Abstract
Results of in vitro and genetic studies have provided evidence for four pathways by which proteins are targeted to the chloroplast thylakoid membrane. Although these pathways are initially engaged by distinct substrates and involve some distinct components, an unresolved issue has been whether multiple pathways converge on a common translocation pore in the membrane. A homologue of eubacterial SecY called cpSecY is localized to the thylakoid membrane. Since SecY is a component of a protein-translocating pore in bacteria, cpSecY likely plays an analogous role. To explore the role of cpSecY, we obtained maize mutants with transposon insertions in the corresponding gene. Null cpSecY mutants exhibit a severe loss of thylakoid membrane, differing in this regard from mutants lacking cpSecA. Therefore, cpSecY function is not limited to a translocation step downstream of cpSecA. The phenotype of cpSecY mutants is also much more pleiotropic than that of double mutants in which both the cpSecA- and ΔpH-dependent thylakoid-targeting pathways are disrupted. Therefore, cpSecY function is likely to extend beyond any role it might play in these targeting pathways. CpSecY mutants also exhibit a defect in chloroplast translation, revealing a link between chloroplast membrane biogenesis and chloroplast gene expression.
The majority of chloroplast-localized proteins are associated with the internal membrane system of the organelle, the thylakoid membrane. Many thylakoid membrane proteins are products of nuclear genes and are synthesized in the cytoplasm; others are products of chloroplast genes and are synthesized in the chloroplast stroma. In either case, newly synthesized proteins must be targeted to their correct position with respect to the membrane. Mechanisms of targeting to the thylakoid lumen have been particularly well studied. All known lumenal proteins are nuclear-encoded and are synthesized with a bipartite targeting sequence (for review see Cline and Henry, 1996). The NH2-terminal segment is a stromal targeting sequence that directs the protein across the chloroplast envelope. Adjacent to the stromal targeting sequence is a cleavable lumenal targeting sequence that resembles the signal sequences that target proteins for translocation across bacterial cytoplasmic membranes. Two energetically and genetically distinct pathways have been described for the translocation of proteins to the thylakoid lumen (for review see Cline and Henry, 1996). One set of lumenal proteins is targeted by a mechanism that requires cpSecA, a chloroplast-localized homologue of the bacterial protein SecA (Yuan et al., 1994; Nohara et al., 1995; Voelker and Barkan, 1995; Voelker et al., 1997). Cytochrome f, a chloroplast-encoded integral thylakoid protein, is also targeted to the membrane via this pathway (Voelker and Barkan, 1995; Mould et al., 1997; Nohara et al., 1997; Voelker et al., 1997). A second set of lumenal proteins is targeted via a pathway that relies upon a trans-thylakoidal ΔpH in vitro (for review see Cline and Henry, 1996) and that involves the hcf106 (Voelker and Barkan, 1995) and tha4 genes (our unpublished results) in vivo. A third pathway has been proposed for several integral membrane proteins that appear to integrate spontaneously in vitro (Kim et al., 1996; Robinson et al., 1996). Finally, the integration of the polytopic membrane protein light-harvesting chlorophyll a/b binding protein (LHCP),1 whose targeting signals lie within the mature portion of the protein (Viitanen et al., 1988), defines a fourth pathway in that it requires neither cpSecA (Voelker et al., 1997) nor hcf106 (Voelker and Barkan, 1995), but does require GTP (Hoffman and Franklin, 1994) and cp54 (Li et al., 1995), a homologue of the signal recognition particle protein SRP54.
These previous experiments indicated that each targeting pathway involves some unique components. It is possible, however, that the different targeting machineries also have shared components. For example, two or more of the pathways may converge on a common translocation pore in the membrane, in analogy to the convergence of signal recognition particle (SRP)-dependent and -independent pathways on Sec61-containing translocons in the endoplasmic reticulum membrane (for review see Rapoport et al., 1996). One likely component of the translocon for proteins that engage cpSecA is cpSecY, a chloroplast- localized SecY homologue, since SecY forms a component of the translocon in the bacterial plasma membrane (for review see Rapoport et al., 1996). Genes encoding cpSecY have been discovered in algal chloroplast genomes (for review see Vogel et al., 1996) and in the Arabidopsis thaliana and spinach nuclear genomes (Laidler et al., 1995; Berghoefer and Kloesgen, 1996). The protein encoded by the Arabidopsis cpSecY cDNA is targeted to chloroplast thylakoid membranes in vitro (Laidler et al., 1995). However, functional studies of cpSecY have not been reported.
To gain insight into the roles of cpSecY in vivo, we have used a reverse genetics strategy to obtain maize mutants with transposon insertions in a nuclear gene encoding cpSecY. As shown below, cpSecY mutants exhibit a severe loss of thylakoid membrane. This phenotype is much more severe and global than that of tha1 mutants, which have exceedingly low levels of cpSecA (Voelker et al., 1997). Therefore, cpSecY function is not limited to a role in translocating proteins that previously engaged cpSecA. Furthermore, cpSecY mutants have a more severe phenotype than double mutants with lesions in both the cpSecA and the ΔpH lumenal targeting pathways, implicating cpSecY in either the cp54-dependent pathway, the “spontaneous” pathway, and/or in an uncharacterized targeting pathway. An unexpected aspect of the cpSecY mutant phenotype is a global defect in chloroplast translation. Thus, the activity of the chloroplast translation machinery may be linked to the biogenesis of chloroplast membranes.
Materials and Methods
Plant Material
Plants with Mu insertions in the gene encoding cpSecY (csy1) were identified at Pioneer Hi-Bred (Johnston, IA)by using a primer complementary to the Mu terminal inverted repeat (Bensen et al., 1995; Meeley and Briggs, 1995) and four primers specific for the maize cpSecY cDNA. Pioneer Hi-Bred generously provided a small number of progeny seed from plants that scored positive in the PCR screen. csy1/+ plants were propagated by crossing with inbred lines and subsequent self-pollination to recover homozygous mutant seedlings.
Also used in this study were the targeting mutants tha1 (Voelker and Barkan, 1995; Voelker et al., 1997), hcf106 (Barkan et al., 1986; Voelker and Barkan, 1995) and tha4. The tha4 and hcf106 mutant phenotypes are very similar (i.e., the ΔpH thylakoid targeting pathway is specifically disrupted [see Fig. 8, Voelker and Barkan, 1995]) but the two genes are not allelic. The hcf106-mum3 allele, a deletion derivative of the original hcf106 mutation (Das and Martienssen, 1995), was used in these experiments; seed was provided by R. Martienssen (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). Some experiments included the albino mutant iojap, which has been described as lacking in plastid ribosomes (Walbot and Coe, 1979) , and the albino mutant w3, which has a defect in the carotenoid biosynthetic pathway (Fong et al., 1983).
Figure 8.
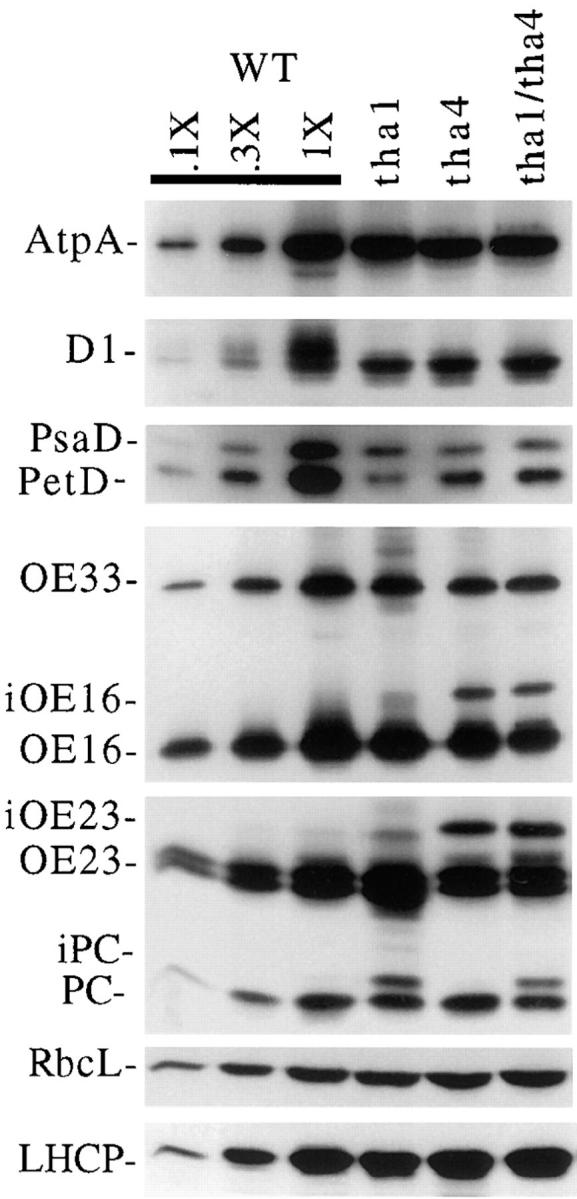
Immunoblot analysis of leaf proteins in tha1/tha4 double mutants. The procedures and antibodies were the same as those described in Fig. 7.
Unless otherwise stated, studies were performed with leaf material obtained from seedlings grown for 9–12 d in a growth chamber (14-h d [400 μE/m2 per s) at 28°C and 10-h nights at 25°C). Samples that were compared in any single experiment were grown and harvested in parallel. Etiolated mutant leaves were obtained as described previously (Voelker et al., 1997).
Cloning of Maize cpSecY cDNAs
A degenerate primer (5′-GAA[GAT]AT[GAT]AT[AGCT]GGCAT-3′) was designed to a conserved amino acid region (met-pro-ile-ile-phe-ser) of cpSecY and then used to prime reverse transcription of maize seedling leaf poly A+ RNA by following the manufacturer's protocol for Superscript™ II reverse transcriptase (GIBCO BRL, Gaithersburg, MD). PCR amplification was performed on the cDNA template using the reverse transcription primer and a second degenerate primer (5′-GT[A/G/C/ T]CC[A/G/C/T]TTCAT[A/T/C]AA[C/T]GC-3′ encoding val-pro-phe-ile-asn-ala). Each 50-μl reaction contained 1 μl of the reverse transcription reaction, 20 pmol of each primer, 20 μM deoxynucleoside triphosphate, 1.5 mM MgCl2, 50 mM KCl, and 10 mM Tris-HCl, pH 8.3. The reactions were incubated at 95°C for 5 min, supplemented with 5 U of Taq DNA polymerase, and then incubated according to the following “touchdown” profile: 10 cycles of 94°C/30 s, 54°/40 s (−1.0°/cycle), 72°/60 s; 10 cycles of 94°/30 s, 44°/40 s (−0.5°/cycle), 72°/60 s; 20 cycles of 94°/30 s, 40°/40 s, 72°/ 60 s. These reactions were subjected to a secondary amplification involving nested degenerate primers that included a 5′ EcoRI site (5′-GGAATTCAT[A/T/C]AA[C/T]GC[A/T/G/C]CA[A/G]AT[A/T/C]GT-3′ encoding ile-asn-ala-gln-ile-val) and a 5′ BamHI site (5′-CGGGATCCAT[G/A/ T]AT[A/G/C/T]GGCAT[A/T/C/G]AC[A/C/T/G]CC-3′ encoding gly-val-met-pro-ile-ile), according to the following conditions: 20 cycles of 95°C/ 30 s, 48°/40 s (−0.5°/cycle), 72°/60 s; 20 cycles of 95°/30 s, 39°/40 s, 72°/60 s). The amplification product was gel-purified and then subcloned into a Bluescript SK+ plasmid. The isolated insert was used to screen a cDNA library constructed from mRNA isolated from the leaves of 2-wk-old greenhouse-grown maize, inbred line B73.
Sequence Alignments
Sequences of SecY homologues from Arabidopsis thaliana (EMBL/GenBank/DDBJ accession number U37247), Spinacia oleracea (accession number Z54351), Escherichia coli (accession number 134413), Synechococcus PCC7942 (accession number 401077), cryptomonas (accession number X62348), and Pyrenomonas salina (accession number X74773) were compared to the maize csy1 cDNA sequence (accession number AF039304). Alignments were calculated using ClustalW 1.7 (Thompson et al., 1994) and BoxShade (Bioinformatics Group, ISREC, Lausanne, Switzerland), with default parameters.
Isolation and Analysis of Plant DNA and mRNA
DNA was extracted and then analyzed as previously described (Voelker et al., 1997). The genomic Southern blot was probed with a partial csy1 cDNA, corresponding to amino acids 205–387 (see Fig. 1) that was radiolabeled by the random hexamer priming method. RNA was extracted and analyzed by Northern hybridization as described in Jenkins et al. (1997). The association of chloroplast mRNAs with polysomes was assayed as described previously (Barkan, 1993). The csy1 transcript was detected with the full-length csy1 cDNA radiolabeled by random hexamer priming. The 16S rRNA and LHCP mRNAs were detected with the radiolabeled DNA probes described in Barkan (1993). The rbcS mRNA was detected with a radiolabeled DNA probe prepared from the cDNA described in Nelson et al. (1984). The atpF probe was described in Jenkins et al. (1997). The rbcL mRNA was detected with a radiolabeled RNA probe prepared by in vitro transcription of a plasmid containing a 570-bp PstI fragment of the maize rbcL gene.
Figure 1.

Deduced amino acid sequence of a maize cDNA encoding a SecY homologue. The Arabidopsis (arab) and spinach proteins are chloroplast localized (Laidler et al., 1995; Berghoefer and Kloesgen, 1996). The Synechococcus (synech) and E. coli sequences are representative of bacterial SecY. Each triangle indicates the site of the Mu transposon insertion in the mutant allele named to its left. Positions of introns are indicated by arrows.
Extraction and Analysis of Protein and Chlorophyll
Leaf proteins were extracted, analyzed on immunoblots, radiolabeled in vivo, and then immunoprecipitated by using the antibodies and methods described previously (Voelker and Barkan, 1995). The relative chlorophyll content of mutant and normal leaves was determined by excising a 35-mg segment from the tip of 10-d-old seedlings, grinding in a mortar and pestle in the presence of 2 ml of 80% acetone, centrifuging at 12,000 g for 1 min, and then measuring the absorbance of the supernatant at 652 nm.
Electron Microscopy
Leaf tissue was fixed overnight at room temperature in 4% glutaraldehyde in 0.05 M potassium phosphate, pH 7.0. After three washes in phosphate buffer, the tissue was postfixed in 2% osmium tetroxide in the same buffer. Samples were dehydrated in an ethanol series and then embedded in Spurr's resin. Sections were stained for 50 min in 5% uranyl acetate and for 10 min in Reynolds lead citrate (Reynolds, 1963). The tissue preparation, sectioning, and electron microscopy was performed by J. Selker (University of Oregon Electron Microscopy Facility, Eugene, OR).
Results
The Maize Genome Contains Several Genes That Are Closely Related to secY
We took advantage of the Trait Utility System for Corn developed at Pioneer Hi-Bred (Meeley and Briggs, 1995) to obtain plants with Mu transposons in a nuclear gene encoding cpSecY. A prerequisite for this approach was knowledge of the nucleotide sequence of the maize cpSecY gene(s). To obtain maize cDNAs encoding cpSecY, the deduced Arabidopsis, spinach, Cryptomonas, and Pyrenomonas cpSecY proteins were compared to identify regions of high conservation. A degenerate primer designed against one conserved region was used to prime reverse transcription of maize leaf polyA+ RNA. A second degenerate primer was then used in conjunction with the first to amplify a PCR product. The amplified cDNA fragment was of the expected size (550 bp) and was related in sequence to known cpSecY genes (data not shown). When the PCR fragment was used to screen a maize leaf cDNA library, several full-length cDNA clones (2.1 kbp) were obtained whose DNA sequence revealed a continuous open reading frame encoding a protein with high identity to plastid and eubacterial SecY proteins (Fig. 1).
Arabidopsis and spinach cpSecY are synthesized with an NH2-terminal extension of ∼120 amino acids, relative to the bacterial and plastid-encoded SecY homologues (Fig. 1). This region has characteristics typical of chloroplast targeting signals (for review see Cline and Henry, 1996). The maize cDNAs encode a protein with an NH2-terminal extension of a similar length and with features of chloroplast targeting signals. Downstream of this region, the deduced maize protein is highly similar to other chloroplast-localized SecY homologues (e.g., 94% similar/86% identical to Arabidopsis cpSecY). As shown below, the phenotypes associated with mutations in the gene encoding the maize cDNA are restricted to the chloroplast. Together, these observations provide strong evidence that the cDNA encodes maize cpSecY. The gene encoding this cDNA was named csy1.
Genomic Southern blots were probed at high stringency with a cDNA fragment encoding a highly conserved region of cpSecY (Fig. 2). Multiple bands were detected when the genomic DNA was digested with enzymes that do not cut within the genomic sequence corresponding to the probe. These results revealed the existence of at least two and perhaps three genes in the maize genome that are closely related to the cDNA clone. To estimate the relative contributions of these different genes to the pool of cpSecY in leaf tissue, the same probe was used to identify clones in a maize leaf cDNA library. 18 independent cDNA clones were obtained. The nucleotide sequences of the 3′ untranslated regions were identical for all 18 clones, although their sites of polyadenylation varied (Fig. 3 c). A probe made from this 3′ untranslated region detected only one band on genomic Southern blots (data not shown), indicating that all of the cDNAs were derived from the same gene. These results strongly suggest that the csy1 gene contributes the vast majority of cpSecY in leaf tissue.
Figure 2.
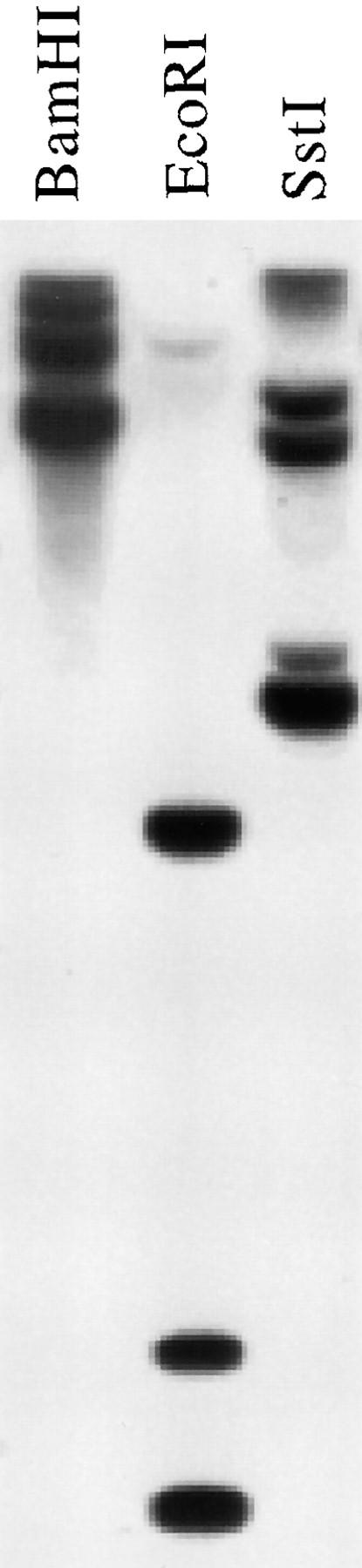
Genomic Southern blot of DNA from the maize inbred line Tx303 probed with the csy1 cDNA. The probe consisted of the cDNA fragment encoding amino acids 208– 381. BamHI and EcoRI do not recognize sites inside the genomic region corresponding to the probe. The filter was washed at high stringency (65°C in 0.3× SSC).
Figure 3.
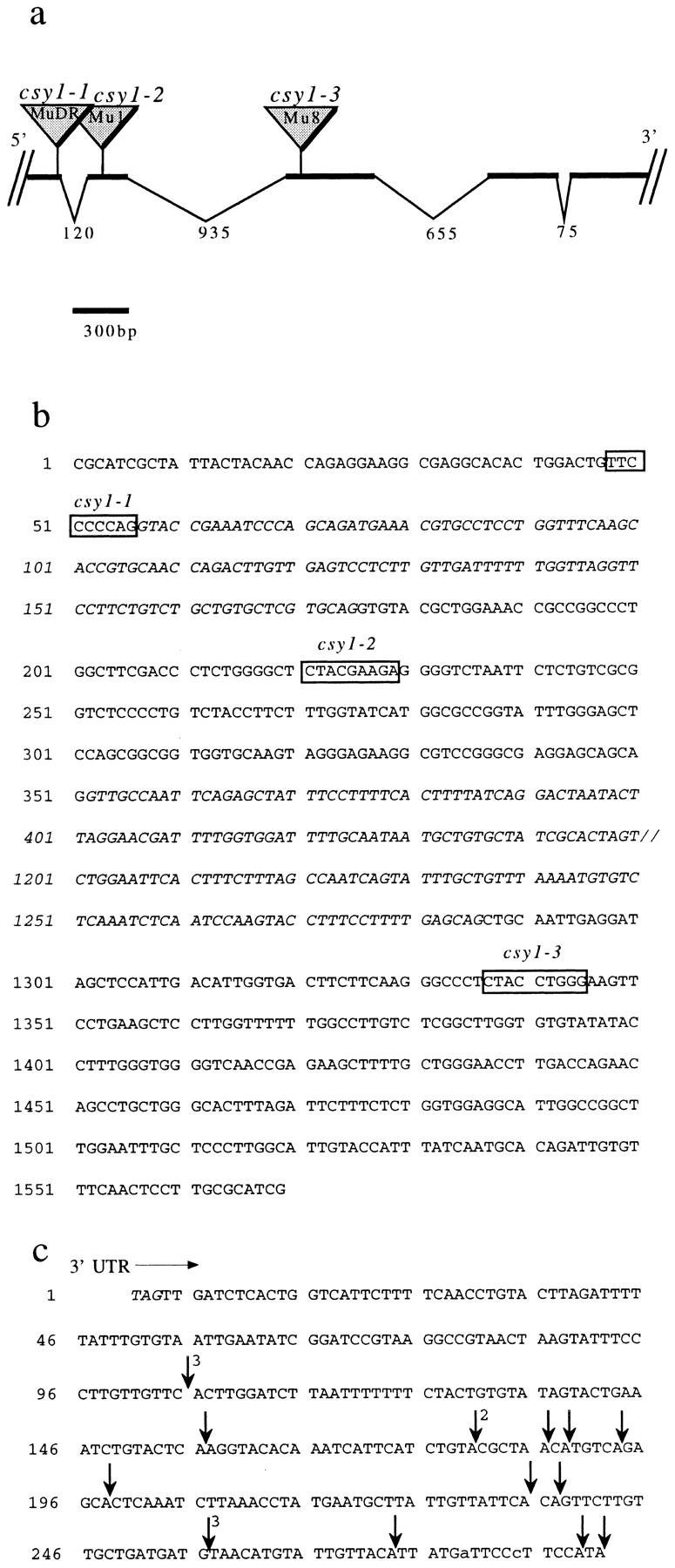
Map and partial nucleotide sequence of csy1. (a) Partial genomic map of csy1. Sites of Mu insertions are indicated. The identity of the Mu family member at each site was deduced based upon the sequence of its terminal-inverted repeat (Chandler and Hardeman, 1992). The insertion sites are shown in the context of the encoded amino acid sequence in Fig. 1. The size of each intron in nucleotides is indicated. (b) Partial nucleotide sequence of csy1. The sequence shown begins 135 nucleotides downstream of the start codon and ends within the third exon. Locations of Mu insertions are shown by boxes marking the nine base pairs that were duplicated upon insertion. Intron sequence is in italics. Only a portion of the sequence of the second intron is shown, with the missing sequence represented by //. The complete nucleotide sequence of the csy1 cDNA and more extensive genomic sequence information has been deposited (EMBL/GenBank/DDBJ accession numbers AF039304 and AF039305). (c) Sites of polyadenylation in 18 independent leaf csy1 cDNA clones. The sequence shown begins at the stop codon. The number of cDNA clones with the same polyadenylation site is indicated in superscript. Clones with identical 3′ ends differed at their 5′ ends, indicating that they represent independent cDNAs.
Mu Insertions Cause Severe Defects in the Expression of csy1
The progeny of maize plants with Mu transposons in csy1 were tentatively identified at Pioneer Hi-Bred in a PCR screen involving csy1-specific primers used in conjunction with a primer corresponding to the Mu terminal inverted repeat (Bensen et al., 1995; Meeley and Briggs, 1995). We confirmed that three of these putative alleles had Mu insertions in csy1. To define the precise position of each insertion, the PCR products resulting from amplification with a Mu primer and a csy1 primer were cloned and then their nucleotide sequences were determined. Fig. 3 shows a partial map of the csy1 gene, including the location of the Mu insertion in each mutant allele. csy1-1 contains a Mu insertion at a 5′ splice junction, in sequences encoding the presumed chloroplast targeting signal. csy1–2 has a Mu insertion in sequences encoding the targeting signal, and csy1–3 has a Mu insertion in sequences encoding the highly conserved portion of cpSecY. As is typical of nuclear-encoded chloroplast proteins, an intron lies between sequences encoding the presumed targeting sequence and sequences encoding the portion of the protein that was derived from the ancestral bacterial gene (refer to Fig. 1).
The insertions in csy1 are genetically linked to the pigment-deficient phenotype shown in Fig. 4. This pigment deficiency did not segregate from csy1 in >50 meioses. Homozygous mutants are nearly albino, with a distinct yellow-green tint. Heterozygous plants appear entirely normal. All three csy1 mutant alleles are similar in phenotype, confirming that the pigment-deficiency results from the Mu insertions in csy1. The chlorophyll content of csy1 mutant seedlings grown under relatively low-intensity light (400 μE/m2 per s) is ∼10% of that of their normal siblings grown in parallel, when normalized to leaf wet weight (data not shown). In contrast, tha1 and hcf106 mutants, with lesions in the cpSecA and ΔpH thylakoid targeting pathways, respectively, accumulate near normal amounts of chlorophyll (Martienssen et al., 1987, and our unpublished observations).
Figure 4.
Pigment deficiency of csy1 mutant seedlings. A csy1 mutant (left), a wild-type sibling (middle), and an iojap mutant (right) were photographed when they were 2-wk old. The iojap mutant is shown to illustrate a true albino mutant. All three csy1 alleles are associated with similar pigmentation defects.
During the development of their first three leaves, csy1 mutant plants grow at the same rate as their wild-type siblings (data not shown). The early normal growth rate indicates that mitochondrial function is not significantly disrupted by these mutations and supports the notion that csy1 function is limited to the chloroplast compartment. The rate of growth of the mutant seedlings began to lag as seed reserves became depleted and they died after the development of four leaves, as is typical of mutations that disrupt photosynthesis in maize (Miles, 1982).
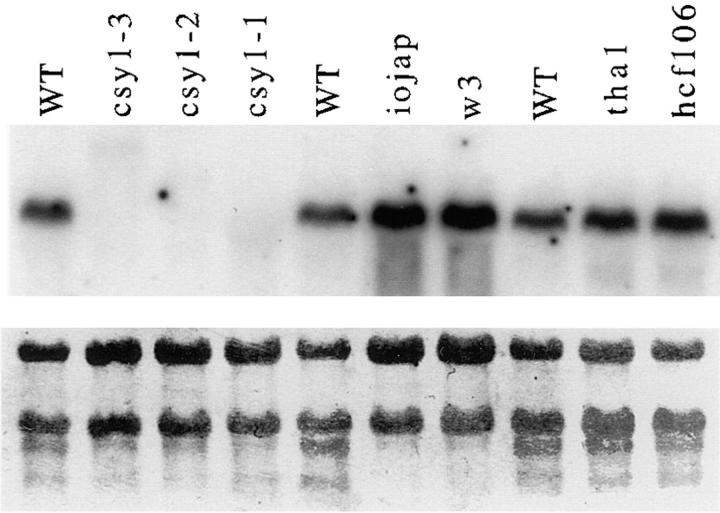
A single csy1 mRNA of ∼2.1 kb was detected in RNA prepared from wild-type leaf tissue (Fig. 5). This mRNA was not detectable in any of the mutant alleles. A low abundance aberrant transcript of ∼3.5 kb in csy1–3 is of the expected size of a chimeric transcript that includes the Mu8 sequences. Two albino mutants, iojap and w3, whose albinism results from different primary defects (Robertson et al., 1978; Walbot and Coe, 1979; Han et al., 1992), and two mutants disrupted in thylakoid protein targeting (tha1 and hcf106), were analyzed in parallel. All of these contain normal levels of csy1 mRNA, indicating that the loss of the mRNA in the csy1 mutants is not a consequence of their pigment deficiency or of a defect in thylakoid protein targeting. The lack of detectable csy1 mRNA in the mutants indicates that the mutations are either null or nearly so.
Figure 5.
RNA gel blot hybridization showing csy1 mRNA in seedling leaf tissue. Total leaf RNA (10 μg) was gel fractionated and then hybridized with a radiolabeled csy1 cDNA probe (top panel). The same filter was stained with methylene blue to visualize rRNAs (bottom panel). Other mutants were analyzed to illustrate that the csy1 mRNA defects do not result from defects in thylakoid protein targeting (tha1 and hcf106), as a consequence of pigment deficiencies (w3 and iojap), or as a consequence of an early block in chloroplast development (iojap).
Chloroplasts in csy1 Mutants Have Little Thylakoid Membrane
The ultrastructure of csy1 mutant chloroplasts was examined by transmission electron microscopy (Fig. 6). Both the bundle sheath and mesophyll chloroplasts in mutant leaves contain little internal membrane. Small internal vesicles were frequently observed (Fig. 6 b and data not shown). Mitochondrial morphology is not altered in the mutants (Fig. 6, a–c).
Figure 6.
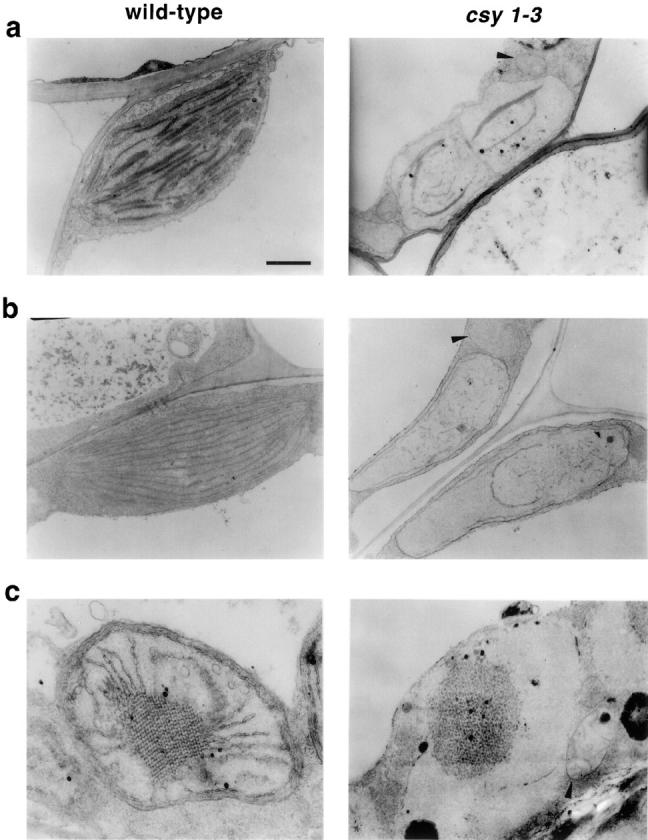
Ultrastructure of csy1 mutant chloroplasts and etioplasts. (a) Mesophyll chloroplasts. (b) Bundle sheath chloroplasts. (c) Etioplasts. Arrowheads point to mitochondria. Chloroplast ultrastructure was similar in all three mutant alleles (data not shown). Bar, 1 μm.
Etioplasts in csy1 mutants are also abnormal in structure (Fig. 6 c). Whereas normal etioplasts contain a crystalline prolamellar body with radiating membranes, csy1 mutant etioplasts contain a condensed but noncrystalline prolamellar body that resembles structures observed in maize mutants defective in chlorophyll biosynthesis (Mascia and Robertson, 1978). However, unlike the etioplasts in chlorophyll-biosynthetic mutants, csy1 mutant etioplasts lack the radiating membranes, indicating that their ultrastructural defects are not simply a consequence of the absence of chlorophyll.
The accumulation of representative proteins from each major thylakoid membrane complex was assayed by immunoblot (Fig. 7 a). Lesions in either the ΔpH or cpSecA lumenal targeting pathway do not significantly reduce the content of the chloroplast ATP synthase or of LHCP (Voelker and Barkan, 1995). In csy1 mutant leaves, however, the abundance of these proteins is ∼10-fold lower than normal. The psaD and petD gene products and the D1 protein are reduced to <10% of their normal levels in csy1 mutants. The abundance of these proteins likely reflects the abundance of other subunits of the photosystem I core complex, the cytochrome b6 f complex, and the photosystem II core complex, respectively, since subunits of these complexes are generally reduced coordinately in mutants in which the availability of any single subunit is limiting (for review see Barkan et al., 1995).
Figure 7.
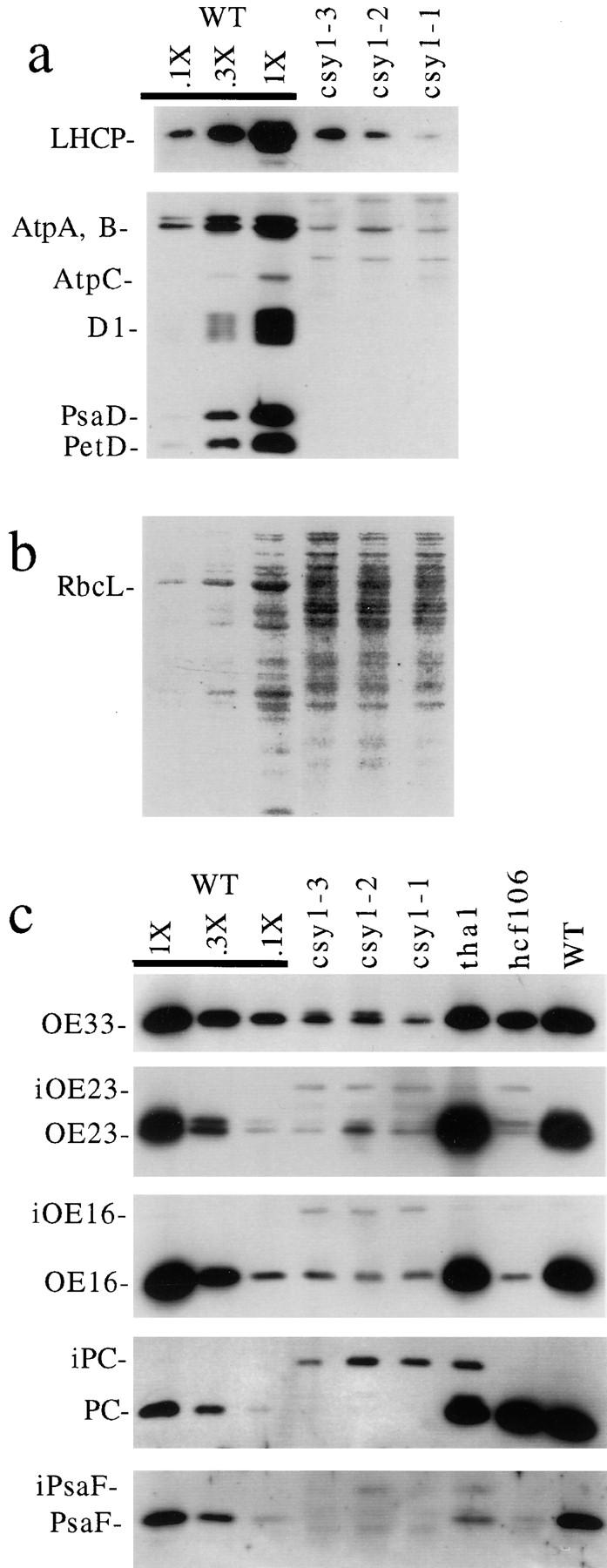
Immunoblot analysis of leaf proteins in csy1 mutants. 5 μg of total leaf protein (or the indicated dilutions of the wild-type [WT] sample) were fractionated by SDS-PAGE and then transferred to nitrocellulose. (a) Accumulation of integral and membrane extrinsic thylakoid proteins. LHCP, PsaD, D1, and PetD were each detected with monospecific antisera. AtpA, AtpB, and AtpC were detected with sera raised against the CF1 complex. (b) The protein on the blot used in a was visualized by staining with Ponceau S. RbcL is the large subunit of Rubisco. (c) Accumulation of lumenal proteins and their precursors. Proteins were detected with the indicated monospecific antisera. iOE23, iOE16, iPC, and iPsaF are of the size predicted for the stromal intermediates of OE23, OE16, PC, and PsaF, respectively. The stromal intermediate of OE33 migrates only slightly slower than the mature form, and can be seen as a doublet in the csy1 mutant samples.
The accumulation of lumenal proteins in csy1 mutant leaves was measured to assess the status of the cpSecA and ΔpH lumenal targeting pathways (Fig. 7 c). A disruption in lumenal targeting is reflected by a decrease in the abundance of the mature protein coupled with an increase in the abundance of its stromal precursor (Voelker and Barkan, 1995). In csy1 mutants, the mature form of three substrates of the cpSecA pathway (the 33-kD subunit of the oxygen evolving complex [OE33], plastocyanin [PC], and PsaF) accumulate to <10% of their normal levels, and proteins corresponding in size to their stromal intermediates accumulate to unusually high levels. Two substrates of the ΔpH pathway, the 23- and 16-kD subunits of the oxygen evolving complex (OE23 and OE16, respectively), behave analogously. These properties are consistent with the possibility that both the ΔpH and cpSecA pathways involve a translocon with a cpSecY subunit. The loss of LHCP is likewise consistent with the idea that cpSecY is required for the integration of proteins that engage cp54. However, it is possible that one or more of these defects is a consequence of the reduction in membrane content, or results because cpSecY is required to target components of other translocons to the membrane.
cpSecY Mutants Are More Severe in Phenotype Than Mutants Simultaneously Disrupted in Both the cpSecA and ΔpH Targeting Pathways
Every aspect of the mutant phenotype investigated was more global and severe in csy1 mutants than in tha1 and hcf106 mutants, suggesting that cpSecY function is not limited to either the cpSecA or the ΔpH thylakoid targeting pathway. It remained possible, however, that cpSecY functions in both pathways. If so, and if it has no additional functions, the csy1 mutant phenotype should resemble the phenotype resulting from simultaneous disruption of both the cpSecA and ΔpH pathways. To address this possibility, tha4/tha1 double mutants were constructed. The tha1 gene encodes cpSecA and the mutation causes at least a 40-fold decrease in cpSecA accumulation (Voelker et al., 1997). The tha4 mutation disrupts the ΔpH lumenal targeting pathway (our unpublished results and Fig. 8) and its phenotype strongly resembles that of the hcf106 mutation (Voelker and Barkan, 1995).
Fig. 8 shows that tha1/tha4 double mutants have a much less severe phenotype than csy1 mutants. For example, LHCP and RbcL, the large subunit of ribulose bisphosphate carboxylase (Rubisco), accumulate normally in the double mutants but are severely reduced in csy1 mutants (refer to Fig. 7). The accumulation of the lumenal proteins OE16 and OE23 (and their stromal precursors) is similar in tha1/tha4 double mutants to that in tha4 single mutants. The accumulation of the lumenal proteins OE33 and PC (and their stromal precursors) is similar in tha1/tha4 double mutants to that in tha1 single mutants. Components of the ATP synthase, photosystem I, photosystem II, and the cytochrome b6f complex also accumulate to similar levels in the double and single mutants (Fig. 8, AtpA, PsaD, D1, and PetD, respectively). Finally, the chlorophyll content of double mutant seedlings is ∼50% of that in their normal siblings, and is similar to that of tha1 and tha4 single mutants (data not shown).
The fact that the csy1 mutant phenotype is much more global than that of tha1/tha4 double mutants suggests that csy1 gene function is not limited to the cpSecA and ΔpH targeting pathways. This finding is consistent with the possibility that cpSecY is part of the translocon for integrating proteins via the cp54 pathway and/or for the proposed spontaneous pathway (for review see Cline and Henry, 1996). However, because the tha4 mutation does not result in the complete loss of tha4 gene product (Walker, M., and A. Barkan, unpublished results), we cannot eliminate the possibility that a complete disruption of both the ΔpH and cpSecA pathways might mimic the csy1 phenotype.
Chloroplast Translation Is Disrupted in cpSecY Mutants
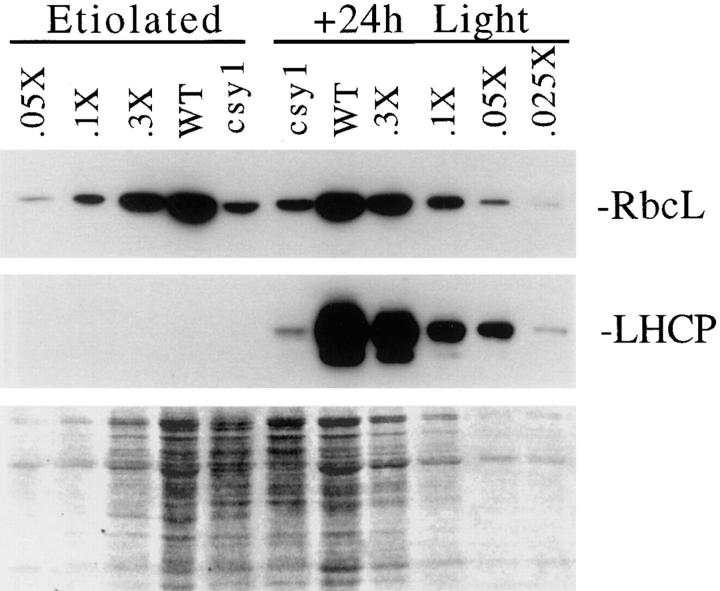
We observed that the level of the large subunit of Rubisco is dramatically reduced in csy1 mutants grown under day– night cycles (Fig. 7 b). To address the possibility that this was due to photooxidative damage, the RbcL subunit was quantified in leaves that had developed in the absence of light, and in the same leaves after 24 h of exposure to light (Fig. 9). In both cases, RbcL abundance was reduced 10-fold in csy1 mutant leaves, indicating that the Rubisco deficiency is not a consequence of photooxidative damage.
Figure 9.
Immunoblot showing loss of the large subunit of Rubisco in both dark- and light-grown csy1 mutant leaves. Protein was extracted from etiolated csy1–2 and wild-type seedling leaf tips. The plants were then exposed to light for 24 h and protein was extracted from leaves of the same plants. Total leaf proteins (5 μg) or the indicated dilutions of the wild-type samples were fractionated by SDS-PAGE, transferred to nitrocellulose, and then probed with antiserum specific for either RbcL or LHCP. LHCP was analyzed to illustrate that the etiolated plants had not been exposed to light, since it is known that LHCP does not accumulate in leaves grown in the absence of light and that it accumulates rapidly after exposure to light (Nelson et al., 1984). (Bottom) Results of staining the filter with Ponceau S, to illustrate total bound proteins.
The dependence of Rubisco accumulation on cpSecY function was unexpected, given cpSecY's presumed role in thylakoid membrane biogenesis. Therefore, the basis for its loss was explored in some detail. Rubisco is composed of two types of subunits, a small subunit encoded by nuclear rbcS genes and a large subunit encoded by the chloroplast rbcL gene. A defect in the synthesis or targeting of either the small or large subunit is sufficient to account for the loss of the entire enzyme, since the two subunits accumulate stoichiometrically when the synthesis of one subunit is blocked (Schmidt and Mishkind, 1983; Spreitzer et al., 1985). We considered the possibilities that: (a) csy1 is required for normal rates of import of proteins into the chloroplast; (b) csy1 is required for normal accumulation of Rubisco mRNAs; and (c) csy1 is required for normal chloroplast translation.
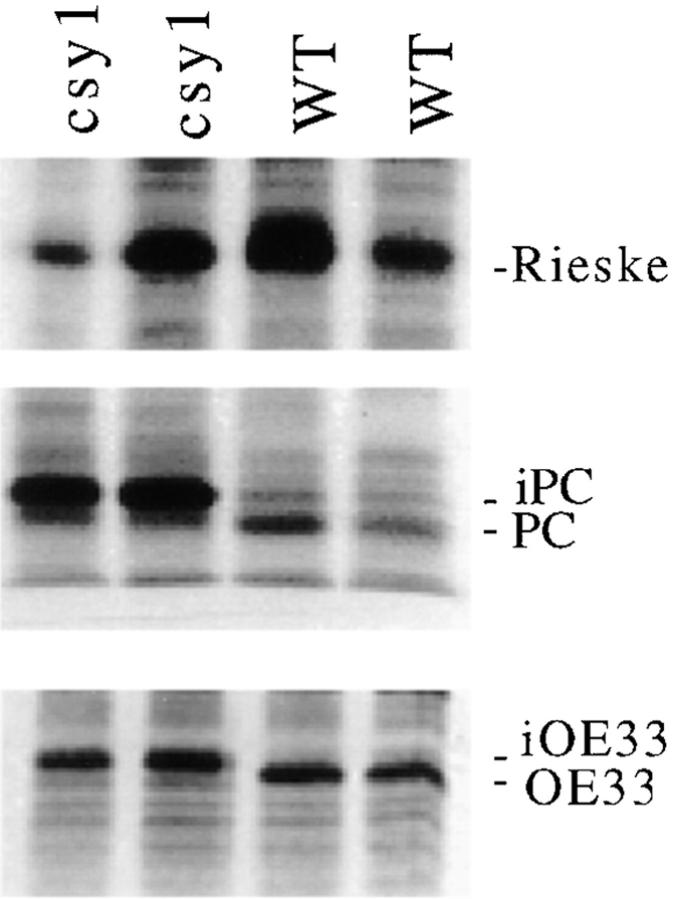
Nuclear-encoded proteins that are imported into the chloroplast engage a common translocation machinery that spans the inner and outer envelope membranes (for review see Schnell, 1995; Cline and Henry, 1996). A defect in import is expected to be reflected by a reduced rate of cleavage of stromal targeting sequences. Rates of processing of three nuclear-encoded chloroplast proteins were monitored by pulse labeling leaf proteins in vivo, immunoprecipitation, and then gel fractionation (Fig. 10). There were no detectable differences between wild-type and csy1 mutant samples in the rates of accumulation of imported forms of the Rieske protein, PC, and OE33. PC and OE33 accumulated in their stromal intermediate form rather than in their mature lumenal forms in csy1 mutants, a predicted consequence of a disruption in the cpSecA-dependent lumenal targeting pathway. These results provide strong evidence that the csy1 gene does not function either directly or indirectly in the import of proteins into the chloroplast. They also show that the low levels of PC and OE33 in csy1 mutants result from an increase in their rates of degradation rather than a decrease in their rates of synthesis.
Figure 10.
Immunoprecipitation of chloroplast proteins from in vivo- labeled leaf extracts. The results of identical analyses of two mutant and two wild-type seedlings are shown. Seedling leaves were radiolabeled with 35[S]methionine/cysteine for 60 min. Protein containing 300,000 cpm was subject to immunoprecipitation with antisera specific for the chloroplast Rieske protein, PC, or OE33. Precipitated proteins were fractionated by SDS-PAGE and then the radioactive proteins were detected by direct autoradiography. iPC and iOE33 are proteins of the size predicted for the stromal intermediates of PC and OE33, respectively. The csy1–2 allele was used for this experiment.
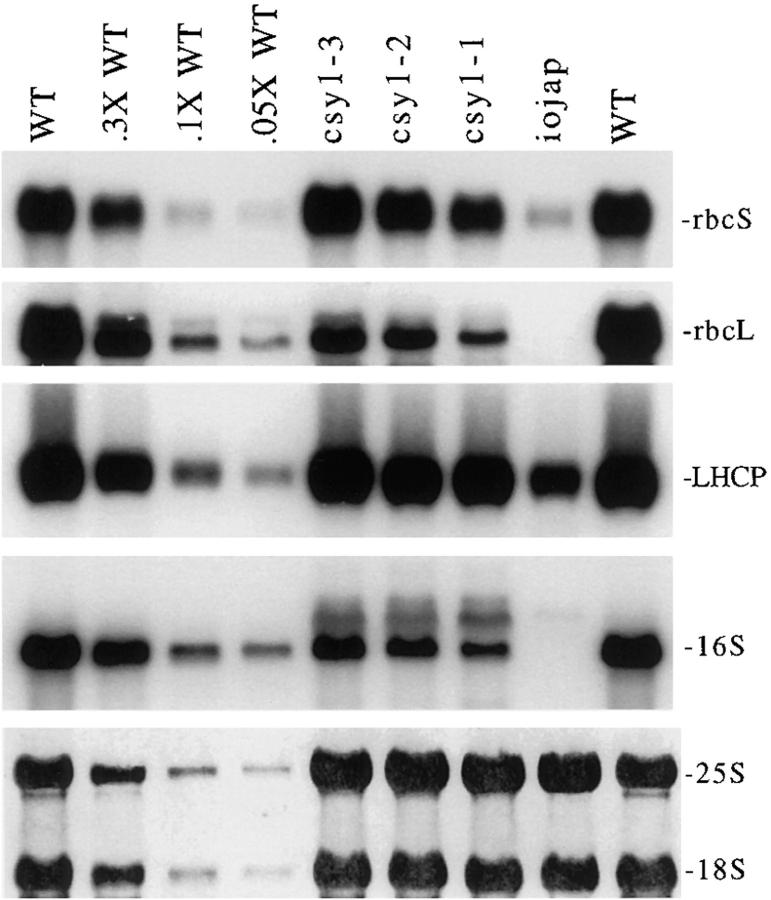
Mutants in which chloroplast development is blocked at an early stage contain reduced levels of LHCP mRNA and of several other nuclear-encoded mRNAs encoding chloroplast proteins (for review see Taylor, 1989). This has given rise to the notion that the transcription of certain nuclear genes is dependent upon a signal from chloroplasts. To determine whether a disruption of this hypothetical signal by csy1 mutations could be responsible for the Rubisco deficiency, the LHCP and rbcS mRNAs were quantified by Northern hybridization (Fig. 11). Both mRNAs accumulate to normal levels in csy1 mutants. Therefore, the signaling between nucleus and chloroplast appears to be intact in csy1 mutants and the loss of Rubisco cannot be attributed to a defect in the accumulation of the rbcS mRNA.
Figure 11.
RNA gel–blot hybridizations of nuclear and chloroplast transcripts in csy1 mutants. 5 μg (or the indicated dilutions of the wild-type sample) of total leaf RNA were gel fractionated and then hybridized with radiolabeled probes specific for the indicated transcripts. Two identical blots were prepared and then probed sequentially for LHCP and rbcL mRNAs, or rbcS mRNA and 16S rRNA. (Bottom) Results of staining one of the filters with methylene blue to visualize the bound rRNAs.
The abundance of the chloroplast rbcL mRNA is reduced four- to fivefold in csy1 mutants (Fig. 11). Since this was not sufficient to account for the 10-fold decrease in Rubisco protein, the status of rbcL mRNA translation was assessed by examining its association with polysomes. Fig. 12 shows that whereas essentially all of the rbcL mRNA is associated with polysomes in normal chloroplasts, only a small proportion is polysome associated in csy1 mutants. The analogous result was obtained for the chloroplast atpF mRNAs (Fig. 12) and psbA mRNA (data not shown). These findings indicate a decreased rate of translation initiation or a stalling of ribosomes at a site early on the mRNAs examined. Previously it was noted that the abundance of the rbcL mRNA is reduced fourfold in maize mutants with defects in chloroplast polysome assembly (Barkan, 1993), and it was proposed that this resulted from increased access of ribonucleases to sites that are ordinarily protected by ribosomes. It seems likely, therefore, that the rbcL mRNA deficiency in csy1 mutants is a consequence of its lack of association with ribosomes rather than an independent manifestation of the csy1 defect.
Figure 12.
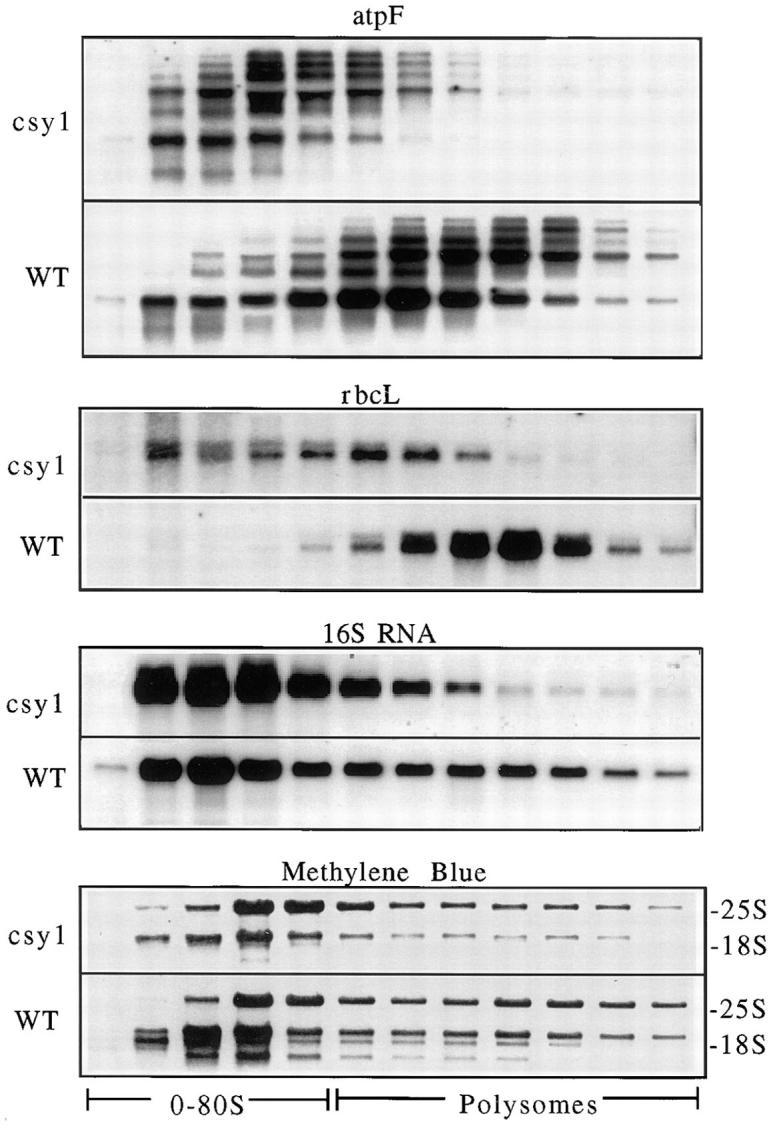
Analysis of chloroplast polysomes in csy1 mutants. Total leaf extracts prepared under conditions that maintain the integrity of polysomes were fractionated in sucrose gradients. An equal proportion of each fraction was analyzed by RNA gel–blot hybridization with the probes indicated. The methylene blue– stained filters show the distribution of cytosolic rRNAs in the gradients.
To obtain a more global picture of the status of the chloroplast translation machinery, 16S rRNA was quantified (Fig. 11) and its association with polysomes was examined (Fig. 12). The content of 16S rRNA is reduced approximately fivefold in csy1 mutants, presumably reflecting a similar decrease in the content of 30S ribosomes. PhosphorImager quantification revealed little difference between mutant and wild-type samples in the proportion of 16S rRNA found in polysomes, although the average polysome size is reduced in the mutants. The distribution of 16S rRNA in the gradient was less dramatically altered than that of the rbcL, atpF, and psbA mRNAs, suggesting that either the translation of only a subset of chloroplast mRNAs is disrupted in csy1 mutants, or that the translational block is global but that the timing of the block (i.e., initiation, early or late in elongation) varies between mRNAs.
Discussion
cpSecY Function Is Not Limited to the Translocation of Proteins That Engage cpSecA
In bacteria, secreted proteins that interact with SecA pass through a translocon consisting of SecY, SecE, and SecG (for review see Rapoport et al., 1996). It seems likely that cpSecY is, by analogy, a component of the thylakoid translocon for proteins that engage cpSecA. The csy1 mutant phenotype is consistent with this notion. The rate of processing of two cpSecA substrates, PC and OE33, is reduced dramatically in csy1 mutants (refer to Fig. 10). In addition, a >10-fold decrease in the accumulation of mature PC and PsaF is accompanied by an increase in the accumulation of their stromal intermediates (refer to Fig. 7). However, we cannot eliminate the possibility that the poor translocation of cpSecA substrates is a consequence of the lack of target membrane. Furthermore, if the signal peptidase itself requires cpSecY to cross the membrane, then the rate of signal-sequence cleavage may not be a reliable indicator of a translocation defect.
It is clear from our results, however, that regardless of whether cpSecY functions in the translocation of proteins that engage cpSecA, it must also play other roles. Previously, we found that tha1 mutants contain little, if any, cpSecA, but nonetheless contain normal amounts of thylakoid membrane (Voelker et al., 1997). tha1 mutants also differ from csy1 mutants in that they contain normal levels of LHCP and Rubisco (Voelker and Barkan, 1995). Therefore, cpSecY function must extend beyond translocating proteins that previously interacted with cpSecA. Our results are consistent with the possibility that cpSecY is an essential component of the thylakoid Sec translocon, whereas cpSecA is merely a facilitator. Alternatively, it seems plausible that cpSecY mediates the translocation of ΔpH-pathway substrates. Indeed, csy1 mutants exhibit a reduced accumulation of mature ΔpH substrates and an increase in the abundance of their stromal precursors. The defects in the ΔpH pathway cannot be attributed to a loss of tha4 and hcf106 gene products because both accumulate to normal levels in csy1 mutants (data not shown). On the other hand, the magnitude of the defect in the ΔpH pathway as reflected by the immunoblot assay appears less severe than that in the cpSecA pathway (refer to Fig. 7), suggesting that cpSecY may not function directly in this pathway. The notion that defects in ΔpH substrate processing are only an indirect effect of the absence of cpSecY is supported by the recent discovery of a bacterial secretory mechanism related to the thylakoid–ΔpH pathway that does not require SecY in vivo (Santini et al., 1998).
To address the possibility that cpSecY function does not extend beyond the ΔpH and cpSecA lumenal targeting pathways, we compared the phenotype of cpSecY mutants with that of tha1/tha4 double mutants (Fig. 8). The double mutants differ from csy1 mutants in that they have a much higher content of chlorophyll and of the core thylakoid membrane complexes, and they are not deficient for LHCP or Rubisco. The much more severe phenotype of csy1 mutants suggests that cpSecY performs some function in addition to any role it might play in the SecA and ΔpH pathways. Sec61p, the endoplasmic reticulum homologue of SecY, functions in the translocation of proteins that engage SRP54 (for review see Rapoport et al., 1996). It seems likely, by analogy, that cpSecY mediates the integration of proteins like LHCP that engage cp54. It is also plausible that cpSecY is required for the integration of proteins typified by CFoII, which differ from SecA, ΔpH, and cp54 substrates in that they are capable of integration into protease-treated thylakoid membranes (Robinson et al., 1996). Since cpSecY is predicted to be an integral membrane protein with only limited exposure to the stroma (Laidler et al., 1995; Berghoefer and Kloesgen, 1996), it may be resistant to proteases.
Does cpSecY Function in the Integration of Proteins into the Inner Envelope Membrane?
Membrane proteins that are synthesized in the cytosol and imported into the chloroplast stroma have two potential destinations: the thylakoid membrane and the inner envelope membrane. Two routes to the inner envelope can be envisioned for such proteins, a “stop transfer” route in which the protein becomes arrested in the inner envelope during import, and a “conservative sorting” route in which the protein is imported into the stroma and then reexported to the inner envelope by an ancestral secretory system. Recent evidence suggests that TIC110, a nuclear- encoded inner envelope protein, passes through the stroma before its insertion into the envelope membrane, consistent with a conservative sorting mechanism (Luebeck et al., 1997). Should a conservative sorting mechanism exist, a chloroplast-localized SecY homologue could well be involved. However, the csy1 gene is clearly not critical for the targeting of all inner envelope proteins because csy1 mutants are not defective for chloroplast import (refer to Fig. 10), a process that requires several integral components of the inner envelope, including TIC110. In addition, the accumulation of TIC110 protein is unaltered in csy1 mutants (data not shown). It remains possible that the product of the csy1 gene functions in the targeting of just a subset of proteins to the inner envelope, or that another of the closely related genes detected by Southern hybridization (refer to Fig. 2) could encode an envelope-localized cpSecY isoform.
Role of cpSecY in the Elaboration of the Thylakoid Membrane
The loss of thylakoid membrane in csy1− chloroplasts indicates that cpSecY plays a role in membrane accumulation itself. In contrast, the absence of cpSecA in tha1 mutants does not interfere with the elaboration of the thylakoid membrane (Voelker et al., 1997). Many steps in chloroplast–lipid synthesis are localized to the inner envelope membrane (for review see Joyard et al., 1991). Thus, a loss of thylakoid membrane could result if cpSecY functions in integrating lipid biosynthetic enzymes into the inner envelope. Alternatively, cpSecY may play a role in the transfer of lipids from the inner envelope to the thylakoid membrane. There is evidence that thylakoid membranes initially arise by invagination of the inner envelope membrane, followed by vesicle formation and vesicle fusion (for review see Hoober et al., 1994; Cline and Henry, 1996). A stromal protein has been identified that promotes the fusion in vitro of vesicles isolated from chromoplasts (Hugueney et al., 1995). If a similar mechanism exists in chloroplasts, integral membrane proteins are likely to be required to capture these vesicles. cpSecY might be needed for the integration of such proteins. It is interesting in this regard that many of the plastids lacking cpSecY contain small membrane vesicles (refer to Fig. 6 and data not shown), as would be expected if vesicle fusion were disrupted.
Role of cpSecY in Chloroplast Translation
An unanticipated aspect of the csy1 mutant phenotype is the loss of the stromal enzyme Rubisco. This is not a consequence of the chlorophyll deficiency per se, since several maize mutants with more severe chlorophyll deficiencies accumulate near normal levels of Rubisco (Harpster et al., 1984). Likewise, the Rubisco deficiency is not a consequence of photooxidative damage since it is equally severe in dark- and light-grown plastids (refer to Fig. 9). The rbcS mRNA accumulates to normal levels in csy1 mutants (refer to Fig. 11) and the import of nuclear-encoded proteins into the mutant chloroplasts appears normal (refer to Fig. 10). These negative results led us to investigate the possibility that csy1 mutants have a defect in chloroplast translation. In fact, csy1− plastids exhibit a dramatic decrease in the average number of ribosomes associated with the rbcL mRNA and with several other chloroplast mRNAs examined (refer to Fig. 12 and data not shown). The fact that the distribution of 16S rRNA in the polysome gradients is not dramatically affected in csy1 mutants (refer to Fig. 12) suggests that the mutation does not cause a global decrease in the translation initiation rate. Thus, the translation defect in csy1 mutants differs from that in cps mutants, which are defective in chloroplast polysome assembly (Barkan, 1993). The small amount of pre-16S rRNA in csy1 mutants (refer to Fig. 11) and in all cps mutants (Barkan, 1993) is likely the result, rather than the cause, of the translational defects (see Discussion in Barkan, 1993).
This translation defect is intriguing in that it suggests a link between chloroplast membrane biogenesis and chloroplast gene expression. One possibility is that the absence of cpSecY results in the prolonged stalling of ribosomes that are translating membrane proteins, thereby reducing the concentration of free ribosomes available for initiation. A proportion of chloroplast ribosomes is associated with thylakoid membranes (Chua et al., 1973; Margulies and Michaels, 1975; Minami and Watanabe, 1984; Breidenbach et al., 1988), and chloroplast ribosomes pause during the translation of several membrane proteins (Kim et al., 1991; Stollar et al., 1994). It is plausible that the release of some pauses is facilitated by interactions between the ribosome and a proteinaceous receptor in the thylakoid membrane. This would be analogous to the release of SRP-mediated ribosome pausing by interactions with the SRP receptor in the endoplasmic reticulum membrane (for review see Walter and Johnson, 1994). An alternative possibility arises from the recent finding that several chloroplast RNA–binding proteins are associated with internal chloroplast membranes (Zerges and Rochaix, 1998). If translational activators are among these proteins, then the absence or mislocalization of these activators in csy1 mutants could cause defects in translation. Consistent with this notion are the observations that several translational activators in yeast mitochondria are tightly associated with the mitochondrial inner membrane (for review see Gillham et al., 1994), and that polysomes containing the rbcL mRNA have been found in association with thylakoid membranes (Minami and Watanabe, 1984; Breidenbach et al., 1988; Klein et al., 1988).
In summary, the data presented here provide strong evidence that cpSecY functions in more than just the cpSecA-dependent movement of proteins across the thylakoid membrane. Our results suggest further that cpSecY functions in some aspect of thylakoid membrane biogenesis other than translocating proteins to the thylakoid lumen. Finally, analysis of csy1 mutants has revealed a link between chloroplast translation and the biogenesis of internal chloroplast membranes. A more detailed description of cpSecY function will require biochemical studies and the use of leaky and/or conditional mutations in genes encoding cpSecY.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 GM48179).
Abbreviations used in this paper
- LHCP
light-harvesting chlorophyll a/b binding protein
- OE16
16-kD subunit of oxygen evolving complex
- OE23
23-kD subunit of oxygen evolving complex
- OE33
33-kD subunit of oxygen evolving complex
- PC
plastocyanin
- Rubisco
ribulose biphosphate carboxylase
- SRP
signal recognition particle
- csy1
chloroplast secY 1
Footnotes
We would like to express our deep gratitude to K. Canada and especially to B. Meeley at Pioneer Hi-Bred for identifying and providing the mutants used in this study, and for being such forthcoming and helpful collaborators. We would also like to thank J. Selker for performing the electron microscopy, Y. Wang for DNA sequencing, R. Voelker and M. Walker for technical advice, and R. Voelker and M. Covington for help with the tha1/tha4 double mutant experiment (all four from University of Oregon, Eugene, OR). Antibodies were provided by S. Merchant (University of California, Los Angeles, CA) (CF1), B. Taylor (CSIRO, Canberra, Australia) (LHCP), D. Malkin (University of California, Berkeley, CA) (PsaF), and K. Keegstra and J. Davila-Aponte (both from Michigan State University, East Lansing, MI) (TIC110). hcf106-mum3 seed was provided by Rob Martienssen (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
Address all correspondence to Alice Barkan, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403. Tel.: (541) 346-5145. Fax: (541) 346-5891.
References
- Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered RNA metabolism. Plant Cell. 1993;5:389–402. doi: 10.1105/tpc.5.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Miles D, Taylor W. Chloroplast gene expression in nuclear, photosynthetic mutants of maize. EMBO (Eur Mol Biol Organ) J. 1986;5:1421–1427. doi: 10.1002/j.1460-2075.1986.tb04378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Voelker R, Mendel-Hartvig J, Johnson D, Walker M. Genetic analysis of chloroplast biogenesis in higher plants. Physiol Plant. 1995;93:163–170. [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. Cloning and characterization of the maize An1gene. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoefer J, Kloesgen RB. Isolation and characterization of a cDNA encoding the SecY protein from spinach chloroplasts. Plant Physiol. 1996;112:863. doi: 10.1074/jbc.270.31.18341. [DOI] [PubMed] [Google Scholar]
- Breidenbach E, Jenni E, Boschetti A. Synthesis of two proteins in chloroplasts and mRNA distribution between thylakoids and stroma during the cell cycle of Chlamydomonas reinhardtii. . Eur J Biochem. 1988;177:225–232. doi: 10.1111/j.1432-1033.1988.tb14366.x. [DOI] [PubMed] [Google Scholar]
- Chandler VL, Hardeman KJ. The Mu elements of Zea mays. . Adv Genet. 1992;30:77–122. doi: 10.1016/s0065-2660(08)60319-3. [DOI] [PubMed] [Google Scholar]
- Chua N-H, Blobel G, Siekevitz P, Palade GE. Attachment of chloroplast polysomes to thylakoid membranes in Chlamydomonas reinhardtii. . Proc Natl Acad Sci USA. 1973;70:1554–1558. doi: 10.1073/pnas.70.5.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu Rev Cell Dev Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- Das L, Martienssen R. Site-selected transposon mutagenesis at the hcf106locus in maize. Plant Cell. 1995;7:287–294. doi: 10.1105/tpc.7.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, F., D.E. Koehler, and J.D. Smith. 1983. Fluridone induction vivipary during maize seed development. In Third International Symposium on Pre-Harvest Sprouting in Cereals. J.E. Kruger and D.E. LaBerge, editors. Westview Press, Boulder, CO. 188–196.
- Gillham NW, Boynton JE, Hauser CR. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- Han C-D, Coe EHJ, Martienssen RA. Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. EMBO (Eur Mol Biol Organ) J. 1992;11:4037–4046. doi: 10.1002/j.1460-2075.1992.tb05497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpster MH, Mayfield SP, Taylor WC. Effects of pigment-deficient mutants on the accumulation of photosynthetic proteins in maize. Plant Molec Biol. 1984;3:59–71. doi: 10.1007/BF00040030. [DOI] [PubMed] [Google Scholar]
- Hoffman NE, Franklin AE. Evidence for a stromal GTP requirement for the integration of a chlorophyll a/b-binding polypeptide into thylakoid membranes. Plant Physiol. 1994;105:295–304. doi: 10.1104/pp.105.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober JK, White RA, Marks DB, Gabriel JL. Biogenesis of thylakoid membranes with emphasis on the process in Chlamydomonas. . Photosynth Res. 1994;39:15–31. doi: 10.1007/BF00027139. [DOI] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, d'Harlingue A, Kuntz M, Camara B. Identification of a plastid protein involved in vesicle fusion and/or membrane protein translocation. Proc Natl Acad Sci USA. 1995;92:5630–5634. doi: 10.1073/pnas.92.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BD, Kulhanek DJ, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Block MA, Douce R. Molecular aspects of plastid envelope biochemistry. Eur J Biochem. 1991;199:489–509. doi: 10.1111/j.1432-1033.1991.tb16148.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Gamble-Klein P, Mullet JE. Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J Biol Chem. 1991;266:14931–14938. [PubMed] [Google Scholar]
- Kim SJ, Robinson DR, Robinson C. An Arabidopsis thalianacDNA encoding PSII-X, a 4.1 kDa component of photosystem II: a bipartite presequence mediates SecA/ΔpH-independent targeting into thylakoids. FEBS (Fed Eur Biochem Soc) Lett. 1996;390:175–178. doi: 10.1016/0014-5793(96)00658-8. [DOI] [PubMed] [Google Scholar]
- Klein RR, Mason HS, Mullet JE. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J Cell Biol. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidler V, Chaddock AM, Knott TG, Walker D, Robinson C. A SecY homolog in Arabidopsis thaliana. . J Biol Chem. 1995;270:17664–17667. doi: 10.1074/jbc.270.30.17664. [DOI] [PubMed] [Google Scholar]
- Li X, Henry R, Yuan J, Cline K, Hoffman NE. A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc Natl Acad Sci USA. 1995;92:3789–3793. doi: 10.1073/pnas.92.9.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebeck J, Heins L, Soll J. A nuclear-coded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J Cell Biol. 1997;137:1279–1286. doi: 10.1083/jcb.137.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies MM, Michaels A. Free and membrane-bound chloroplast polyribosomes in Chlamydomonas reinhardtii. . Biochem Biophys Acta. 1975;402:297–308. doi: 10.1016/0005-2787(75)90267-1. [DOI] [PubMed] [Google Scholar]
- Martienssen, R., A. Barkan, A. Scriven, and W. Taylor. 1987. Identification of a nuclear gene involved in thylakoid structure. In Plant Membranes: Structure, Function, Biogenesis. C. Leaver and H. Sze, editors. Alan R. Liss, Inc., New York. 181–192.
- Mascia PN, Robertson DS. Studies of chloroplast development in four maize mutants defective in chlorophyll biosynthesis. Planta. 1978;143:207–211. doi: 10.1007/BF00387791. [DOI] [PubMed] [Google Scholar]
- Meeley RB, Briggs S. Reverse genetics for maize. Maize Genet. . Coop Newsletter. 1995;69:67. [Google Scholar]
- Miles, D. 1982. In Methods in Chloroplast Molecular Biology. R. Hallick, M. Edelman, and N.-H. Chua, editors. Elsevier, New York. 75–106.
- Minami E-I, Watanabe A. Thylakoid membranes: the translation site of chloroplast DNA-regulated thylakoid polypeptides. Arch Biochem Biophys. 1984;235:562–570. doi: 10.1016/0003-9861(84)90230-3. [DOI] [PubMed] [Google Scholar]
- Mould R, Knight JS, Bogsch E, Gray JC. Azide-sensitive thylakoid membrane insertion of chimeric cytochrome fpolypeptides imported by isolated pea chloroplasts. Plant J. 1997;11:1051–1058. doi: 10.1046/j.1365-313x.1997.11051051.x. [DOI] [PubMed] [Google Scholar]
- Nelson T, Harpster MH, Mayfield SP, Taylor WC. Light-regulated gene expression during maize leaf development. J Cell Biol. 1984;98:558–564. doi: 10.1083/jcb.98.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohara T, Nakai M, Goto A, Endo T. Isolation and characterization of the cDNA for pea chloroplast SecA: Evolutionary conservation of the bacterial-type SecA-dependent protein transport within chloroplasts. FEBS (Fed Eur Biochem Soc) Letters. 1995;364:305–308. doi: 10.1016/0014-5793(95)00415-6. [DOI] [PubMed] [Google Scholar]
- Nohara T, Asai T, Nakai M, Sugiura M, Endo T. Cytochrome fencoded by the chloroplast genome is imported into thylakoids via the SecA-dependent pathway. Biochem Biophys Res Commun. 1997;224:474–478. doi: 10.1006/bbrc.1996.1051. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eucaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque strain in electron microscopy. J Cell Biol. 1963;17:208. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, D.S., I.C. Anderson, and M.D. Bachmann. 1978. Pigment-deficient mutants: Genetic, biochemical, and developmental studies. In Maize Breeding and Genetics. D.B. Walden, editor. Wiley–Interscience Publication, New York. 461–494.
- Robinson D, Karnauchov I, Herrmann RG, Kloesgen RB, Robinson C. Protease-sensitive thylakoidal import machinery for the Sec-, delta pH - and signal recognition particle-dependent protein targeting pathways, but not for CFoII integration. Plant J. 1996;10:149–155. [Google Scholar]
- Santini C-L, Ize B, Chanal A, Mueller M, Giordano G, Wu L-F. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. . EMBO (Eur Mol Biol Organ) J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G, Mishkind M. Rapid degradation of unassembled ribulose 1,5-bisphosphate carboxylase small subunits in chloroplasts. Proc Natl Acad Sci USA. 1983;80:2632–2636. doi: 10.1073/pnas.80.9.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ. Shedding light on the chloroplast protein import machinery. Cell. 1995;83:521–524. doi: 10.1016/0092-8674(95)90090-x. [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ, Goldschmidt-Clermont M, Rahire M, Rochaix J-D. Nonsense mutations in the Chlamydomonaschloroplast gene that codes for the large subunit of ribulosebisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA. 1985;82:5460–5464. doi: 10.1073/pnas.82.16.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollar NE, Kim J-K, Hollingsworth MJ. Ribosomes pause during the expression of the large ATP synthase gene cluster in spinach chloroplasts. Plant Physiol. 1994;105:1167–1177. doi: 10.1104/pp.105.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. Regulatory interactions between nuclear and plastid genomes. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:211–233. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position–specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitanen PV, Doran ER, Dunsmuir P. What is the role of the transit peptide in thylakoid integration of the light-harvesting chlorophyll a/b protein? . J Biol Chem. 1988;263:15000–15007. [PubMed] [Google Scholar]
- Voelker R, Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO (Eur Mol Biol Organ) J. 1995;14:3905–3914. doi: 10.1002/j.1460-2075.1995.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R, Mendel-Hartvig J, Barkan A. Transposon-disruption of a maize nuclear gene, tha1, encoding a chloroplast SecA homolog: in vivorole of cp-SecA in thylakoid protein targeting. Genetics. 1997;145:467–478. doi: 10.1093/genetics/145.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H, Fischer S, Valentin K. A model for the evolution of the plastid sec apparatus inferred from secYgene phylogeny. Plant Molec Biol. 1996;32:685–692. doi: 10.1007/BF00020209. [DOI] [PubMed] [Google Scholar]
- Walbot V, Coe JEH. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. . Proc Natl Acad Sci USA. 1979;76:2760–2764. doi: 10.1073/pnas.76.6.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Yuan J, Henry R, McCaffery M, Cline K. SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science. 1994;266:796–798. doi: 10.1126/science.7973633. [DOI] [PubMed] [Google Scholar]
- Zerges W, Rochaix JD. Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. . J Cell Biol. 1998;140:101–110. doi: 10.1083/jcb.140.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]