Abstract
The Pseudomonas syringae type III effector AvrRpt2 promotes bacterial virulence on Arabidopsis thaliana plants lacking a functional RPS2 gene (rps2 mutant plants). To investigate the mechanisms underlying the virulence activity of AvrRpt2, we examined the phenotypes of transgenic A. thaliana rps2 seedlings constitutively expressing AvrRpt2. These seedlings exhibited phenotypes reminiscent of A. thaliana mutants with altered auxin physiology, including longer primary roots, increased number of lateral roots, and increased sensitivity to exogenous auxin. They also had increased levels of free indole acetic acid (IAA). The presence of AvrRpt2 also was correlated with a further increase in free IAA levels during infection with P. syringae pv. tomato strain DC3000 (PstDC3000). These results indicate that AvrRpt2 alters A. thaliana auxin physiology. Application of the auxin analog 1-naphthaleneacetic acid promoted disease symptom development in PstDC3000-infected plants, suggesting that elevated auxin levels within host tissue promote PstDC3000 virulence. Thus, AvrRpt2 may be among the virulence factors of P. syringae that modulate host auxin physiology to promote disease.
Keywords: disease, indole acetic acid, pathogen, virulence, host physiology
To successfully colonize and cause disease, a pathogen must modify normal signaling and physiological processes of its host to render host tissue suitable for pathogen growth and disease symptom production. These modifications likely include the suppression of general antimicrobial defenses and the release of water and/or nutrients from plant cells, which require the coordinate action of many pathogen virulence factors (1–4). Central to the virulence of the bacterial plant pathogen Pseudomonas syringae, the causal agent of leaf spotting diseases, is the type III secretion system, which delivers bacterial proteins, referred to as type III effectors, directly into host cells (3, 5, 6). P. syringae mutants defective in type III secretion are unable to grow or cause disease on susceptible plants (7, 8), demonstrating the importance of type III effectors in P. syringae pathogenesis. Because most effectors are secreted into plant cells during infection, they are hypothesized to promote disease by modulating host physiology and signaling events from within plant cells. Many of the P. syringae type III effectors characterized to date have been demonstrated to suppress host defense responses (3, 9, 10). However, the function of many other effectors during pathogenesis is not well understood.
We are interested in elucidating the role of the P. syringae type III effector protein AvrRpt2 during pathogenesis. AvrRpt2 was initially identified based on its ability to trigger disease resistance on Arabidopsis thaliana plants carrying the resistance gene RPS2 (11, 12). AvrRpt2 also promotes pathogen virulence on host plants lacking a functional RPS2 gene (i.e., rps2 mutant plants) (13–17).
The mechanisms by which AvrRpt2 promotes P. syringae virulence are not well understood. A favored hypothesis is that AvrRpt2 promotes pathogen growth by suppressing host defenses. However, AvrRpt2 virulence activity seems to occur by a mechanism that functions independently or downstream of the defense hormone salicylic acid (SA) (14), suggesting that AvrRpt2 may promote pathogenesis by altering some other aspect of host cell physiology or signaling. Structural and functional analyses have revealed that AvrRpt2 is a cysteine protease that functions to cleave several host proteins, including RIN4 (18, 19). This protease activity is required for AvrRpt2 to elicit RPS2-mediated activation of defense (20, 21) and also may be responsible, at least in part, for AvrRpt2 virulence activity (19, 22). It is important to note that AvrRpt2 promotes virulence independently of RIN4, suggesting that AvrRpt2 has virulence targets other than RIN4 (22, 23).
To gain additional insight into the possible mechanisms underlying AvrRpt2 virulence activity, we examined the phenotypes of transgenic A. thaliana plants expressing the avrRpt2 gene in the absence of pathogen infection. The transgenic plants exhibit several phenotypes reminiscent of plant mutants with altered auxin responses, indicating that AvrRpt2 alters Arabidopsis auxin physiology. We also investigated alterations in host auxin physiology during P. syringae infection. Our findings indicate that P. syringae actively alters host auxin physiology as a strategy to increase virulence.
Results
Transgenic Seedlings Expressing avrRpt2 Exhibit Altered Auxin Physiology.
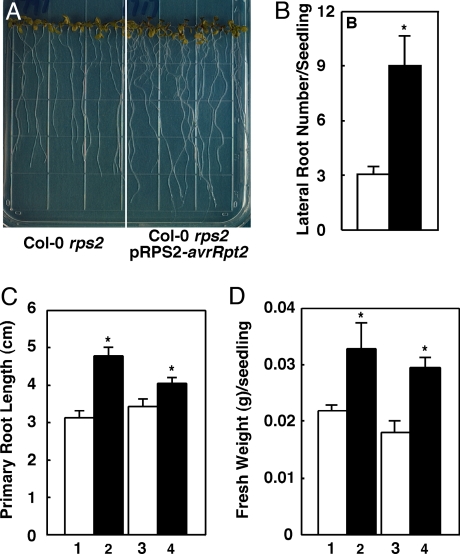
To gain insight into the function of AvrRpt2 in plant cells, we examined several independent transgenic Arabidopsis lines expressing avrRpt2 that exhibit increased susceptibility to P. syringae (13, 14), including two independent lines of Col-0 rps2-expressing avrRpt2 under the control of the RPS2 promoter and two independent lines of No-0 rps2-expressing avrRpt2 under the control of the Cauliflower Mosaic Virus 35S promoter (13). Soil-grown mature plants of all four lines expressing avrRpt2 are phenotypically similar to their nontransgenic progenitors and plants transformed with empty vector before inoculation with P. syringae (13). However, when grown on tissue culture plates, avrRpt2-transgenic seedlings were significantly different from control seedlings. Light-grown seedlings of all four transgenic lines developed more lateral roots (Fig. 1 A and B) and slightly, but significantly, longer primary roots (Fig. 1 A and C). The transgenic seedlings also had higher fresh weight than the nontransgenic control seedlings (Fig. 1D). Additionally, the roots of transgenic seedlings exhibited a slight defect in gravitropism (Fig. 1A and data not shown). When germinated and grown in the dark, the transgenic seedlings exhibited less pronounced apical hooks and more open cotyledons than control seedlings (data not shown).
Fig. 1.
Phenotypes of avrRpt2-transgenic seedlings. (A) Seven-day-old light-grown Col-0 rps2 seedlings (Left) and Col-0 rps2-transgenic seedlings expressing pRPS2-avrRpt2 (Right). (B) Number of lateral roots per seedling of 7-day-old Col-0 rps2 seedlings (empty bar) and Col-0 rps2-transgenic seedlings expressing pRPS2-avrRpt2 (filled bar). Each bar represents the mean number of lateral roots of a minimum of 10 seedlings ± SEM. (C and D) Primary root length (C) and fresh weight (D) of 7-day-old seedling populations of indicated genotypes. 1, Col-0 rps2 parent; 2, Col-0 rps2 pRPS2-avrRpt2 parent; 3, F2 seedlings that do not carry the pRPS2-avrRpt2 transgene; 4, F2 seedlings carrying the pRPS2-avrRpt2 transgene. Each bar represents the mean primary root length of a minimum of 10 seedlings ± SEM. For data shown in A and B, similar results were obtained in experiments using seedlings from a second independent Col-0 rps2 pRPS2-avrRpt2-transgenic line, as well as in experiments using seedlings of two independent No-0 rps2 p35S-avrRpt2-transgenic lines, nontransgenic No-0 rps2 seedlings, or No-0 rps2 seedlings transformed with the pMON10098 empty vector (data not shown). For data shown in C and D, similar results were obtained in an experiment using No-0 rps2 p35S-avrRpt2-transgenic seedlings, No-0 rps2-nontransgenic seedlings, and an F2 population of a No-0 rps2 population segregating for the p35S-avrRpt2 transgene (see Materials and Methods) (data not shown). The asterisks indicate a significant difference with P < 0.05, when compared with data from control seedlings, based on the results of an unpaired one-tailed Student's t test.
To confirm that the phenotypes of avrRpt2-transgenic seedlings described earlier are because of the expression of the avrRpt2 transgene, we examined the phenotypes of two independent F2 populations segregating for the avrRpt2 transgene (see Materials and Methods). In both cases, the average primary root length of avrRpt2-transgenic seedlings was significantly longer, and the average fresh weight of avrRpt2-transgenic seedlings was significantly higher than that of their nontransgenic siblings (Fig. 1 C and D and data not shown). The observation of these phenotypes in seedlings from multiple independent transgenic lines, and specifically in F2 seedlings carrying the avrRpt2 transgene from two independent segregating populations, indicates that the phenotypes of avrRpt2-transgenic seedlings are because of the presence of the avrRpt2 transgene.
Alterations in lateral root initiation, primary root length, gravitropic responses, and apical hook formation have been reported for seedlings of Arabidopsis mutants with altered auxin physiology (24–27). Thus, our finding that avrRpt2-transgenic seedlings exhibit phenotypes reminiscent of plants with altered auxin levels or responses suggests that AvrRpt2 alters host auxin physiology.
Transgenic Seedlings Expressing avrRpt2 Exhibit Increased Sensitivity to Exogenously Applied Auxin.
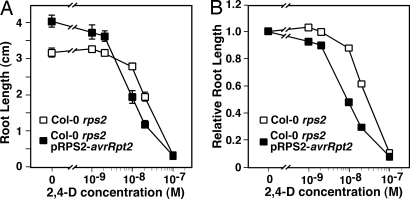
To further examine auxin responses in the avrRpt2-transgenic seedlings, we studied the sensitivity of these seedlings to a range of concentrations of the auxin analog 2,4-Dichlorophenoxyacetic acid (2,4-D). As shown in Fig. 2, elongation of the primary roots of nontransgenic Col-0 rps2 seedlings was inhibited on plates containing 2,4-D at concentrations >1 × 10−8 M. Although Col-0 rps2 pRPS2-avrRpt2 seedlings grew longer primary roots on plates lacking 2,4-D, their primary roots were significantly shorter than those of control seedlings on plates containing 2,4-D at concentrations of ≥1 × 10−8 M. The increased auxin sensitivity of avrRpt2-transgenic plants is more apparent when inhibition of root growth is expressed relative to root length in the absence of 2,4-D (Fig. 2B). We observed similar results in root inhibition studies for the other three avrRpt2-transgenic lines. Thus, avrRpt2-transgenic seedlings are more sensitive to the inhibitory effects of 2,4-D. This increased sensitivity seems to be specific to auxin because avrRpt2-transgenic seedlings exhibited normal sensitivities to methyl jasmonate, cytokinin, and 1-aminocyclopropane-1-carboxylic acid (the metabolic precursor of ethylene) (data not shown).
Fig. 2.
The avrRpt2-transgenic seedlings exhibit increased sensitivity to the auxin analog 2,4-D. Seeds of Col-0 rps2 plants (open squares) and Col-0 rps2-transgenic plants expressing pRPS2-avrRpt2 (filled squares) were germinated and grown on 1/2 × MS plates containing the indicated concentrations of the auxin analog 2,4-D for 7 days and then measured for root length. (A) Length of primary roots of seedlings on plates containing the indicated concentration of 2,4-D. Data points represent the average root length of a minimum of 10 seedlings ± SEM. (B) Data from A expressed as relative root length, normalizing the average length of seedlings grown on 2,4-D to average length of seedlings grown in the absence of 2,4-D. Similar results were obtained in a second independent experiment when testing a second, independent Col-0 rps2 pRPS2-avrRpt2-transgenic line, as well as in experiments using No-0 rps2 p35S-avrRpt2 and control seedlings (data not shown).
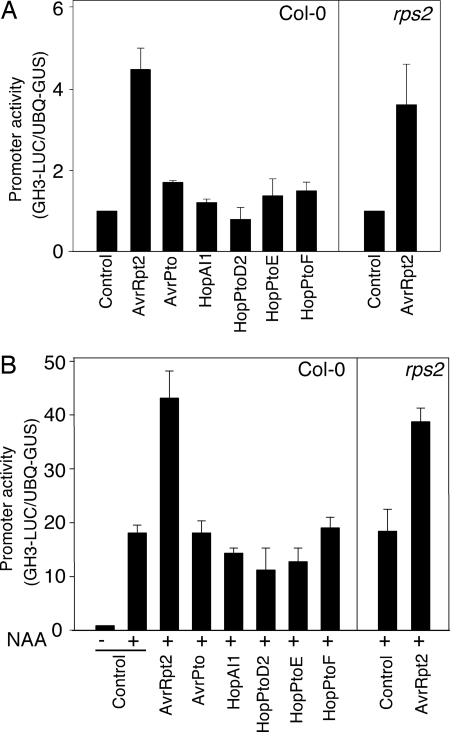
Auxin induces the expression of many auxin-responsive genes, including genes belonging to the Aux/IAA and GH3 gene families (28). To investigate whether the increased auxin sensitivity of avrRpt2-transgenic seedlings is associated with enhanced induction of auxin-responsive genes, we transiently expressed avrRpt2 and an auxin-responsive GH3-LUC reporter gene (29) in Arabidopsis mesophyll protoplasts and monitored expression of GH3-LUC (Fig. 3) in the presence or absence of exogenous auxin. The reporter gene activation assay was performed 6 h after DNA transfection, before the initiation of avrRpt2/RPS2-dependent cell death in wild-type Arabidopsis protoplasts. In the transfected protoplasts, the GH3-LUC reporter was strongly induced by treatment with 1-naphthaleneacetic acid (NAA) (Fig. 3B). In the absence of NAA, the expression of GH3-LUC was elevated in protoplasts expressing avrRpt2, and the induction of GH3-LUC upon addition of NAA was further enhanced by AvrRpt2 (Fig. 3 A and B). These data indicate that the increase in auxin sensitivity conferred by the AvrRpt2 protein is associated with an increase in auxin-induced gene expression. It is worth noting that enhanced expression of GH3-LUC was observed in both wild-type RPS2 and rps2 mutant protoplasts, indicating that the ability of AvrRpt2 to enhance auxin sensitivity in plant cells is independent of RPS2 function (Fig. 3A).
Fig. 3.
AvrRpt2 promotes auxin-induced gene expression in Arabidopsis protoplasts. Col-0 or Col-0 rps2-101c protoplasts were cotransfected with constructs encoding the indicated type III effector and a GH3-LUC reporter (29) and incubated for 3 h before treatment with 1 μM NAA. Expression of GH3-LUC was assayed 3 h after NAA treatment. The activities of GH3-LUC were detected in the absence (A) and presence (B) of NAA.
To assess whether this activity is specific to AvrRpt2, we tested several other well characterized type III effectors in this assay, including AvrPto, HopAI1, HopPtoD2, HopPtoE, and HopPtoF. None of the other effectors tested stimulated expression of GH3-LUC in the absence of NAA (Fig. 3A) or enhanced the auxin-dependent induction of this reporter (Fig. 3B).
Free IAA Levels Are Elevated in avrRpt2-Transgenic Seedlings.
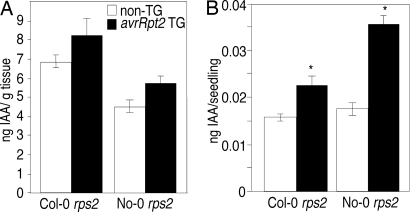
We used gas chromatography/mass spectrometry to measure free IAA levels in shoot and root tissues of avrRpt2-transgenic seedlings (see Materials and Methods). Although we reproducibly detected higher levels of free IAA in the shoots of avrRpt2-transgenic seedlings than in control seedlings, this difference was not significant when the amount of free IAA was normalized by fresh weight of the tissue samples (Fig. 4A). However, given that IAA is not present uniformly within seedlings, but is concentrated in young, developing leaves (30), normalizing the amount of free IAA per fresh weight of shoot tissue may underestimate any localized increase in free IAA in transgenic seedlings. When calculated on a per-seedling basis, the average free IAA level in the shoot tissue of avrRpt2-transgenic seedlings was significantly higher (≈1.2-fold) than that of the control seedlings (Fig. 4B). We also observed elevated free IAA levels in the root tissue of avrRpt2-transgenic plants in several experiments (data not shown). Thus, the presence of AvrRpt2 may be associated with a small, but reproducible, increase in free IAA levels in transgenic seedlings.
Fig. 4.
The avrRpt2-transgenic seedlings contain higher levels of free IAA. Free IAA levels were measured in shoot tissue of 7-day-old transgenic seedlings expressing avrRpt2 (filled bars) and nontransgenic (empty bars) seedlings. Results from experiments with Col-0 rps2 and Col-0 rps2-transgenic seedlings expressing pRPS2-avrRpt2 and No-0 rps2 and No-0 rps2-transgenic seedlings expressing p35S-avrRpt2 are shown. (A) Free IAA levels were normalized by fresh weight of the seedlings. (B) Data shown in A, with free IAA levels normalized for the number of seedlings assayed. Each bar represents the mean of three independent measurements ± SEM. The asterisks indicate a significant difference with P < 0.05, when compared with control tissue, based on the results of an unpaired one-tailed Student's t test. Similar results were obtained in two independent experiments, as well as in free IAA measurements of the root tissue of these seedlings (data not shown).
AvrRpt2 Promotes the Accumulation of Free IAA in PstDC3000-Infected Plants.
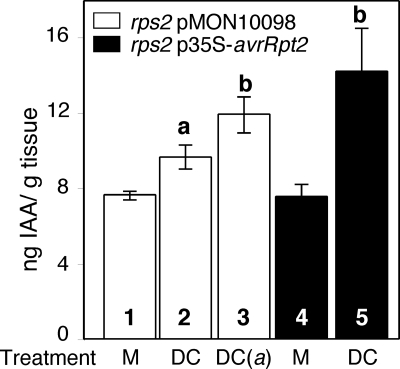
To determine whether AvrRpt2 also is associated with an increase in IAA levels during pathogenesis, we measured free IAA in mature plants infected with PstDC3000. Two days after infection, free IAA levels were significantly and reproducibly higher in PstDC3000-infected plants than in uninfected (data not shown) or mock-treated plants (Fig. 5, bars 1 and 2). These results are consistent with previous studies demonstrating that free IAA levels increase in Arabidopsis plants after infection with P. syringae (31, 32). Free IAA accumulation was significantly higher in plants infected with P. syringae expressing avrRpt2 (Fig. 5, bars 2 and 3). We did not observe differences in IAA levels between mature, mock-infected avrRpt2-transgenic and control plants (Fig. 5, bars 1 and 4), suggesting that the transgenic plants have reestablished auxin homeostasis by this stage (33). However, we observed an increase in free IAA levels in infected avrRpt2-transgenic plants when compared with uninfected (data not shown) or mock-treated avrRpt2-transgenic plants (Fig. 5, bars 4 and 5). Therefore, the presence of AvrRpt2, either when delivered by PstDC3000 or expressed in transgenic plants, resulted in significantly larger increases in free IAA levels during infection. Because AvrRpt2 does not cause significantly increased growth of PstDC3000 on rps2 plants within the first 2 days after inoculation (13), it is unlikely that the promotion of IAA accumulation by AvrRpt2 in these experiments is simply because of a difference in bacterial population size in the infected plants. These results suggest that AvrRpt2 modulates free IAA levels within the host during PstDC3000 pathogenesis.
Fig. 5.
The presence of AvrRpt2 is associated with enhanced increases in free IAA levels in PstDC3000-infected plants. Free IAA levels in rosette leaves of 3- to 4-week-old No-0 rps2 p35S-avrRpt2 plants (filled bars) or No-0 rps2 plants transformed with the pMON10098 empty vector (empty bars) 2 days after mock inoculation with 10 mM MgCl2 containing 0.02% L-77 (M, bars 1 and 4), inoculation with PstDC3000 (DC, bars 2 and 5), or PstDC3000(avrRpt2) [DC(a), bar 3] at a concentration of ≈5 × 108 cfu/ml in 10 mM MgCl2 containing 0.02% Silwet L-77. Each bar represents the mean of three replicates ± SEM. a, Significant difference when compared with the values obtained for uninoculated or mock-inoculated samples. b, Significant difference from the value obtained for No-0 rps2 pMON10098 plants inoculated with DC3000 (based on the results of an unpaired one-tailed Student's t test; P < 0.05). Similar results were obtained in a second independent experiment.
Exogenous Auxin Promotes Disease Development.
Auxin has been clearly shown to promote disease caused by pathogens that induce plant cell proliferation, such as Agrobacterium tumefaciens (crown gall disease) and Pseudomonas savastanoi (knot disease) (34). However, a role for auxin in A. thaliana–P. syringae interactions is not well established. Recent work by Navarro et al. (35) demonstrates that down-regulation of auxin signaling is a component of the plant basal defense response and that stimulation of auxin signaling may promote disease susceptibility. The observations that free IAA levels increase during P. syringae infection (Fig. 5) (31) and that AvrRpt2 promotes a further increase in free IAA are consistent with these results. Thus, P. syringae may use virulence factors (such as AvrRpt2) to increase free IAA levels during infection as a strategy to promote disease susceptibility in host tissue. We therefore predict that plants exposed to elevated levels of auxin would exhibit enhanced susceptibility to P. syringae.
We tested the effect of auxin on P. syringae pathogenesis by applying auxin analogs during infection. When wild-type Col-0 or No-0 plants were treated with 1 × 10−5 M or 5 × 10−5 M NAA at the time of PstDC3000 inoculation, they developed significantly more severe disease symptoms, including an increase in the number of water-soaked lesions and chlorosis, than plants inoculated with PstDC3000 alone (Fig. 6 and data not shown). The leaves of control plants treated only with NAA became epinastic, but did not develop chlorosis or necrosis (data not shown), indicating that the increased severity of disease symptoms was not because of phytotoxic effects of NAA application. Interestingly, despite the increase in symptom severity, we were unable to detect an increase in bacterial growth in NAA-treated plants at 2 and 4 days after infection (data not shown). Similar results were obtained when we treated plants with 5 × 10−6 M 2,4-D (data not shown) or when NAA was applied at 24 and 48 h after infection and also were observed by Navarro et al. (35). Thus, auxin promotes disease symptom development during infection by P. syringae.
Fig. 6.
Plants treated with auxin analogs exhibit altered susceptibility to PstDC3000 infection. Four- to five-week-old wild-type Col-0 plants were dip-inoculated in PstDC3000 at a concentration of ≈5 × 108 cfu/ml in 10 mM MgCl2 containing 0.02% Silwet L-77. The plant on the Right was simultaneously treated with 5 × 10−5 M NAA at the time of infection. The photo was taken 4 days after inoculation. Similar results were observed in more than three independent experiments when plants were treated with 5 × 10−6 M 2,4-D (data not shown) and in experiments using No-0 plants (data not shown).
Discussion
We examined the phenotypes of uninfected transgenic A. thaliana plants expressing the P. syringae type III effector protein, AvrRpt2, to gain insight into the effect of this virulence factor on host physiology. Four independent transgenic lines expressing avrRpt2 exhibited seedling phenotypes associated with altered auxin physiology, including longer primary roots, increased number of lateral roots, reduced gravitropic responses, and increased sensitivity to exogenous auxin (Figs. 1, 2, and 3 and data not shown). These phenotypes may be because of enhanced accumulation of free IAA in transgenic seedlings (Fig. 4). We further found that the presence of AvrRpt2 during PstDC3000 infection is correlated with enhanced increases in free IAA levels in infected plants (Fig. 5), and that application of auxin during infection promoted disease development in infected plants (Fig. 6). Thus, AvrRpt2 may promote pathogen virulence by altering host auxin physiology.
We previously demonstrated that AvrRpt2 suppresses the expression of several SA-dependent PR genes in A. thaliana (14). However, suppression of SA-mediated defenses does not seem to be the primary mechanism by which AvrRpt2 promotes P. syringae virulence because AvrRpt2 virulence activity functions independently or downstream of SA in both A. thaliana and tomato (14, 16). Our observations that AvrRpt2 alters host auxin physiology and that auxin promotes PstDC3000 virulence suggest that modulation of host auxin physiology is an alternative mechanism by which AvrRpt2 promotes P. syringae virulence. The mechanism by which AvrRpt2 modulates IAA physiology is not presently known, nor is it clear whether AvrRpt2 cysteine protease activity is required for this function. It is possible that cleavage of one or more host proteins by AvrRpt2 (19) leads to alterations in IAA homeostasis or signaling within the host.
A local increase in free IAA levels at the site of P. syringae infection could potentially change many aspects of plant cell signaling or physiology to promote P. syringae virulence. For instance, increased auxin levels could stimulate cell wall loosening and changes in membrane permeability (36, 37), resulting in the leakage of water and/or nutrients into the apoplastic space. Auxin also up-regulates the production of ethylene (38), which has been proposed to mediate pathogen-induced tissue damage and thus promote disease symptom production (39). Additionally, an increase in free IAA may promote stomatal opening (40, 41), potentially facilitating bacterial escape to the leaf surface and promoting pathogen transmission (15). Auxin also has been implicated in the suppression of plant defense (42–45). Because inhibition of IAA signaling is a component of basal and SA-mediated defense responses in A. thaliana (35, 46), gaining control of host auxin physiology may be crucial for P. syringae to overcome host defense mechanisms. Alternatively (or additionally), auxin may directly promote pathogen fitness under the stressful conditions encountered within the host tissue (47).
Modulation of host hormone physiology may be a common virulence strategy among plant pathogens. In addition to AvrRpt2, several other virulence factors have been implicated in modulating auxin levels or signaling (2). The P. syringae phytotoxin coronatine, a molecular mimic of the endogenous plant hormone jasmonate (48–51), also affects auxin homeostasis (50, 52). Additionally, the Xanthomonas campestris pv. vesicatoria type III effector AvrBs3 induces the expression of several auxin-induced genes in susceptible pepper plants (53). Interestingly, a number of Pseudomonas and Xanthomonas strains have been reported to produce free IAA in culture (34, 54–56). Signaling mediated by the plant hormone abscisic acid (ABA) also is targeted by P. syringae, and one or more type III effectors may promote pathogen virulence by manipulating ABA homeostasis (57). Future studies investigating the roles of auxin, ABA, and other plant hormones during pathogenesis should provide additional insight into the physiology of plant disease and the strategies used by plant pathogens to manipulate their hosts.
Materials and Methods
Bacterial Strains and Plant Lines.
The Pseudomonas syringae pv. tomato strain PstDC3000 has been described in ref. 58. The isogenic P. syringae strains PstDC3000(avrRpt2) and PstDC3000 carrying the empty vector pVSP61 were described by Chen et al. (13). The wild-type A. thaliana ecotypes Col-0 and No-0, the Col-0 rps2-101C and No-0 rps2-201C mutants, the transgenic lines expressing avrRpt2, and the corresponding lines carrying the pMON10098 empty vector control used in this study are described in Chen et al. (13). Plants were grown under short-day conditions (8-h light/16-h dark) at 21°C with 75–80% humidity.
The F2 population segregating for the pRPS2-avrRpt2 transgene resulted from crossing a Col-0 rps2 pRPS2-avrRpt2-transgenic line with Col-0 rps2 rpm1 plants (14). The No-0 rps2 population segregating for the p35S-avrRpt2 transgene is the T2 population from which the homozygous No-0 rps2 p35S-avrRpt2-transgenic line was isolated (13). Genomic DNA of seedlings was extracted, and plants carrying the avrRpt2 transgene were identified by PCR using the avrRpt2-specific primers, ZYC1 and ZYC2 (13).
Germination and Root Length Measurements.
Sterilized A. thaliana seeds were planted onto the surface of plant tissue culture plates containing 1/2 × Murashige and Skoog (MS) (59) salt mixture, B-5 vitamins, 1% sucrose, and 0.8% agar and then incubated at 4°C for 2 days before transfer to a growth chamber. Plants were grown under constant light at 24°C. To measure seedling root length, the plates were oriented vertically so the roots grew down along the surface of the media. After the seedlings were grown under 24°C for 7 days, they were photographed by using a digital camera and measured by using National Institutes of Health Image 1.61 software.
Auxin and Cytokinin Responsiveness Assays.
2,4-D (Sigma—Aldrich) and NAA (Sigma–Aldrich) were dissolved in 0.1 N sodium hydroxide and adjusted to a concentration of 1 × 10−3 M in water. The cytokinin kinetin (N6-furfuryladenine; ICN Biomedicals ) was dissolved in water and adjusted to a concentration of 2 × 10−3 M. After being filter-sterilized, the solutions were used as stocks to make 1/2 × MS plates containing different concentrations of 2,4-D or kinetin. Sterilized seeds were planted onto these plates and germinated as described earlier. Primary root length was measured on the seventh day of growth at 24°C.
Type III Effector Constructs and Arabidopsis Protoplast Transient Assay.
The effector genes were cloned as PCR fragments into a plant expression vector with an HA epitope tag at the C terminus (9, 29). The protoplast transient expression assay was conducted as described in ref. 9. Briefly, 2 × 104 protoplasts were transfected with 20 μg of DNA, including both the type III effector and GH3-LUC reporter (29) constructs. UBQ10-GUS also was included in the transfection as an internal control. Three hours after incubation, the transfected protoplasts were treated with 1 μM NAA for another 3 h. The GH3-LUC and UBQ10-GUS activities were detected by a luminometer and fluorometer. The promoter activity was presented as a LUC/GUS ratio.
Plant Inoculation and Measurement of in Planta Bacterial Growth.
Four- to five-week-old A. thaliana plants were dip-inoculated with the indicated PstDC3000 strains at a concentration of ≈5 × 108 cfu/ml in 10 mM MgCl2 containing 0.02% Silwet L-77 (OSi Specialties) as described in ref. 11. In all experiments, the control treatment was mock-inoculated with 10 mM MgCl2 containing 0.02% Silwet L-77. In the experiments in which plants were treated with exogenously applied auxin analogs, NAA or 2,4-D was added to the inoculation solutions at the indicated final concentrations. In experiments in which auxin analogs were applied 1 or 2 days after inoculation, infected plants were dipped into a solution of 10 mM MgCl2 containing 0.02% Silwet L-77 with NAA or 2,4-D at the indicated final concentrations. To measure bacterial populations within leaf tissue, one or two leaves were removed from each infected plant at the indicated time points, and the bacteria were quantified as described in ref. 49.
Measurement of Free IAA Levels in Seedlings and PstDC3000-Infected Plants.
Seedlings of avrRpt2-transgenic lines and of the control plants were grown vertically under light for 7 days on 1/2 × MS plates as described earlier. Tissue was harvested by cutting the seedlings to separate the roots from the shoots, combining the respective tissues into 0.3-g samples, and freezing them in liquid nitrogen. The samples were stored at −80°C before IAA analysis.
Three- to four-week-old No-0 rps2 plants and No-0 rps2 p35S-avrRpt2 plants were dip-inoculated with PstDC3000 or PstDC3000(avrRpt2) at a concentration of ≈5 × 108 cfu/ml in 10 mM MgCl2 containing 0.02% Silwet L-77 or mock-inoculated in 10 mM MgCl2 containing 0.02% Silwet L-77. Whole rosettes were harvested before inoculation (uninoculated) and 48 h after mock, PstDC3000, or PstDC3000(avrRpt2) inoculation. Tissue from several plants was combined to generate ≈0.3-g fresh weight samples, which were immediately frozen in liquid nitrogen and stored at −80°C until used for analysis of IAA levels.
The 0.3-g samples of Arabidopsis tissue were ground in a solution of 65% isopropanol and 0.2 M imidazole buffer (pH 7.0) by using 4 ml/g extraction buffer containing [13C6]-IAA as an internal standard. The free IAA was extracted and measured by using the methods described in ref. 60, with the following modifications. The homogenized solution was diluted 1:10 with double deionized water before running over the amino anion-exchange column, and all evaporations were carried out in a rotary evaporator with a two-stage vacuum pump. The amount of free IAA in leaf tissue was calculated by using the isotope-dilution equation (61).
Acknowledgments
We thank Jin Li for characterization of the avrRpt2-transgenic seedlings, Dr. Lana Barkawi and Dr. Julie Tilman for assisting with IAA measurements, Amy Szumlanski for help with experiments, Drs. Melisa Lim and Jason Reed for helpful discussion, and Neva Laurie-Berry and Lionel Navarro for helpful comments. This work was supported by U.S. Department of Agriculture Grant 2002-35319-12729, a David and Lucille Packard Fellowship for Science and Engineering (to B.N.K.), National Science Foundation Grants DBI 00-77769 and MCB 06-43854 (to J.D.C.) and MCB 0446109 (to J.S.), U.S. Department of Agriculture-National Research Initiative Grant 2005-35318-16197 (to J.D.C.), and National Institutes of Health Grant R01 GM70567 (to J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Alfano JR, Collmer A. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunkel BN, Chen Z. In: The Prokaryotes. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. New York: Springer; 2006. [Google Scholar]
- 3.Mudgett MB. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- 4.Abramovitch RB, Anderson JC, Martin GB. Nat Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis GR, Van Gijsegem F. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 6.Alfano JR, Collmer A. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 7.Lindgren PB, Peet RC, Panopoulos NJ. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niepold F, Anderson D, Mills D. Proc Natl Acad Sci USA. 1985;82:406–410. doi: 10.1073/pnas.82.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 10.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Annu Rev Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 11.Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu GL, Katagiri F, Ausubel FM. Mol Plant–Microbe Interact. 1993;6:434–443. doi: 10.1094/mpmi-6-434. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN. Mol Plant–Microbe Interact. 2000;13:1312–1321. doi: 10.1094/MPMI.2000.13.12.1312. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, Kloek AP, Cuzick A, Moeder W, Tang D, Innes RW, Klessig DF, McDowell JM, Kunkel BN. Plant J. 2004;37:494–504. doi: 10.1111/j.1365-313x.2003.01984.x. [DOI] [PubMed] [Google Scholar]
- 15.Guttman DS, Greenberg JT. Mol Plant–Microbe Interact. 2001;14:145–155. doi: 10.1094/MPMI.2001.14.2.145. [DOI] [PubMed] [Google Scholar]
- 16.Lim MT, Kunkel BN. Mol Plant–Microbe Interact. 2005;18:626–633. doi: 10.1094/MPMI-18-0626. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Blumer SE, Sundin GW. J Bacteriol. 2005;187:8088–8103. doi: 10.1128/JB.187.23.8088-8103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Mol Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- 19.Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. Proc Natl Acad Sci USA. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Axtell MJ, Staskawicz BJ. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 21.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 22.Lim MT, Kunkel BN. Plant J. 2004;40:790–798. doi: 10.1111/j.1365-313X.2004.02251.x. [DOI] [PubMed] [Google Scholar]
- 23.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodward AW, Bartel B. Ann Bot (London) 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- 26.Lincoln C, Britton JH, Estelle M. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobbie L, Estelle M. Plant J. 1995;7:211–220. doi: 10.1046/j.1365-313x.1995.7020211.x. [DOI] [PubMed] [Google Scholar]
- 28.Hagen G, Guilfoyle T. Plant Mol Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- 29.Kovtun Y, Chiu WL, Zeng W, Sheen J. Nature. 1998;395:716–720. doi: 10.1038/27240. [DOI] [PubMed] [Google Scholar]
- 30.Ljung K, Bhalerao RP, Sandberg G. Plant J. 2001;28:465–474. doi: 10.1046/j.1365-313x.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- 31.O'Donnell PJ, Schmelz EA, Moussatche P, Lund ST, Jones JB, Klee HJ. Plant J. 2003;33:245–257. doi: 10.1046/j.1365-313x.2003.01619.x. [DOI] [PubMed] [Google Scholar]
- 32.Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. Plant J. 2004;39:790–808. doi: 10.1111/j.1365-313X.2004.02168.x. [DOI] [PubMed] [Google Scholar]
- 33.Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G. Plant Mol Biol. 2002;49:249–272. [PubMed] [Google Scholar]
- 34.Yamada T. Annu Rev Phytopathol. 1993;31:253–273. doi: 10.1146/annurev.py.31.090193.001345. [DOI] [PubMed] [Google Scholar]
- 35.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 36.Buchanan BB, Gruissem W, Jones RL. Biochemistry and Molecular Biology of Plants. Rockville, MD: Am Soc Plant Physiol; 2000. [Google Scholar]
- 37.Tian Q, Uhlir NJ, Reed JW. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abel S, Nguyen MD, Chow W, Theologis A. J Biol Chem. 1995;270:19093–19099. doi: 10.1074/jbc.270.32.19093. [DOI] [PubMed] [Google Scholar]
- 39.Bent AF, Innes RW, Ecker JR, Staskawicz BJ. Mol Plant–Microbe Interact. 1992;5:372–378. doi: 10.1094/mpmi-5-372. [DOI] [PubMed] [Google Scholar]
- 40.Dietrich P, Sanders D, Hedrich R. J Exp Bot. 2001;52:1959–1967. doi: 10.1093/jexbot/52.363.1959. [DOI] [PubMed] [Google Scholar]
- 41.Irving HR, Gehring CA, Parish RW. Proc Natl Acad Sci USA. 1992;89:1790–1794. doi: 10.1073/pnas.89.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinette D, Matthysse AG. J Bacteriol. 1990;172:5742–5749. doi: 10.1128/jb.172.10.5742-5749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rezzonico E, Flury N, Meins F, Jr, Beffa R. Plant Physiol. 1998;117:585–592. doi: 10.1104/pp.117.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinshi H, Mohnen D, Meins FJ. Proc Natl Acad Sci USA. 1987;84:89–93. doi: 10.1073/pnas.84.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. J Biol Chem. 2007;282:10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Curr Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 47.Bianco C, Imperlini E, Calogero R, Senatore B, Amoresano A, Carpentieri A, Pucci P, Defez R. Arch Microbiol. 2006;185:373–382. doi: 10.1007/s00203-006-0103-y. [DOI] [PubMed] [Google Scholar]
- 48.Bender CL, Alarcon-Chaidez F, Gross DC. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooks DM, Bender CL, Kunkel BN. Mol Plant Pathol. 2005;6:629–639. doi: 10.1111/j.1364-3703.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- 50.Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL. Plant J. 2005;42:207–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- 51.Laurie-Berry N, Joardar V, Street IH, Kunkel BN. Mol Plant–Microbe Interact. 2006;19:789–800. doi: 10.1094/MPMI-19-0789. [DOI] [PubMed] [Google Scholar]
- 52.Thilmony R, Underwood W, He SY. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 53.Marois E, Ackerveken GVD, Bonas U. Mol Plant–Microbe Interact. 2002;15:637–646. doi: 10.1094/MPMI.2002.15.7.637. [DOI] [PubMed] [Google Scholar]
- 54.Ansari MM, Sridhar R. Folia Microbiol (Prague) 2000;45:531–537. doi: 10.1007/BF02818722. [DOI] [PubMed] [Google Scholar]
- 55.Fett WF, Osman SF, Dunn MF. Applied Environ Microbiol. 1987;53:1839–1845. doi: 10.1128/aem.53.8.1839-1845.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y. Mol Plant–Microbe Interact. 1998;11:156–162. doi: 10.1094/MPMI.1998.11.2.156. [DOI] [PubMed] [Google Scholar]
- 57.de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whalen M, Innes R, Bent A, Staskawicz B. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 60.Chen K, Miller A, Patterson G, Cohen J. Plant Physiol. 1988;86:822–825. doi: 10.1104/pp.86.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ilic N, Normanly J, Cohen J. Plant Physiol. 1996;111:781–788. doi: 10.1104/pp.111.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]