materials and Methods
Plasmid Construction.
Recombinant plasmids used to generate the various Gag proteins are described in SI Methods.
VLP Release Assay.
293T cells (3 × 106) in 100-mm plates were transfected with 2 μg of pCAGGS/Gag plasmids, alone or with 4 μg of pCR3.1/YFP, pCR3.1/YFP–WWP1 (or a derivative), or various amounts of YFP–VPS4(E228Q). VLPs were pelleted by ultracentrifugation of 8 ml of 0.22-μm-filtered culture supernatants, collected 24 h after transfection over a 4-ml 20% sucrose cushion for 2 h at 100,000 × g. VLP and cell lysates were analyzed by Western blotting.
Gag Ubiquitination Assay.
293T cells (5 × 105) in six-well plates were cotransfected with 1 μg of pCAGGS/Gag-derived plasmids, 500 ng of pHA–ubiquitin, and 1 μg of pCR3.1/YFP, pCR3.1/YFP–WWP1, or pCR3.1/YFP–WWP1–C890S. Twenty-four hours after transfection, cells were lysed and cleared of cellular debris by microcentrifugation. Gag proteins were then immunoprecipitated with αPFV serum and protein G–Sepharose beads and analyzed by Western blotting.
Western Blot Analysis.
Virion and cell lysates and immunoprecipitates were separated on polyacrylamide gels and transferred to nitrocellulose membranes. Gag and HA–ubiquitin were detected by using αPFV serum and a monoclonal αHA antibody, respectively, followed by peroxidase-conjugated secondary antibodies and chemiluminescent substrate reagents.
Fluorescence Microscopy.
HeLa cells (4 × 105) on 35-mm poly-d-lysine-coated glass bottom dishes (Mattek) were cotransfected with plasmids expressing CFP-fused and unfused Lck–Gag at a 1:1 ratio, either alone or along with 200 ng of pCR3.1/YFP–WWP1–WW. Twenty-four hours after transfection, cells were fixed in paraformaldehyde and visualized by deconvolution microscopy by using an Olympus IX70-based DeltaVision suite (Applied Precision) and a ×100 objective, as described in ref. 22.
Electron Microscopy.
DF-1 cells were transfected with plasmids expressing GFP–VPS4(E228K) along with Lck–Gag or MLV GagPol along with pdsRED and fixed in paraformaldehyde. After fluorescence imaging, cells were fixed in glutaraldehyde, postfixed in osmium tetroxide, dehydrated in ethanol, critical point dried, and sputter coated with platinum. Cells were identified by their location on a finder grid and imaged by using a Hitachi S4700 field emission SEM (University of Missouri Electron Microscopy Core Facility).
Supplementary Material
Acknowledgments
We thank Scott Eastman and members of the P.D.B. Laboratory for advice and Axel Rethwilm (University of Wurtzburg, Wurtzburg, Germany) for reagents. This work was supported by National Institutes of Health Grants R01 AI052774 and AI05111 (to P.D.B.) and a Wellcome Trust grant (to M.O.M.). P.D.B. is a Scientist of the Elisabeth Glaser Pediatric AIDS Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708002104/DC1.
References
- 1.Hurley JH, Emr SD. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katzmann DJ, Odorizzi G, Emr SD. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz PD. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 4.Morita E, Sundquist WI. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 5.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Proc Natl Acad Sci USA. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Serrano J, Zang T, Bieniasz PD. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 7.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 8.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 9.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demirov DG, Ono A, Orenstein JM, Freed EO. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. J Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Serrano J. Traffic. 2007;8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- 13.Vogt VM. Proc Natl Acad Sci USA. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putterman D, Pepinsky RB, Vogt VM. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 15.Ott DE, Coren LV, Chertova EN, Gagliardi TD, Schubert U. Virology. 2000;278:111–121. doi: 10.1006/viro.2000.0648. [DOI] [PubMed] [Google Scholar]
- 16.Ott DE, Coren LV, Sowder RC, II, Adams J, Nagashima K, Schubert U. J Virol. 2002;76:3038–3044. doi: 10.1128/JVI.76.6.3038-3044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidecker G, Lloyd PA, Fox K, Nagashima K, Derse D. J Virol. 2004;78:6636–6648. doi: 10.1128/JVI.78.12.6636-6648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patnaik A, Chau V, Wills JW. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, Bennink JR, Krausslich HG, Yewdell JW. Proc Natl Acad Sci USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shehu-Xhilaga M, Ablan S, Demirov DG, Chen C, Montelaro RC, Freed EO. J Virol. 2004;78:724–732. doi: 10.1128/JVI.78.2.724-732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harty RN, Brown ME, McGettigan JP, Wang G, Jayakar HR, Huibregtse JM, Whitt MA, Schnell MJ. J Virol. 2001;75:10623–10629. doi: 10.1128/JVI.75.22.10623-10629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakurai A, Yasuda J, Takano H, Tanaka Y, Hatakeyama M, Shida H. Microbes Infect. 2004;6:150–156. doi: 10.1016/j.micinf.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Yasuda J, Hunter E, Nakao M, Shida H. EMBO Rep. 2002;3:636–640. doi: 10.1093/embo-reports/kvf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasuda J, Nakao M, Kawaoka Y, Shida H. J Virol. 2003;77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. Proc Natl Acad Sci USA. 2001;98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouamr F, Melillo JA, Wang MQ, Nagashima K, de Los Santos M, Rein A, Goff SP. J Virol. 2003;77:11882–11895. doi: 10.1128/JVI.77.22.11882-11895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heidecker G, Lloyd PA, Soheilian F, Nagashima K, Derse D. J Virol. 2007;81:9769–9777. doi: 10.1128/JVI.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano J, Perez-Caballero D, Bieniasz PD. J Virol. 2004;78:5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 34.Bishop N, Horman A, Woodman P. J Cell Biol. 2002;157:91–101. doi: 10.1083/jcb.200112080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottwein E, Jager S, Habermann A, Krausslich HG. J Virol. 2006;80:6267–6275. doi: 10.1128/JVI.02177-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spidel JL, Craven RC, Wilson CB, Patnaik A, Wang H, Mansky LM, Wills JW. J Virol. 2004;78:10606–10616. doi: 10.1128/JVI.78.19.10606-10616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn R, Hicke L. J Biol Chem. 2001;276:25974–25981. doi: 10.1074/jbc.M104113200. [DOI] [PubMed] [Google Scholar]
- 38.Watson H, Bonifacino JS. Mol Biol Cell. 2007;18:1781–1789. doi: 10.1091/mbc.E06-10-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNatt MW, McKittrick I, West M, Odorizzi G. Mol Biol Cell. 2007;18:697–706. doi: 10.1091/mbc.E06-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ. Mol Biol Cell. 2007;18:707–720. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medina G, Zhang Y, Tang Y, Gottwein E, Vana ML, Bouamr F, Leis J, Carter CA. Traffic. 2005;6:880–894. doi: 10.1111/j.1600-0854.2005.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. Dev Cell. 2003;5:709–722. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- 43.Patton GS, Morris SA, Chung W, Bieniasz PD, McClure MO. J Virol. 2005;79:6392–6399. doi: 10.1128/JVI.79.10.6392-6399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stange A, Mannigel I, Peters K, Heinkelein M, Stanke N, Cartellieri M, Gottlinger H, Rethwilm A, Zentgraf H, Lindemann D. J Virol. 2005;79:5466–5476. doi: 10.1128/JVI.79.9.5466-5476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldwin DN, Linial ML. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Muller JG, Rethwilm A. J Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanke N, Stange A, Luftenegger D, Zentgraf H, Lindemann D. J Virol. 2005;79:15074–15083. doi: 10.1128/JVI.79.24.15074-15083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eastman SW, Linial ML. J Virol. 2001;75:6857–6864. doi: 10.1128/JVI.75.15.6857-6864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Low A, Datta S, Kuznetsov Y, Jahid S, Kothari N, McPherson A, Fan H. J Virol. 2007;81:3685–3692. doi: 10.1128/JVI.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamenova SD, Dunn R, Adler AS, Hicke L. J Biol Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.