Abstract
Compartmentalization of proteins into subcellular organelles in eukaryotic cells is a fundamental mechanism of regulating complex cellular functions. Many proteins of Plasmodium falciparum merozoites involved in invasion are compartmentalized into apical organelles. We have identified a new merozoite organelle that contains P. falciparum rhomboid-1 (PfROM1), a protease that cleaves the transmembrane regions of proteins involved in invasion. By immunoconfocal microscopy, PfROM1 was localized to a single, thread-like structure on one side of the merozoites that appears to be in close proximity to the subpellicular microtubules. PfROM1 was not found associated with micronemes, rhoptries, or dense granules, the three identified secretory organelles of invasion. Release of merozoites from schizonts resulted in the movement of PfROM1 from the lateral asymmetric localization to the merozoite apical pole and the posterior pole. We have named this single thread-like organelle in merozoites, the mononeme.
Keywords: malaria
Apicomplexa are named for a set of secretory organelles (rhoptries, micronemes, and dense granules) found at the apical end of these parasites, the end that invades cells in the vertebrate host (1). These organelles were first identified by their distinct morphological appearances in transmission electron microscopy (2–4). Many parasite proteins required for invasion of erythrocytes are segregated into the micronemes (5, 6). For example, Plasmodium falciparum apical membrane antigen 1 (AMA1) is held in the micronemes in merozoites inside of erythrocytes. Release from the erythrocyte triggers the movement of AMA1 to the cell surface where it functions in invasion (7, 8). Similarly, rhoptries sequester a different set of proteins and their contents are released onto the erythrocyte surface (4, 9), presumably to break the local cytoskeleton and to initiate formation of the parasitophorous vacuole. Taken together, these observations provide evidence for compartmentalization of parasite proteins into distinct organelles with related functions in invasion. Presumably, such compartmentalization provides a mechanism for orchestrating the timing of delivery or activity of the proteins during the complex process of invasion.
Apicomplexan rhomboid proteases have been implicated to play an important role in host cell invasion (10, 11). PfROM1 substrates have been identified in P. falciparum by using a mammalian cell-based proteolytic assay (12) that identified various micronemal proteins as potential substrates including AMA1. Because AMA1 has been implicated as essential for invasion, separation of PfROM1 from this substrate within the parasite may be important to prevent premature cleavage. Studies defining the spatiotemporal distribution of PfROM1 in P. falciparum merozoites are, therefore, likely to contribute to a clearer understanding of its role in erythrocyte invasion. Expression of hemagglutinin (HA)-tagged P. falciparum rhomboid-1 (PfROM1) localizes it to a new subcellular compartment distinct from other known merozoite organelles. PfROM1 is present in an asymmetric compartment that appears to be in close proximity to the subpellicular microtubules and similarly runs along the length of the merozoite. We have named this organelle mononeme (Greek: mono, single; neme, thread). Here, we present a detailed subcellular localization of the mononeme in comparison with other known merozoite organelles.
Results
Localization of Transgenic PfROM1 in P. falciparum Merozoites.
The full-length P. falciparum ROM1 gene was confirmed to consist of four exons by RT-PCR with a cDNA ORF of 837 bp encoding a 278-aa protein [supporting information (SI) Figs. 7 and 8]; GenBank accession no. EU180604. PfROM1 with an amino-terminal triple-hemaglutinin (HA) tag (HA-PfROM1) was cloned under the control of its own promoter elements and expressed episomally under drug selection (5 nM WR99210) in the 3D7 clone of P. falciparum. HA-PfROM1 expression was confirmed by using a HA-specific monoclonal antibody (mAb) on immunoblot and in immunofluorescence assay (IFA). By immunoblot, anti-HA mAb detected a single protein of ≈35 kDa, the predicted size of HA-tagged PfROM1, in the transgenic HA-ROM1 3D7 parasite extract (Fig. 1A, lane 1). No reactivity was observed with the parental 3D7 clone (Fig. 1A, lane 2). In IFA, the anti-HA mAb stained mature stages of the transgenic parasites starting from the early schizont stage of two to four nuclei. At this stage, staining was throughout the cell with no clear recognizable pattern (Fig. 1B). However, in mature transgenic schizonts with distinctly separated merozoites, termed segmenters, the staining was highly organized and appeared asymmetrically along one side of the merozoite. The pattern of HA-PfROM1 staining included a bulbous area toward the merozoite apical end and a stalk-like region that extended down one side of the merozoite toward the posterior (Fig. 1C). The fact that PfROM1 is expressed at the merozoite stage, the stage that invades erythrocytes, is consistent with this protease playing a role in the process of invasion. No staining was observed with the parental 3D7 parasites, indicating that there is no cross-reactivity of the anti-HA-specific mAb with parasite proteins (data not shown). Few unstained parasites were observed in the transgenic population (data not shown) that may have deleted part of the episomal promoter or the HA-PfROM1 gene itself.
Fig. 1.
Anti-HA antibodies specifically recognize the HA-PfROM1 protein in transgenic HA-PfROM1 3D7 parasites. (A) Western blot analysis of mature parasite extract probed with anti-HA antibody. Lane 1 shows transgenic HA-PfROM1–3D7 parasite protein extract and lane 2 shows parental 3D7 protein extract. Numbers on the left indicate positions of molecular weight markers in kilodaltons. The arrowhead shows the position of HA-PfROM1 detected in the transgenic parasite extract. (B) IFA analysis of HA-PfROM1 in a developing schizont (4 nuclei stage). (C) Merozoites in a segmenter probed with anti-HA rat mAb 3F10 (green) and the nuclear stain (blue, Upper) or with anti-AMA1 (red, Lower). Note in the individual merozoites anti-HA mAb staining appears to be localized on one side of the nucleus, with characteristic bulbous region (arrowheads) that is toward the apical end of the merozoites. The stalk region runs along one side of the nucleus. (Scale bar, 1 μm.)
Distinct Localization Pattern of PfROM1 Identifies the Mononeme.
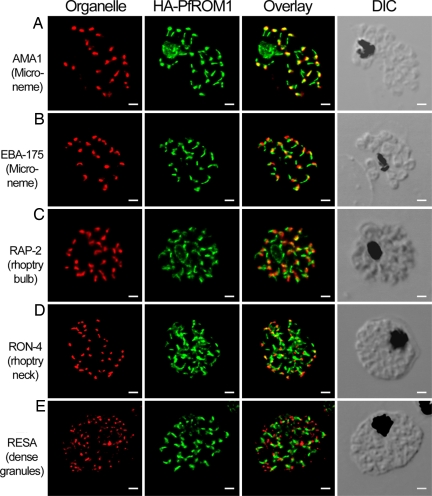
We compared the subcellular localization of HA-PfROM1 with that of proteins in other defined cellular organelles and structures in P. falciparum, including those proteins found in the apical secretory organelles: micronemes, rhoptries, and dense granules. As summarized in Fig. 2, HA-PfROM1 has a subcellular localization distinct from known micronemal markers PfAMA1 and PfEBA175 (Erythrocyte Binding Antigen-175) (Fig. 2 A and B, respectively). This is in contrast with a report that assigned PfROM1 to be exclusively in micronemes (13). The construct used in the previous P. falciparum study (13) lacked 76 aa encoded by the first two exons of PfROM1 that included the first transmembrane domain of PfROM1. This deletion may have affected its localization. HA-PfROM1 staining also differs from the rhoptry bulb marker PfRAP2 (rhoptry-associated protein-2; Fig. 2C) or the rhoptry neck protein PfRON4 (Fig. 2D). Similarly, HA-PfROM1 staining distinctly differed with the dense granule marker, RESA (ring-infected erythrocyte surface antigen; Fig. 2E). Thus, these results indicate that HA-PfROM1 is localized in a subcellular compartment distinct from the micronemes, rhoptries, and dense granules.
Fig. 2.
HA-PfROM1 is not localized to known apical secretory organelles: rhoptries, micronemes, and dense granules. Staining with antibodies against different secretory organelle markers (red) is shown in the left-hand column. (A) AMA1, a micronemal marker; (B) EBA175RII, another micronemal marker; (C) RAP2, a rhoptry bulb marker; (D) RON4, a rhoptry neck marker; (E) RESA, a dense granule marker. Corresponding costaining with rat anti-HA mAb 3F10 (green) together with the overlay between the red and green channels is shown in the subsequent columns. The right-hand column shows the corresponding differential interference contrast microscopy (DIC) images. (Scale bar, 1 μm.)
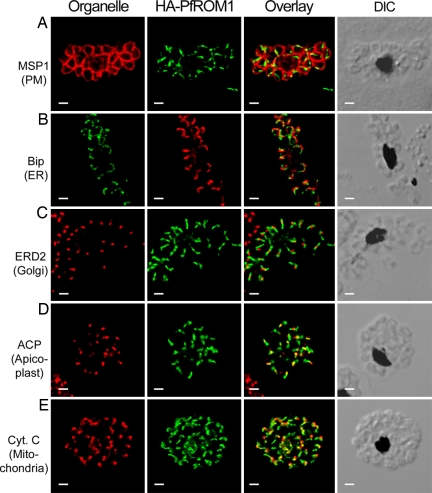
Using antibodies to the merozoite surface protein-1 (MSP1), a protein that is located in the merozoite plasma membrane (Fig. 3A), we demonstrated that HA-PfROM1 staining is intracellular, not colocalizing with the plasma membrane. The staining pattern of HA-PfROM1 is distinct from endoplasmic reticulum (anti-Bip, Ig binding protein of the ER; Fig. 3B) and Golgi (anti-PfERD2, ER lumen protein retaining receptor 2, a cis-Golgi marker; Fig. 3C). These organelles are asymmetrically organized on the same lateral side of the merozoite as HA-PfROM1. Further, HA-PfROM1 staining is distinct from the more round-appearing apicoplast (anti-ACP, acyl-carrier protein; Fig. 3D), but always on the same side of the nucleus as the apicoplast. The apicoplast appears to be localized at the distal end of HA-PfROM1 or in the middle of HA-PfROM1, but the staining of the two organelles does not overlap.
Fig. 3.
HA-PfROM1 does not localize with markers of different known subcellular organelles. Staining with antibodies against different subcellular organelle markers is shown in the left-hand column. (A) MSP1, a merozoite surface marker; (B) Bip, an ER marker; (C) ERD2, a cis-Golgi marker; (D) ACP, an apicoplast marker; (E) cytochrome c, a mitochondrial marker. Corresponding costaining with rat anti-HA mAb 3F10 (A and C–E) or mouse anti-HA mAb 2C16 (B) together with the overlay between the red and green channels is shown in the subsequent columns. The right-hand column shows the corresponding DIC images. (Scale bar, 1 μm.)
HA-PfROM1 Is in Close Proximity to the Merozoite Subpellicular Microtubules.
P. falciparum merozoites have three longitudinally oriented microtubules on one side of the merozoite (14). To determine whether the asymmetric location of PfROM1 is associated with microtubules, we determined the staining pattern of the microtubules with antibodies specific for chicken brain α-tubulin and its relation to HA-PfROM1. This antitubulin antibody has been demonstrated to specifically react with P. falciparum α-tubulin expressed in asexual blood stage development (15). HA-PfROM1 was observed to be localized in close proximity to longitudinal subpellicular microtubules of the merozoite (Fig. 4) with extensive staining overlap between them.
Fig. 4.
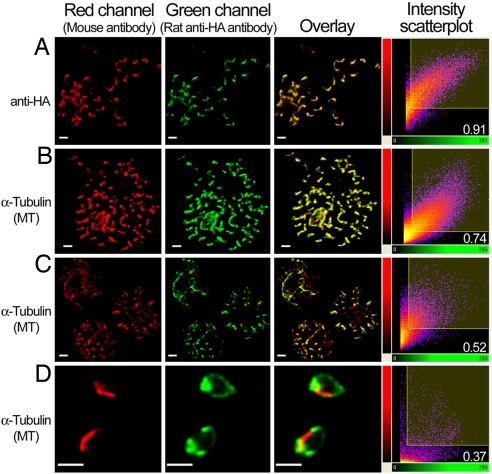
Quantitative assessment of the colocalization between HA-PfROM1 and microtubule staining. (A) Staining with mouse anti-HA mAb 2C16 and rat anti-HA mAb 3F10. (B–D) Staining with mouse anti-microtubule mAb DM1A and rat anti-HA mAb of mature segmenters, immature schizonts, and free merozoites, respectively. Scatter 2D plots of voxel intensities in red and green channels observed for the respective pictures are shown in the right-hand column. Intensity threshold in both the channels was automatically determined by excluding dark pixels by using the algorithm in “Coloc” function of the image analysis software, Imaris (Bitplane) as described in ref. 33. Pearson's correlation coefficient was determined for voxel intensity correlation between the red and green channels and is displayed in the lower-right-hand corner. (Scale bar, 1 μm.)
Specificities of the anti-HA and anti-α-tubulin antibodies was verified to rule out the possibility of potential cross-reactivities resulting in their apparent colocalizations. As seen in Fig. 1A and SI Fig. 9A, anti-HA antibodies only react with the transgenic parasites expressing the HA-PfROM1 (35 kDa), excluding the possibility that it is cross-reacting with microtubules. In addition, the anti-α-tubulin antibody detects only parasite α-tubulin in monomeric (50 kDa) or higher polymeric forms (100–150 kDa) in the transgenic HA-PfROM1 3D7 parasite and does not detect the 35-kDa HA-PfROM1 protein (SI Fig. 9B), indicating its specificity and lack of cross-reactivity with HA-PfROM1. The staining of PfROM1 and microtubules no longer overlap in merozoites released from infected erythrocytes, indicating that costaining observed in the segmenters is due to their close proximity and not cross-reactivity (see below).
To quantify the extent of colocalization between HA-PfROM1 and the microtubules, confocal images were analyzed by the “Coloc” module of the Imaris software package (Bitplane). Results of the correlation analysis between the staining intensities observed for the two antibodies were expressed as Pearson correlation coefficients. Colocalization of HA-PfROM1 and microtubules was calibrated to images of merozoites within segmenters stained with two different HA-specific mAbs labeled with green and red fluorescent dyes (Fig. 4A and data not shown) that had a correlation coefficient of 0.922 ± 0.007 (mean ± SE) in three experiments. A representative image of the microtubules and HA-PfROM1 staining of merozoites within segmenters is shown in Fig. 4B. The correlation coefficient of the staining overlap between them (Fig. 4B and data not shown) was 0.691 ± 0.027 (mean ± SE) in four different mature schizonts and free merozoite clusters analyzed. Further, in less mature schizonts, the two antibodies overlap less (Fig. 4C, coefficient of correlation of 0.52) than in segmenters (Fig. 4B). After the merozoites are released on rupture of infected erythrocyte, HA-PfROM1 traffics to the posterior pole of the merozoite and is also found in the apical end of the merozoite. The microtubules are still localized longitudinally along the length of the merozoites on one side of the merozoite, no longer overlapping with the HA-PfROM1 (Fig. 4D). Thus, HA-PfROM1 and microtubule colocalization observed in merozoites within the segmenters appears to be due to the close proximity between the two, probably within 200 nm, the resolution limit of confocal microscopy.
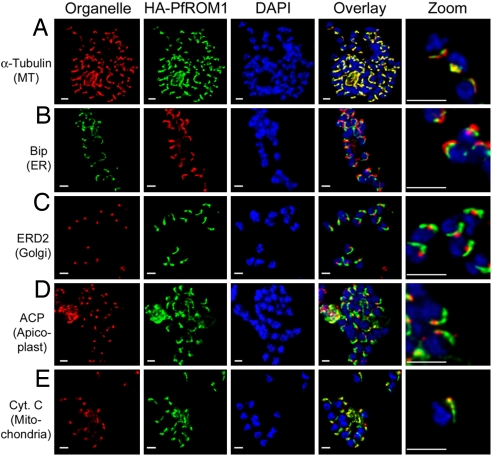
As shown by electron microscopy, the mitochondrion and the apicoplast are aligned asymmetrically in the P. falciparum merozoites along the same side as the microtubules (16, 17). We found that, in addition to HA-PfROM1, other organelles in P. falciparum merozoites are also aligned on the same side of the parasite nucleus as the microtubules (Fig. 5A), the ER (Fig. 5B), the Golgi (Fig. 5C), the apicoplast (Fig. 5D), and the mitochondrion (Fig. 5E). This lateral asymmetry in P. falciparum merozoites suggests that the microtubules may serve as a reference axis for the organization of different organelles, conferring an apparent lateral polarization to the merozoites. A role for the microtubules in the spatial distribution of organelles has also been suggested in higher eukaryotes (18, 19). The functional significance for maintaining a lateral asymmetry of subcellular organellar organization in P. falciparum merozoite biology is not clearly understood.
Fig. 5.
Asymmetric organization of organelles confers a lateral polarity in P. falciparum merozoites. Staining with antibodies against different subcellular organelle markers is shown in the left-hand column. (A) α-Tubulin antibodies staining the merozoite subpellicular microtubules. (B) Bip, an ER marker. (C) ERD2, a cis-Golgi marker. (D) ACP, an apicoplast marker. (E) Cytochrome c, a mitochondrial marker. Corresponding costaining with rat anti-HA mAb 3F10 (A and C–E) or mouse anti-HA mAb 2C16 (B) together with nuclear staining with DAPI (blue) and an overlay of all channels is shown in subsequent columns. The right-hand column shows a digital zoom of merozoites showing that the red and green staining occurs on the same side of the nucleus. (Scale bar, 1 μm.)
Release of Merozoites from Infected Erythrocytes Triggers the Movement of PfROM1 to the Merozoite Surface.
In preparations of merozoites that were released from the infected erythrocytes, HA-PfROM1 localization was found to be dramatically different from the observed lateral asymmetric staining pattern observed in merozoites within segmenters. HA-PfROM1 was circumferentially spread around the entire merozoite surface with marked concentrations at the apical and posterior ends of the merozoite (Fig. 6). HA-PfROM1 staining appeared to be colocalized, in part, with the PfAMA1 staining that translocates from micronemes to the parasite surface on release of micronemal contents during invasion (Fig. 6A).
Fig. 6.
HA-PfROM1 is secreted on the merozoite surface and concentrates at the posterior pole of the merozoite. Staining of free merozoites with antibodies against a micronemal marker (A) or with a rhoptry marker at the apical end (B) is shown in the left-hand column (red). Corresponding costaining with rat anti-HA mAb 3F10 (green) together with the overlay between the red and green channels is shown in subsequent columns. The right-hand column shows the corresponding DIC image. Arrow heads point to the merozoite posterior pole. (Scale bar, 1 μm.)
Discussion
We provide evidence for an organelle that is distinct from other invasion-related organelles of P. falciparum. This organelle extends along one side of the merozoite from a point just posterior to the nucleus to the apical end of the merozoite. We have named this single thread-like organelle, the mononeme. This organelle contains a protease, P. falciparum rhomboid-1 (PfROM1), that cleaves proteins in their transmembrane region. All rhomboids recognize a canonical sequence in the transmembrane regions of their substrate proteins (20), and in addition, individual rhomboids recognize specific sequences in the transmembrane region (12, 21) that confer substrate specificity on their proteolytic activity. Of the rhomboids in P. falciparum merozoites, only PfROM1 has been shown to cleave PfAMA1 (12) albeit in an in vitro assay. In merozoites, PfAMA1 is located in micronemes and thus separated from PfROM1. However, as shown here, both the protease and PfAMA1 appear on the merozoite plasma membrane during the normal process of parasite release from the infected erythrocytes. The merozoite initially binds to the erythrocytes without any apparent orientation and then rapidly reorients to form a junction between the apical region of the parasite and erythrocyte. The initial attachment may be mediated by PfAMA1 (22), and if so, AMA1 must be cleaved rapidly to allow apical reorientation. In addition, as the merozoite enters the erythrocyte, the AMA1 remaining on the merozoite surface is clipped by the parasite sheddase. It is also possible that PfROM1 clips other micronemal proteins during the invasion process. Whatever the merozoite target protein of PfROM1, the separation of PfROM1 from micronemal proteins is likely to be critical for invasion.
Toxoplasma gondii ROM1, the orthologue of PfROM1, is located in the Golgi, secretory vesicles, and in micronemes (10; L. D. Sibley, unpublished data). PfROM1 was also thought to be located in micronemes (13), based on data localizing a PfROM1 construct that was missing two 5′ exons which encode one of the transmembrane domains of PfROM1 (SI Figs. 7, 8, and 10). We have shown that the location of the full-length PfROM1 is separate from AMA1 and EBA175, two proteins found in micronemes (17, 23). The PfROM1 construct used here was similar to that used for localization of T. gondii ROM1 (10). The structure of the organelle containing PfROM1 is thread-like along one side of the merozoite, unlike the apical, punctuate appearance of micronemes. The difference in location of ROM1 in T. gondii and P. falciparum may reflect a difference in the cell biology of the invasion process, the location of the parasite receptors that are used in invasion, and the location of the targets of ROM1. For example, the extracellular form of T. gondii can survive outside the cell while it crawls over the next host cell, whereas P. falciparum merozoites invade immediately after release from the infected erythrocyte. Thus, the mononeme separates the protease, PfROM1, from its substrate until the protease activity is needed during invasion.
PfROM1 by immunoconfocal studies appears to colocalize with the microtubules. As the resolution of confocal microscopy is ≈200 nm (in the x-y plane), it is impossible to know the exact spatial proximity of the two organelles to each other. We speculate that, on release of the merozoite from the erythrocyte, the mononeme may be moved along microtubules by microtubular motors to the apical end to fuse with the merozoite plasma membrane. Testing this speculation will require resolution of the exact location of PfROM1 and of the microtubules and the microtubular motor during merozoite release and invasion. After merozoites are released from infected erythrocytes, PfROM1 and microtubules are no longer colocalized.
Materials and Methods
PfROM1 Epitope Tagging and Generation of Transgenic Parasites.
Based on sequence homology with T. gondii ROM1 (SI Fig. 10), we predicted the P. falciparum orthologue to be composed of two PlasmoDB annotated genes PF11_0149 and PF11_0150. This was confirmed by RT-PCR performed on total RNA extracted from mature asexual blood stage parasites, using TRIzol (Invitrogen) as described earlier (24), using the following primer pairs: forward primer 5′-TTTTTTCTGTTTATCCTTATTATTATACTTG-3′ and reverse primer 5′-ATATATATATGTGTACATTAATTTATTTGCG-3′. The cDNA amplicon on sequencing showed an ORF of 837 bp from four exons (SI Figs. 7 and 8). A PfROM1 transgene with triple-hemaglutinin (HA) epitope tag inserted after the initiation codon of the PfROM1 cDNA sequence together with 1.6 kb of 5′ upstream untranslated region was cloned into the p3D7 transfection plasmid (25). For detailed cloning and transfection methodology see the SI Materials and Methods. Parasites were cultured in RPMI medium 1640 supplemented with 0.5% Albumax (Invitrogen) according to standard in vitro culture techniques (26).
Antibodies.
The following sets of antibodies were used in this study: anti-HA epitope tag antibodies [rat anti-HA mAb 3F10 (Roche) and mouse anti-HA mAb 2C16 (BioVision)]; merozoite surface: rabbit anti-MSP119 affinity-purified antibodies [from S. Singh, Malaria Vaccine Development Branch/National Institute of Allergies and Infectious Diseases/National Institutes of Health (MVDB/NIAID/NIH)]; micronemal markers: mouse anti-AMA1 monoclonal antibody (mAb) 1G6 (from C. Long, MVDB/NIAID/NIH) and rabbit anti-EBA175RII affinity-purified antibodies (from D. Narum, MVDB/NIAID/NIH); rhoptry bulb markers: mouse anti-RAP2 mAb 3H7 and anti-Rhop3 mAb 3E10 (from A. Saul, MVDB/NIAID/NIH); rhoptry neck marker: anti-RON4 mAb 24C6 (from J. F. Dubremetz, University of Montpellier, France); dense granule marker: rabbit anti-RESA antibodies (from R. Anders, La Trobe University, Melbourne, Australia); ER and cis-Golgi markers: polyclonal rat anti-PfBip and polyclonal rabbit anti-ERD2 antibodies, respectively (27–29) (from MR4, American Type Culture Collection, deposited by J. Adams); apicoplast marker: polyclonal rabbit anti-ACP antibodies (from G. McFadden, University of Melbourne, Australia); microtubule marker: mouse anti-chicken brain α-tubulin mAb DM1A (Sigma); and mitochondria marker: sheep anti-rabbit cytochrome c affinity-purified antibodies (Sigma).
Secondary antibodies highly cross-adsorbed against other species and coupled to Alexa fluorophores (Molecular Probes) were used to enhance detection specificity of respective primary antibodies as described below. Goat anti-rat IgG-Alexa 488: anti-HA (mAb 3F10), and anti-PfBip antibodies; goat anti-mouse-Alexa 594: anti-HA (mAb 2C16), anti-RAP2 (mAb 3H7), anti-Rhop3 (mAb3E10), and anti-tubulin (mAb DM1A); goat anti-mouse-Alexa 568: anti-AMA1 (mAb 1G6), and anti-RON4 (mAb 24C6); goat anti-rabbit-Alexa 594: anti-EBA175RII, anti-MSP119, anti-PfERD2, and anti-PfACP; goat anti-rabbit-Alexa 568: anti-RESA antibodies; and donkey anti-sheep-Alexa 568: anti-cytochrome c antibodies.
Immunoblot Analysis.
Mature schizonts (≥8 nuclei) staged from asexual blood stage parasites were subjected to saponin lysis (30). Parasite extract equivalent to ≈2 × 106 schizonts were loaded per lane of the 4–12% gradient Bis-Tris gel under reducing conditions. On electrophoretic separation, proteins were transferred to a nitrocellulose membrane and subjected to immunoblot analysis as described earlier (31). In brief, the blot was blocked with 5% skimmed milk. Anti-HA mAbs 3F10 (Roche) or 2C16 (BioVision) was used at 1:1,000 dilution to probe the immunoblot strips. Detection was done by using the West-Pico kit (Pierce), as per manufacturer's instructions.
Immunofluorescence Assay (IFA).
IFA analysis was performed as per standard protocol described earlier (32). In brief, parasite thin blood smears were air-dried and fixed with cold acetone. All primary and secondary antibody incubations were carried out at 37°C separated by extensive washing with PBS with 0.05% Tween-20 (Bio-Rad) and subsequently stained with nuclear staining dye DAPI (Invitrogen). The smears were mounted under coverslips by using Vectashield hard-set mounting medium, and stored at 4°C until image acquisition through confocal microscopy was performed.
Confocal Microscopy and Image Analysis.
A Leica SP2 confocal microscope and software was used for image acquisition. All images were collected by using a PL APO ×100/1.4 oil immersion objective and a confocal zoom of ×6. Fluorophores were excited with an Argon laser (488 nm), a diode laser at 561 nm, and a HeNe laser at 594 nm. A 405-nm diode laser was used for DAPI excitation. All images were collected as 3D data sets (z-stacks) with a step size of 0.12 nm between the successive optical sections. The photomultiplier (PMT) gains and detector slit positions for each channel were adjusted to minimize any potential cross-talk between the channels. This assured acquisition of a specific signal in each channel. Deconvolution of all image stacks was performed by using Huygens Essential (version 300p5, Scientific Volume Imaging) to improve the maximum resolution of the data, as well as to minimize background noise. Deconvolved images were saved and analyzed through Imaris image analysis software (version 5.7.2, Bitplane). For the ease of presentation, all images in this study are displayed as maximum projection of the 3D image stacks.
Colocalization analysis was performed by using “Coloc” module Imaris (Bitplane), as described in ref. 33, allowing a distinction between actual colocalization versus random association between signals from two different labels in the entire 3D volume of each image file. The result of such colocalization analysis is expressed as a Pearson's correlation coefficient generated for each colocalization experiment.
Supplementary Material
Acknowledgments
We thank Dr. Susan K. Pierce [Laboratory of Immunogenetics, National Institute of Allergies and Infectious Disease (NIAID), National Institutes of Health (NIH)] for thoughtful discussions on the manuscript; Drs. Owen Schwartz, Juraj Kabat, and Meggan Czapiga (Biological Imaging, Research Technologies Branch, NIH) for advice and help on confocal microscopy; Dr. David Sibley (Washington University, St. Louis, MO), for sharing unpublished information about T. gondii ROM-1 localization. We thank Drs. Sanjay Singh, Carole Long, David Narum, and Allan Saul (Malaria Vaccine Development Branch, NIAID, NIH); Dr. Jean-François Dubremetz (University of Montpellier, Montpellier, France); Dr. Geoff McFadden (University of Melbourne, Australia); Dr. Robin Anders (La Trobe University, Melbourne, Australia); and MR4 (American Type Culture Collection) for generous gifts of the antibodies used in this study. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequence reported in this paper has been deposited in the GenBank database (accession no. EU180604).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709999104/DC1.
References
- 1.Levine ND, Corliss JO, Cox FE, Deroux G, Grain J, Honigberg BM, Leedale GF, Loeblich AR, III, Lom J, Lynn D, et al. J Protozool. 1980;27:37–58. doi: 10.1111/j.1550-7408.1980.tb04228.x. [DOI] [PubMed] [Google Scholar]
- 2.Garnham PC, Bird RG, Baker JR. Trans R Soc Trop Med Hyg. 1960;54:274–278. doi: 10.1016/0035-9203(60)90075-4. [DOI] [PubMed] [Google Scholar]
- 3.Aikawa M. Exp Parasitol. 1971;30:284–320. doi: 10.1016/0014-4894(71)90094-4. [DOI] [PubMed] [Google Scholar]
- 4.Aikawa M, Miller LH, Johnson J, Rabbege J. J Cell Biol. 1978;77:72–82. doi: 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams JH, Hudson DE, Torii M, Ward GE, Wellems TE, Aikawa M, Miller LH. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- 6.Treeck M, Struck NS, Haase S, Langer C, Herrmann S, Healer J, Cowman AF, Gilberger TW. J Biol Chem. 2006;281:31995–32003. doi: 10.1074/jbc.M606717200. [DOI] [PubMed] [Google Scholar]
- 7.Howell SA, Withers-Martinez C, Kocken CH, Thomas AW, Blackman MJ. J Biol Chem. 2001;276:31311–31320. doi: 10.1074/jbc.M103076200. [DOI] [PubMed] [Google Scholar]
- 8.Howell SA, Hackett F, Jongco AM, Withers-Martinez C, Kim K, Carruthers VB, Blackman MJ. Mol Microbiol. 2005;57:1342–1356. doi: 10.1111/j.1365-2958.2005.04772.x. [DOI] [PubMed] [Google Scholar]
- 9.Sam-Yellowe TY, Shio H, Perkins ME. J Cell Biol. 1988;106:1507–1513. doi: 10.1083/jcb.106.5.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brossier F, Jewett TJ, Sibley LD, Urban S. Proc Natl Acad Sci USA. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowse TJ, Soldati D. Trends Parasitol. 2005;21:254–258. doi: 10.1016/j.pt.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Baker RP, Wijetilaka R, Urban S. PLoS Pathog. 2006;2:e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger TW, Blackman MJ. J Cell Biol. 2006;174:1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bannister LH, Mitchell GH. Ann Trop Med Parasitol. 1995;89:105–111. doi: 10.1080/00034983.1995.11812940. [DOI] [PubMed] [Google Scholar]
- 15.Fowler RE, Fookes RE, Lavin F, Bannister LH, Mitchell GH. Parasitology. 1998;117:425–433. doi: 10.1017/s003118209800328x. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins J, Fowler R, Krishna S, Wilson I, Mitchell G, Bannister L. Protista. 1999;150:283–295. doi: 10.1016/S1434-4610(99)70030-1. [DOI] [PubMed] [Google Scholar]
- 17.Bannister LH, Hopkins JM, Dluzewski AR, Margos G, Williams IT, Blackman MJ, Kocken CH, Thomas AW, Mitchell GH. J Cell Sci. 2003;116:3825–3834. doi: 10.1242/jcs.00665. [DOI] [PubMed] [Google Scholar]
- 18.Kelly RB. Cell. 1990;61:5–7. doi: 10.1016/0092-8674(90)90206-t. [DOI] [PubMed] [Google Scholar]
- 19.Cole NB, Lippincott-Schwartz J. Curr Opin Cell Biol. 1995;7:55–64. doi: 10.1016/0955-0674(95)80045-x. [DOI] [PubMed] [Google Scholar]
- 20.Urban S, Freeman M. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 21.Urban S, Wolfe MS. Proc Natl Acad Sci USA. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Infect Immun. 2004;72:154–158. doi: 10.1128/IAI.72.1.154-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sim BK, Toyoshima T, Haynes JD, Aikawa M. Mol Biochem Parasitol. 1992;51:157–159. doi: 10.1016/0166-6851(92)90211-2. [DOI] [PubMed] [Google Scholar]
- 24.Kyes S, Pinches R, Newbold C. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- 25.Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, Cowman AF, Batchelor A. Mol Microl. 2004;52:159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 26.Flores MVC, Berger-Eiszele SM, Stewart TS. Parasitol Res. 1997;83:734–736. doi: 10.1007/s004360050330. [DOI] [PubMed] [Google Scholar]
- 27.Kumar N, Koski G, Harada M, Aikawa M, Zheng H. Mol Biochem Parasitol. 1991;48:47–58. doi: 10.1016/0166-6851(91)90163-z. [DOI] [PubMed] [Google Scholar]
- 28.Elmendorf HG, Haldar K. EMBO J. 1993;12:4763–4773. doi: 10.1002/j.1460-2075.1993.tb06165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noe AR, Fishkind DJ, Adams JH. Mol Biochem Parasitol. 2000;108:169–185. doi: 10.1016/s0166-6851(00)00198-5. [DOI] [PubMed] [Google Scholar]
- 30.Orjih AU. Lancet. 1994;343:295. doi: 10.1016/s0140-6736(94)91142-8. [DOI] [PubMed] [Google Scholar]
- 31.Singh S, Kennedy MC, Long CA, Saul AJ, Miller LH, Stowers AW. Infect Immun. 2003;71:6766–6774. doi: 10.1128/IAI.71.12.6766-6774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margos G, Bannister LH, Dluzewski AR, Hopkins J, Williams IT, Mitchell GH. Parasitology. 2004;129:273–287. doi: 10.1017/s0031182004005657. [DOI] [PubMed] [Google Scholar]
- 33.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.