Abstract
Peroxisomal membrane protein (Pmp)26p (RnPex11p), a major constituent of induced rat liver peroxisomal membrane, was found to contain a COOH-terminal, cytoplasmically exposed consensus dilysine motif with the potential to bind coatomer. Biochemical as well as immunocytochemical evidence is presented showing that peroxisomes incubated with preparations of bovine brain or rat liver cytosol recruit ADP-ribosylation factor (ARF) and coatomer in a strictly guanosine 5′-O-(3-thiotriphosphate)–dependent manner. Consistent with this observation, ldlF cells expressing a temperature-sensitive mutant version of the ε-subunit of coatomer exhibit elongated tubular peroxisomes possibly due to impaired vesiculation at the nonpermissive temperature. Since overexpression of Pex11p in Chinese hamster ovary wild-type cells causes proliferation of peroxisomes, these data suggest that Pex11p plays an important role in peroxisome biogenesis by supporting ARF- and coatomer-dependent vesiculation of the organelles.
Regarding the biogenesis of peroxisomes various concepts have been postulated. Based on morphological data that demonstrated close spatial relationships between peroxisomes and the ER, Novikoff and Novikoff (44) favored the idea that new peroxisomes are formed by budding and fission from the ER. However, a series of biochemical studies initiated by de Duve and coworkers (36, 51) in the early 1970s, focusing on the biogenetic route of catalase as a peroxisomal marker, did not reveal an ER involvement in the import of catalase. Subsequent findings of Goldman and Blobel (19) and particularly the group of Lazarow and coworkers (52, 54) confirmed this view demonstrating that peroxisomal matrix proteins, such as catalase, urate oxidase, and enzymes of the peroxisomal β-oxidation pathway, are synthesized on free polyribosomes and thus are imported posttranslationally. Accordingly, Goldman and Blobel (19) postulated a model in which peroxisomal matrix proteins are found in nascent peroxisomes, whereas peroxisomal membrane proteins (Pmps) are sorted to the peroxisomes via the ER. However, when Fujiki et al. (16) demonstrated that Pmp22p, a major constituent of the peroxisomal membrane is synthesized on membrane-free polyribosomes, these authors no longer considered ER involvement but, instead, proposed an autonomous mechanism of peroxisome formation (37).
Very recently, this view has again been questioned by studies on Pex15p, a phosphorylated peroxisomal membrane protein (Pmp)1 of Saccharomyces cerevisiae and a chimeric product of Pex3p and catalase in the yeast Hansenula polymorpha (4, 14). Overexpression of both of these polypeptides was reported to cause remarkable induction of multimembrane layers reminiscent of the ER. These observations again may ascribe a distinct role to the ER in the biogenesis of peroxisomes.
As our knowledge on membrane vesiculation has increased considerably within the last few years, we have learned that, with only a few exceptions (34), protein coats are required to shape membranes into vesicles. Therefore, irrespective of whether formation of new peroxisomes occurs by an autonomous or endomembrane system-derived process, both most likely may implicate coat protein– mediated membrane vesiculation. The protein coats identified so far are the clathrin coat involved in endocytosis and vesiculation of the trans-Golgi net work (50) as well as the coat proteins (COP) I and COP II (56, 57, 60). Although still a matter of debate, COP I–coated vesicles are believed to mediate anterograde intra-Golgi transport and retrograde Golgi to ER transport, whereas COP II–coated vesicles bud from the ER and likely transport cargo to the ER-Golgi intermediate compartment (ERGIC; references 3, 9, 59). Assembly and membrane binding of clathrin is mediated by adaptor proteins that, like clathrin, are recruited from a soluble pool (50). Formation of COP I– and COP II–coated vesicles, on the other hand, requires binding of ADP-ribosylation factor 1 (ARF1) and SAR1, respectively, small GTPases that initiate coat assembly in their GTP-bound state (5, 61).
We have investigated the structure and function of major components of rat liver peroxisomal membranes and the mechanism of their membrane integration (10, 24, 29, 32, 49). In the course of these studies we noticed by thyroid hormone induction of a membrane protein, Pmp26p, and concommittant abundance of organelles with a diameter of <0.2 μm (24, 30). Since these observations indicated a function of Pmp26p in peroxisome biogenesis and proliferation, we cloned and sequenced the Pmp26p-cDNA from a rat liver cDNA library. Here we describe the molecular structure and membrane topology of Pmp26p. Interestingly, we found a COOH-terminal dilysine motif exposed toward the cytoplasm. Such motifs have been shown to be involved in binding coatomer and may help to generate vesicles for the retrieval of ER resident transmembrane polypeptides from the ERGIC back to the ER (8, 38). In studies with purified rat liver peroxisomes and permeabilized hepatocytes, we show a GTP-γS–dependent recruitment of ARF and coatomer to the peroxisomal membrane as well as a change in peroxisome morphology of a cell line defective in one of the coatomer subunits. Our results suggest that ARF and coatomer are involved in peroxisome biogenesis.
Materials and Methods
Molecular Biology
Isolated peroxisomal membranes of clofibrate-induced rat liver peroxisomes were subjected to SDS-PAGE and the polypeptides blotted onto polyvinylidene difluoride (PVDF) membranes. Pex11p was excised from the membrane and the tryptic fragments were analyzed by HPLC and microsequencing. The following peptide sequences were identified: (1) AF(I)QATEQSIQATDLVPR; (2) NLETSVSTGR; (3) SVGLTSGIN; (4) NFE(C)DILIPLN. According to the sequence of peptide 1, two degenerated splitting oligonucleotide primers were synthesized and used for reverse transcription–PCR (RT-PCR). A 51-bp fragment was amplified that exactly contained the sequence related to those five amino acid residues (SIQAT) not encoded by the primers. A 35-mer oligonucleotide of the 51-bp fragment was further used to screen a rat liver cDNA library. Eight positive clones were identified and the cDNA cloned behind the T7 promoter into pBluescriptSK(−) phagemid using the BamHI and HindIII restriction sites.
The DNA sequence encoding a human c-myc epitope (EQKLISEEDL) was introduced into the cDNA at the 3′-end directly behind the triplet for the last COOH-terminal amino acid residue yielding Pex11p-Cmyc. This construct in which the COOH-terminal KXKXX motif present in Pex11p is masked by the c-myc epitope was made by PCR using oligonucleotide primers containing BamHI restriction sites. The digested PCR fragment was cloned behind the cytomegalovirus promoter of the eukaryotic expression vector pcDNA3. The intracellular location of Pex11p-Cmyc was studied after stable expression of the construct in AT3 mouse hepatocytes (2) by immunofluorescence using a monoclonal anti-myc antibody.
Isolation of Subcellular Organelles and Membrane Topology of Pex11p
Male Wistar rats weighing 120–150 g were treated with clofibrate (0.5%) or trifluoroacetate (TFA, 1%), both added to the chow for 10 consecutive days. Liver peroxisomes and mitochondria were isolated by isopycnic centrifugation of the light mitochondrial fractions in 14–45% wt/vol Nycodenz density gradients containing an inverse sucrose gradient as described previously (25). Purified microsomal and Golgi vesicles were isolated by sucrose density gradient centrifugation as described (68). The activities of glucosylceramide synthase and sphingomyelin synthase, two marker enzymes of the Golgi apparatus, were measured as described (35).
To analyze the orientation of Pex11p within the peroxisomal membrane, isolated peroxisomes were resuspended in 0.25 M sucrose, 100 mM Pipes-NaOH, pH 7.6, and incubated for 30 min at 0°C with subtilisin added in a concentration of 0.5% of the peroxisomal protein. This concentration was previously shown not to affect peroxisomal content proteins, such as catalase, which becomes degraded only at protease concentrations >5% (10). The protease was blocked with 1 mM PMSF and the fragmentation of Pex11p analyzed by SDS-PAGE and immunoblotting. Antisera against Pex11p and Pmp69p were raised by immunizing rabbits with Pex11p and Pmp69p excised from SDS–polyacrylamide gels after separating peroxisomal membrane proteins. The antisera raised against the COOH termini of the polypeptides were obtained by coupling the COOH-terminal octapeptide of Pex11p and the COOH-terminal undecapeptide of Pmp69p to keyhole limpets hemocyanin.
Binding of Coatomer and ARF to Rat Liver Peroxisomes
Cytosol from bovine brain was isolated as described (70) except that fresh unfrozen tissue was used. The same procedure was applied for the isolation of cytosol from rat liver. These cytosol preparations usually had a protein content of ∼40 mg/ml. To study binding of coatomer and ARF, the organelles (250 μg) were incubated with 20 mg bovine brain cytosol (10 mg rat liver cytosol) and 20–50 μM GTP-γS (guanosine 5′-O-[3-thiotriphosphate]) or GDP-βS (guanosine 5′-O-[2-thiodiphosphate]) in a total volume of 3 ml essentially as described (39). After incubations, peroxisomes were recovered by centrifugation at 15,000 g for 10 min and subjected to SDS-PAGE and immunoblotting. The various coatomer subunits were identified using specific antibodies raised against the individual coatomer subunits α-, β′-, γ-, δ-, ε-, and ζ-COP (18, 23, 65, 66). β-COP was visualized using the monoclonal antibody M3A5 (12). Binding of ARF to peroxisomes was demonstrated by an antibody raised against recombinant ARF1 (17, 48). Coatomer binding to mitochondria or microsomes from rat liver was performed as described for peroxisomes except reisolation of microsomes was carried out by flotation rather than sedimentation. After incubation of peroxisomes with bovine brain or rat liver cytosol in the presence of GTP-γS or GDP-βS, organelles were recovered either by pelleting at 12,000 g for 15 min or by centrifugation in a 14–45% Nycodenz density gradient at 80,000 g for 1 h. Fractions were diluted and pelleted by high speed centrifugation and analyzed by immunoblotting for both ARF (anti-ARF1 antiserum diluted 1:5,000) and coatomer (anti–α-COP antiserum diluted 1:1,000).
Binding of coatomer and ARF to protease-pretreated peroxisomes was studied by incubating peroxisomes for 60 min at 12°C with increasing concentrations of subtilisin, as indicated, in order to digest the NH2- and COOH-terminal cytoplasmic extensions of Pex11p. Controls remained untreated or were treated with the same concentrations of subtilisin previously inactivated by 1 mM PMSF. After digestion, the reisolated peroxisomes were incubated with rat liver cytosol (10 mg) in the presence of GTP-γS or GDP-βS (50 μM). Peroxisomes were identified by the polyclonal anti-Pex11p antiserum, whereas coatomer and ARF binding was detected by anti–α-COP and anti-ARF 1 antiserum, respectively.
Rat liver hepatocytes of clofibrate-treated animals were prepared as described previously (26). The cells (50 mg wet weight) were resuspended in 200 μl of 0.25 M sucrose, 10 mM Hepes-KOH, pH 7.4, and treated with 10 hemolytic units of preactivated streptolysin O (SLO) at 4°C for 3 min as described (6, 53). After permeabilization the cells were incubated with an ATP regenerating system (53), rat liver cytosol (20 mg/ml) and GTP-γS or GDP-βS (50 μM) in a total volume of 500 μl for 30 min at 37°C. The recovered cells were homogenized in sucrose medium and after removal of nuclei and debris by centrifugation at 1,000 g for 3 min, the postnuclear supernate was separated on a linear 14–45% Nycodenz gradient.
Binding of Coatomer to Synthetic Peptides
The coupling of peptides to thiopropyl–Sepharose was performed via an NH2-terminal cysteine residue and followed quantitatively. Equal molar amounts of each coupled peptide were used in the coatomer binding assays. For these assays 2 nmol of coupled peptide were incubated with 500 μg bovine brain cytosol in a total volume of 250 μl of high salt buffer (50 mM Hepes-KOH, pH 7.4, 300 mM NaCl, 1 mM EDTA, 0.5% NP-40), for 2 h at 4°C. After incubation, the Sepharose beads were washed twice with high salt buffer and three times with 25 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, and finally with 25 mM Tris-HCl, pH 7.4, by low speed centrifugation. The bound material was eluted with 4% SDS, 5% mercaptoethanol, 10 mM Tris-HCl, pH 6.8, and analyzed for the presence of α- and β′-COP by SDS-PAGE and immunoblotting.
Light and Electron Microscopy
CHO wild-type cells and mutant ldlF cells were grown as described (21, 22, 53). Immunofluorescence staining of peroxisomes was performed as described previously using anti-Pmp69 peptide antiserum (53).
Tissue samples of liver induced with clofibrate or trifluoroacetate were fixed by perfusion and embedded in LR-White (41). Ultrathin sections were incubated for 24 h with the polyclonal anti-Pex11p antiserum affinity purified against a COOH-terminal Pex11p tail peptide. The first antibody was visualized by 14-nm protein A–gold on sections that were stained with uranyl acetate and lead citrate.
For preembedding immunogold labeling, isolated peroxisomes were primed with bovine brain cytosol in the presence of GTP-γS or GDP-βS as described above. The organelles were washed and incubated with the first antibodies at 37°C for 30 min and at 10°C for 4 h and with the gold- labeled second antibodies for 2 h at 4°C. Washing was performed by pelleting the organelles at 15,000 g for 10 min onto a 55% wt/wt sucrose cushion and resuspending the pelleted organelles in 0.25 M sucrose, 25 mM glycylglycine, pH 7.4, 1 mM EDTA. The final pellets were fixed for 30 min on ice with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.6 containing 0.05% CaCl2, rinsed in buffer and embedded in agar. The agar- embedded specimens were processed by the osmium-tannic acid postfixation method as described (62).
Transmission electron microscopy after DAB-staining of catalase and Epon embedding was done as previously described (20).
Results
Molecular Characterization and Membrane Topology of Pex11p
We cloned the cDNA encoding Pex11p from a rat liver λgt10 cDNA library. The cDNA-derived polypeptide sequence contains 246 amino acid residues with a calculated molecular weight of 27,922 D (Fig. 1). Hydropathy analysis (33) as well as the use of the ProteinPredict program (55) revealed two putative transmembrane spanning sections in Pex11p (amino acid residues 85–104 and 218–238) the first exhibiting the characteristics of an amphipathic α-helix with the polar residues of Arg93, Tyr96, and Asp100 positioned to one side of the helix. The presence of transmembrane domains within Pex11p is compatible with its complete resistance to extraction by sodium carbonate (24), a frequently used criterium indicating membrane integration. Computer search revealed amino acid sequence homology to the peroxin Pex11p, represented by Pmp27p of Saccharomyces cerevisiae and Pmp30/31p of Candida boidinii (15, 58), with a similarity of 49% and an identity of 25% between Pmp26p and ScPmp27p. This weak but significant identity is spread over the entire molecule, indicating a certain degree of evolutionary conservation of the gene. Although there may be differences between Pmp26p and yeast Pex11p, such as membrane topology (40) and topogenic signals (see below), the obvious homology in amino acid sequence suggests that Pmp26p is the rat ortholog of the Pex11p family and therefore, we will name it Pex11p (RnPex11p; reference 11).
Figure 1.
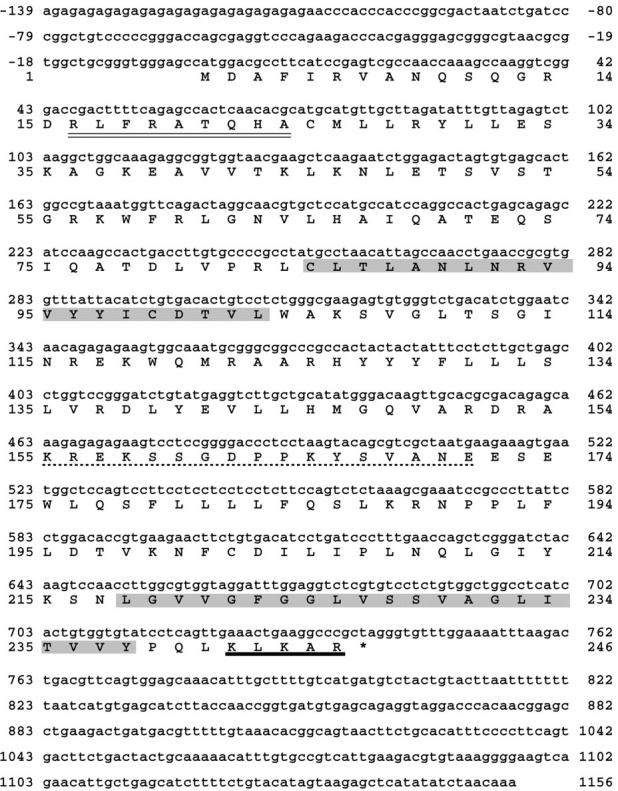
Nucleotide and nucleotide-derived amino acid sequence of rat liver Pex11p-cDNA. Two putative transmembrane spans, as revealed by the Kyte-Doolittle algorithm (33) and the ProteinPredict program (55) are shaded. The consensus PTS2 sequence and the consensus-like sequence of a mPTS are underlined by a double and a single broken line, respectively. The COOH-terminal KXKXX dilysine motif is underlined in bold. These sequence data are available from GenBank/EMBL/DDBJ under accession number AJ224120.
By two different approaches we studied the membrane topology of Pex11p. First, the molecule was tagged at the COOH terminus with a c-myc epitope and the tagged polypeptide, Pex11p-Cmyc, expressed in AT3 cells (results not shown). Peroxisomal localization was demonstrated by colocalizing Pex11p-Cmyc with Pmp69p, which served as a peroxisomal membrane marker. When these cells were differentially permeabilized by digitonin, a monoclonal anti-myc antibody visualized the protein tag. Since digitonin effectively permeabilizes the plasma membrane but leaves peroxisomes impermeable for the antibody (64), this result indicates that the myc epitope in Pex11p may be exposed toward the cytoplasm.
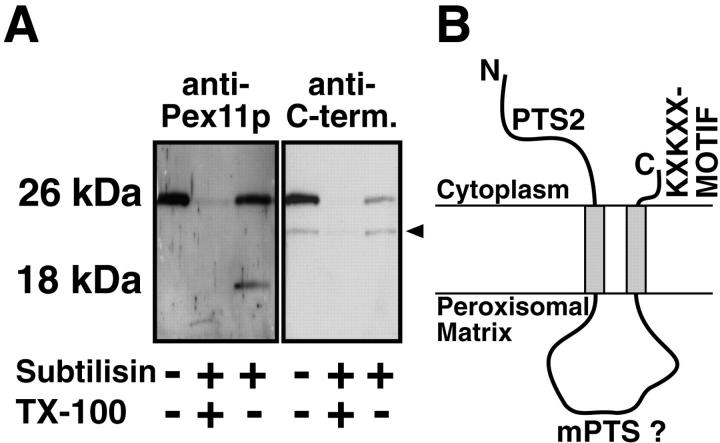
Second, intact, isolated peroxisomes were treated with low concentrations of exogenously added subtilisin, and the membrane-associated cleavage products were analyzed by SDS-PAGE and Western blotting. Using a polyclonal antiserum against Pex11p, we observed the uncleaved polypeptide as well as one major fragment of 18 kD. An antiserum directed against the COOH-terminal octapeptide of Pex11p only recognized the uncleaved protein (Fig. 2 A). A membrane-bound 18-kD fragment is expected if (a) only the first predicted transmembrane span is functional and the NH2 terminus is exposed to the cytoplasm or (b) both transmembrane spans are functional and both termini are oriented cytoplasmically. Since in the former case the proteolytic fragment should also be recognized by the anti–COOH-terminal peptide antibody, which was not the case, these proteolysis experiments, in agreement with the immunofluorescence data on the epitope-tagged Pex11p, suggest that Pex11p is inserted into the membrane by two transmembrane domains and that both the NH2 and COOH termini face the cytoplasm (Fig. 2 B).
Figure 2.
Membrane topology of Pex11p. (A) Treatment of intact isolated rat liver peroxisomes (200 μg) with subtilisin (1 μg) shortened Pex11p to a 18-kD fragment that is recognized by the anti-Pex11p antiserum raised against the entire Pex11p (anti-Pex11p) but not by an antibody directed against its COOH-terminal peptide (anti-C-term.). The characteristic fragmentation of Pex11p is only obtained with intact peroxisomes and does not occur in the presence of Triton X-100 (TX-100). The antipeptide antiserum cross-reacted with an unidentified 23-kD peroxisomal polypeptide (arrowhead). (B) Proposed membrane topology of Pex11p. The polypeptide is inserted into the peroxisomal membrane by two hydrophobic transmembrane spans exposing the NH2 and COOH termini to the cytoplasm. mPTS, peroxisomal membrane targeting signal.
The amino acid residues of positions 16–24 contain the exact consensus sequence of peroxisomal targeting signal 2 (PTS2), originally described in rat liver peroxisomal prethiolase (46) and later on in several other peroxisomal matrix enzymes of different species (67). The presence of a PTS2 in ScPmp27p remains to be proven, because the published consensus motif does not fit with amino acid residue 8 (Y instead of H/Q) of this peptide sequence. In addition to the PTS2, Pex11p contains a sequence in a loop domain between the two transmembrane spans that is strikingly similar but not identical to a peroxisomal membrane targeting signal (mPTS) recently identified in Pmp47p of Candida boidinii (13).
Rat Liver Peroxisomes Bind Coatomer and ARF
As revealed by the sequence data, the COOH terminus of Pex11p carries a dilysine motif of the general type KXKXX. Nilsson et al. (43) first described this motif and demonstrated its function in ER retention of integral membrane proteins. The motif that consists of two lysine residues positioned COOH-terminally at −3 and at either −4 or −5 was subsequently shown to be essential for coatomer binding to Golgi membranes, facilitating the retrieval of ER resident proteins from the ER-Golgi intermediate compartment back to the ER (7, 8, 38).
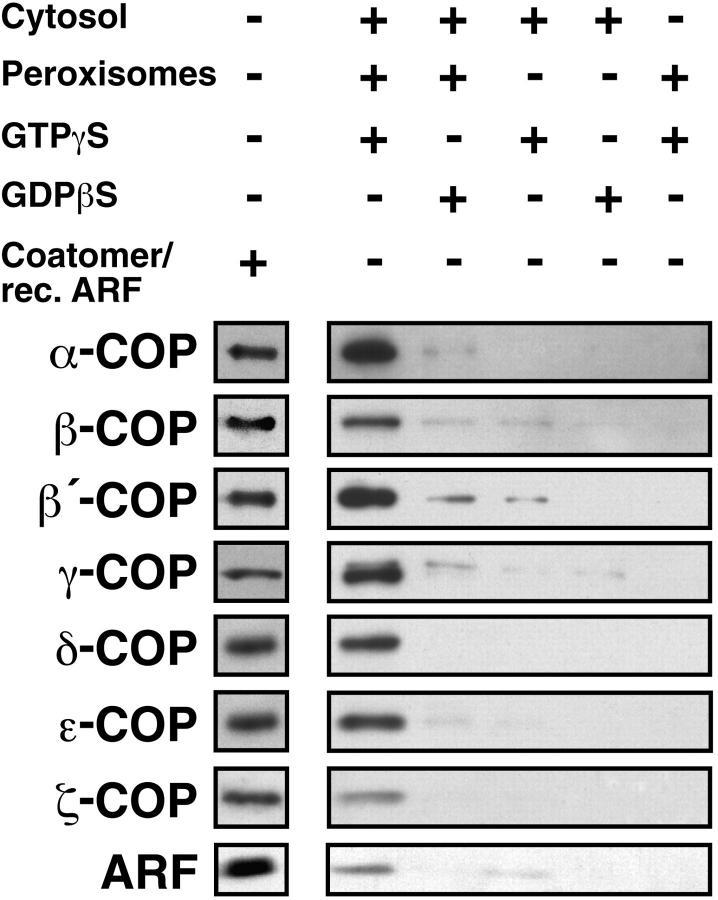
The presence of this motif in a peroxisomal membrane protein at a site accessible from the cytoplasm prompted us to investigate the ability of peroxisomes to bind coatomer. To this end, we incubated highly purified peroxisomes in the presence of bovine brain cytosol and GTP-γS. Control incubations were performed without cytosol and by replacing GTP-γS for GDP-βS. The reisolated peroxisomes were analyzed by Western blotting for binding of the various coatomer subunits. As shown in Fig. 3, specific association of coatomer subunits was only detected when both cytosol and GTP-γS were present in the incubations. All seven coatomer subunits were found to be associated with the peroxisomal membrane.
Figure 3.
GTP-γS–dependent binding of coatomer and ARF to isolated rat liver peroxisomes. Peroxisomes (250 μg) were incubated with bovine brain cytosol (20 mg) in the presence of GTP-γS or GDP-βS (50 μM). Controls were run without peroxisomes and/or without cytosol. After incubation the organelles were recovered by centrifugation and 10-μg aliquots were subjected to SDS-PAGE and immunoblotting using monospecific antibodies against the seven coatomer subunits and ARF 1. The immunoreactivities of the different anti-COP antibodies against isolated bovine brain coatomer and recombinant ARF 1 are shown on the separate lane.
The observed GTP-γS dependence of coatomer binding to peroxisomes suggested an involvement of ARF. ARF belongs to the family of small GTPases and was shown to trigger Golgi-derived vesicle budding and uncoating (61). Therefore, we tested binding of ARF to peroxisomes by Western blotting of organelles reisolated from incubations with cytosol and GTP-γS or GDP-βS. Bound ARF, which was identified using an antiserum directed against ARF 1, was recognized on reisolated peroxisomes only in those incubations that contained both cytosol and GTP-γS (Fig. 3). These peroxisomes exactly cosedimented with coatomer and ARF upon isopycnic centrifugation in a Nycodenz density gradient suggesting tight binding of coatomer and ARF to the organelles.
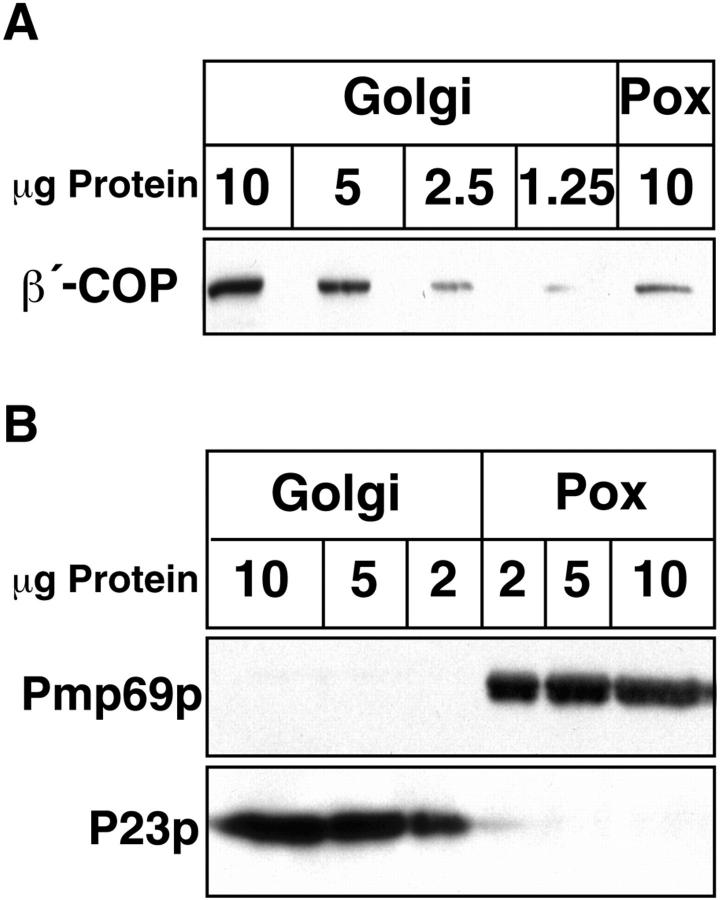
Most critical for the interpretation of these results is the degree of purity of the isolated peroxisomes, particularly with respect to Golgi contamination. According to a previously published protocol we isolated highly purified peroxisomes with catalase activity enriched ∼50–55-fold over the crude homogenate. In these peroxisomal preparations, using a sensitive radioactive assay, galactosyltransferase activity was not detectable (25). Based on the activities of two other Golgi markers, glucosylceramide synthase and sphingomyelin synthase, Golgi contamination of this peroxisomal fraction was calculated to be negligible (Table I). Nevertheless, we analyzed the fraction for Golgi contamination by an additional immunological approach. First, we compared the efficiency in coatomer binding of Golgi membranes and peroxisomes. With equal amounts of Golgi and peroxisomal protein in the assay, Golgi membranes bind approximately five times more coatomer than peroxisomes (Fig. 4 A). If this peroxisomal coatomer binding activity was entirely due to contaminating Golgi, contamination must account for ∼20% of the protein present in the peroxisomal preparation. Second, the peroxisomal fraction was analyzed by Western blotting for its content of P23, a major constituent of Golgi membranes and COP I–coated Golgi-derived vesicles (63). As shown in Fig. 4 B, the anti-P23 antibody reliably identifies its antigen to a level of below 2 μg of Golgi protein. In contrast, 10 μg of peroxisomal protein did not yield any visible P23 signal.
Table I.
Activity of Glucosylceramide- and Sphingomyelin-Synthase in Purified Rat Liver Golgi and Peroxisome Fractions
| Peroxisomes | Golgi | |||
|---|---|---|---|---|
| GlcCer-Synthase [nmol min−1 mg−1] | ND | 765 × 10−3 | ||
| SM-Synthase [nmol min−1 mg−1] | 0.141 × 10−3 | 29 × 10−3 |
Figure 4.
Analysis of Golgi contamination in isolated rat liver peroxisomal fractions. (A) Indicated amounts of isolated Golgi membranes and peroxisomes (Pox) primed with bovine brain cytosol in the presence of GTP-γS were subjected to SDS-PAGE and immunoblotting using an anti–β′-COP antibody. Note that the signal obtained with 10 μg of peroxisomes equals that of ∼2.5 μg of Golgi membranes. In B varying amounts of unprimed Golgi membranes and peroxisomes (Pox) were separated by SDS-PAGE and visualized by immunoblotting using antisera directed against Golgi P23 and Pmp69p, respectively. Note that the anti-P23 antiserum reliably recognizes 2 μg and even less of Golgi membranes and that even with 10 μg of peroxisomes no P23 immunoreaction product is observed.
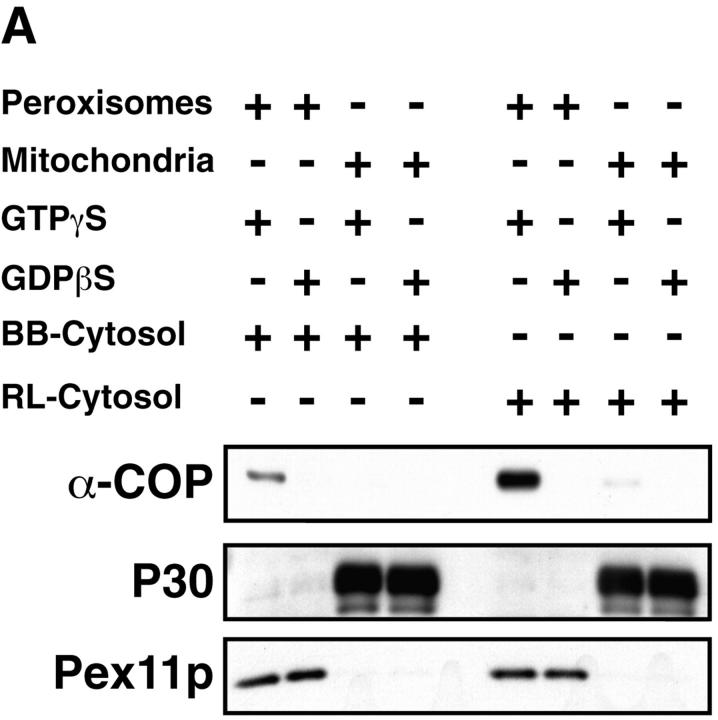
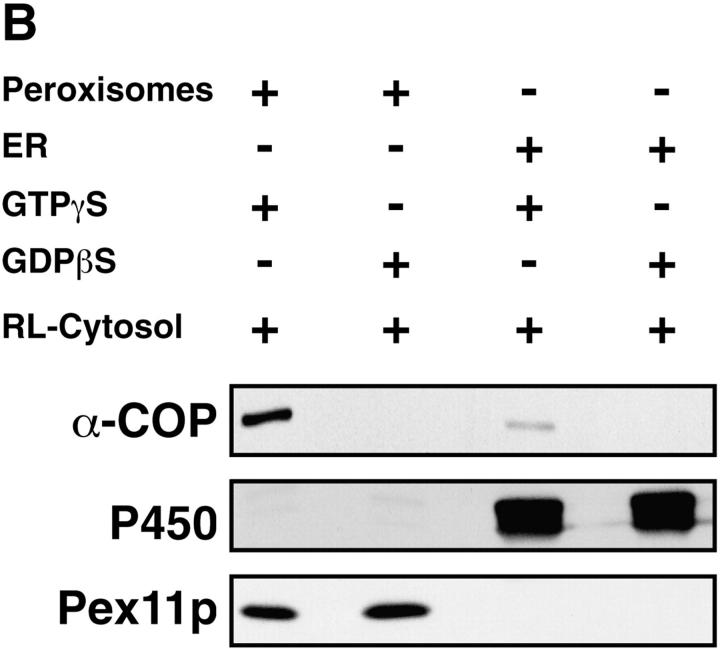
GTP-γS–dependent binding of coatomer and ARF to isolated peroxisomes was also achieved using cytosol from rat liver instead of bovine brain. Comparison of the efficiency of the two cytosol preparations revealed that peroxisomes bind eight times more coatomer (Fig. 5 A) and ARF (not shown) from rat liver cytosol than from an equal amount of bovine brain cytosol. That this recruitment of coatomer is specific for peroxisomes is further demonstrated by analyzing the coatomer-binding activities of isolated mitochondria and ER-derived vesicles. First, highly purified isolated rat liver mitochondria do not bind coatomer (Fig. 5 A). Second, based on the content of cytochrome P 450, our peroxisomal preparation contains ∼3% of contaminating ER-derived microsomes. Compared with the same amount of peroxisomes, however, microsomes bind coatomer six times less efficiently (Fig. 5 B). Thus, the microsomal contamination cannot account for the activity observed in peroxisomes.
Figure 5.
Comparison of the coatomer binding activity of isolated rat liver peroxisomes, mitochondria, and microsomes. Peroxisomes or mitochondria (250 μg) were incubated with bovine brain (BB) or rat liver (RL) cytosol (10 mg) in the presence of GTP-γS or GDP-βS (50 μM). Coatomer binding was analyzed in aliquots of the reisolated organelles separated on SDS-PAGE by Western blotting using anti–α-COP antiserum. Peroxisomes and mitochondria were detected by anti-Pex11p and anti-P30 (30-kD inner mitochondrial membrane proteins) antiserum, respectively. Densitometric evaluation of the α-COP signal revealed that peroxisomes bind eight times more α-COP from rat liver than from bovine brain cytosol. Note that isolated mitochondria do not bind coatomer. (B) Peroxisomes and microsomes (250 μg) were incubated with rat liver cytosol (10 mg) in the presence of GTP-γS and GDP-βS (50 μM) and aliquots of the organelles, recovered by flotation, were analyzed for coatomer binding as above. Densitometric evaluation revealed that microsomes, detected by a polyclonal anti-P450 antiserum, bind coatomer six times less efficiently than peroxisomes.
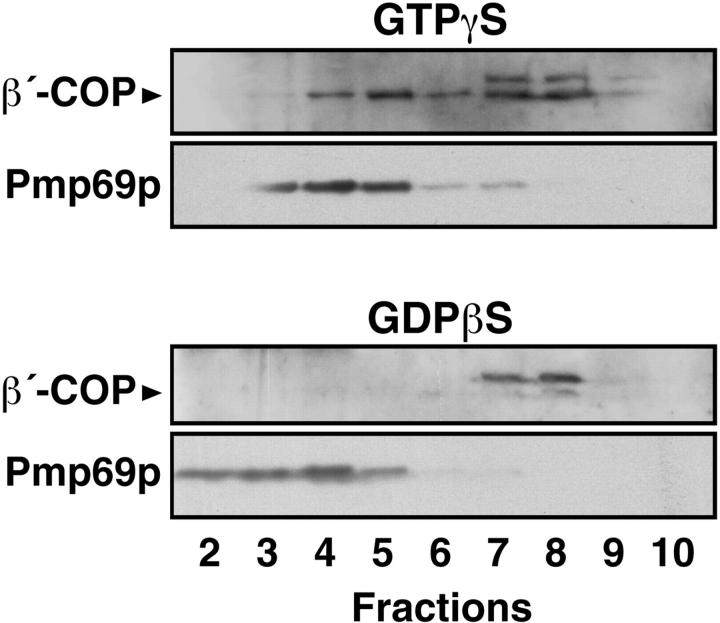
In vitro binding of coatomer to peroxisomes clearly raises the question as to the physiological significance of this observation. Does binding also occur in vivo? To answer this question, we used an SLO-permeabilized cell system in order to allow access of nucleotides and exogenous cytosolic components to the peroxisomal compartment. To this end, permeabilized primary rat hepatocytes were incubated with rat liver cytosol and GTP-γS or GDP-βS and the postnuclear supernate was prepared from the cell homogenate and subjected to Nycodenz density gradient centrifugation. Subsequently, the gradients were analyzed for the distribution of β′-COP and Pmp69p, as markers for coatomer and peroxisomes, respectively (Fig. 6). Whereas in the presence of GDP-βS no coatomer binding is observed, two peak fractions (4, 5 and 7, 8) exhibit binding in the presence of GTP-γS. In fractions 4 and 5 the two markers clearly colocalize suggesting peroxisomal coatomer binding. The second peak fraction of β′-COP represents Golgi-bound coatomer.
Figure 6.
GTP-γS–dependent colocalization of coatomer and peroxisomes in SLO-permeabilized hepatocytes after density gradient fractionation. Permeabilized hepatocytes were incubated with rat liver cytosol and GTP-γS or GDP-βS for 30 min at 37°C. After homogenization of the recovered cells the postnuclear supernate was fractionated on a linear Nycodenz density gradient. The distribution of coatomer and peroxisomes was visualized by Western blotting using rabbit anti–β′-COP and anti-Pmp69p peptide antisera, respectively. Note that the anti–β′-COP antiserum in the Golgi fractions (fractions 7 and 8) recognizes a cross-reaction product of molecular weight slightly higher than β′-COP.
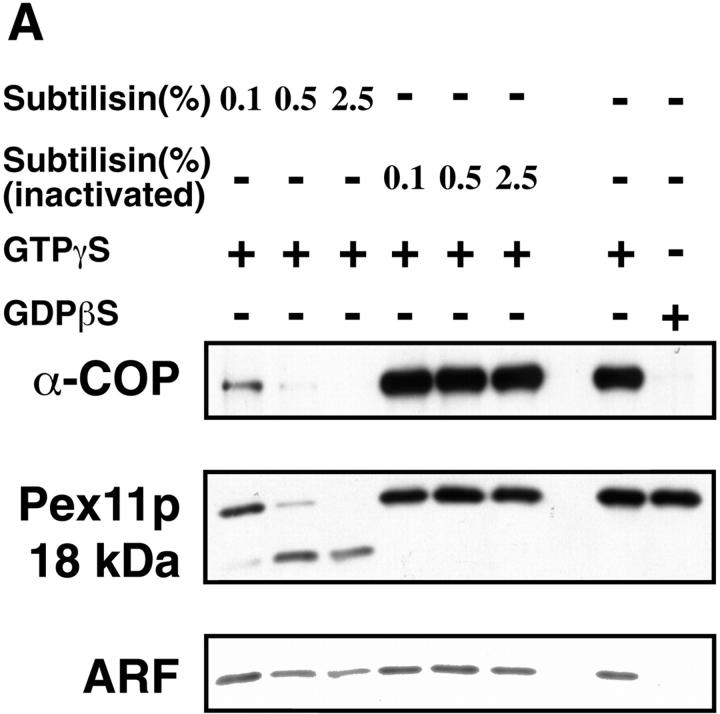
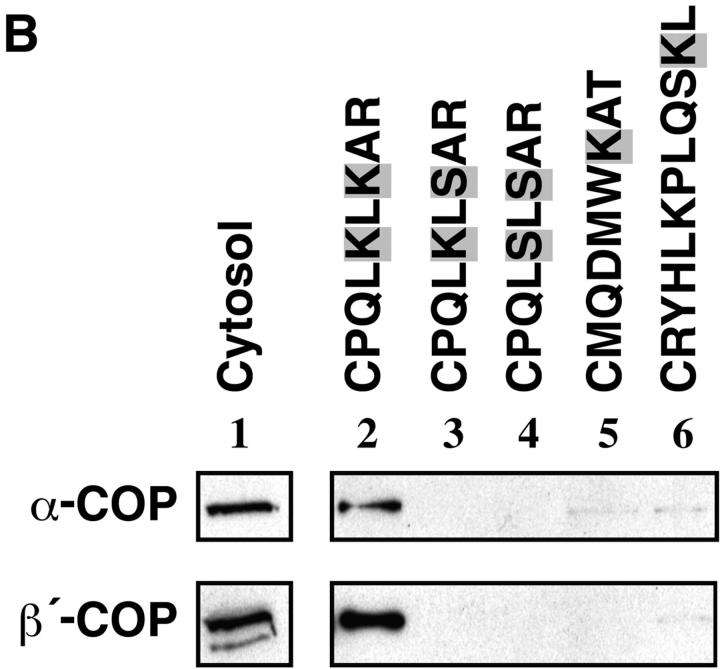
To obtain information about the nature of membrane factors involved in peroxisomal coatomer binding, peroxisomes were treated with various concentrations of subtilisin before incubation with rat liver cytosol. Whereas treatment with PMSF-inactivated subtilisin did not influence ARF and coatomer binding, protease treatment, which severely diminished the concentration of immunoreactive Pex11p, considerably reduced it (Fig. 7 A). These data indicate that at least one peroxisomal membrane factor present on the outer surface is involved in binding of ARF and coatomer. Since Pex11p was supposed to mediate coatomer binding, we studied the interaction of the peroxisomal KXKXX motif with coatomer by means of an immobilized synthetic COOH-terminal tail peptide of Pex11p (CPQLKLKAR). As shown in Fig. 7 B (lane 2), this peptide specifically recognizes and binds coatomer, as revealed by the presence of α- and β′-COP. Only faint amounts of COP I components were detected when the peptide was mutated by substituting lysine residues for serine (Fig. 7, lanes 3 and 4). As controls, the COOH-terminal octapeptide of ScPmp27p (CMQDMWKAT) that contains one lysine residue at position −3, and an undecapeptide representing the COOH-terminal tail of rat peroxisomal acyl-CoA oxidase carrying a PTS1 (CRYHLKPLQSKL) were unable to bind coatomer (Fig. 7, lanes 5 and 6). In lane 1, 10% of the cytosol present in a binding assay is analyzed. These results provide evidence that the COOH-terminal tail of Pex11p is directly involved in coatomer binding.
Figure 7.
Involvement of the dilysine motif of Pex11p in peroxisomal coatomer binding. (A) Binding of coatomer and ARF to peroxisomes (250 μg) pretreated with increasing concentrations of subtilisin, as indicated, was performed by incubations with rat liver cytosol (10 mg) in the presence of GTP-γS or GDP-βS (50 μM). Controls remained untreated or were treated with the same concentrations of subtilisin previously inactivated by 1 mM PMSF. Peroxisomes were identified by the polyclonal anti-Pex11p antiserum that recognizes both the uncleaved Pex11p as well as the 18-kD proteolytic fragment (see legend to Fig. 2), whereas coatomer and ARF binding was detected by anti– α-COP and anti-ARF 1 antiserum, respectively. Note that proteolytic degradation of Pex11p correlates with a decrease in binding of both α-COP and ARF. (B) Selective binding of coatomer to the COOH-terminal Pex11p tail peptide containing the dilysine motif. The synthetic peptide (lane 2) and two mutated versions of it (lanes 3 and 4) as well as the COOH-terminal octapeptide of ScPmp27p (lane 5) and an undecapeptide containing the nine COOH-terminal amino acid residues of rat peroxisomal acyl-CoA oxidase (lane 6) were coupled to thiopropyl–Sepharose via an NH2 terminally attached cysteine residue. After incubation with bovine brain cytosol and extensive washing of the Sepharose, bound coatomer was released by SDS and visualized by SDS-PAGE and immunoblotting using monospecific antisera directed against α- and β′-COP. An aliquot (50 μg) of cytosol used in these incubations was loaded onto the gel as a control (lane 1).
Specific localization of Pex11p to the peroxisomal membrane was also shown by postembedding immunoelectron microscopy of rat liver sections obtained from animals treated with peroxisomal proliferators (Fig. 8, A and B). Compared with untreated liver in which peroxisomes are only scarcely labeled, the labeling density increased remarkably after induction of the peroxisomal compartment. Whereas Pex11p in most peroxisomes exhibited a rather uniform membrane distribution, in some others it appeared to be concentrated in clusters (Fig. 8 B). Based on the idea of coatomer-mediated peroxisomal vesiculation, clustering of Pex11p might precede budding. We also analyzed in vitro peroxisomal coatomer binding by preembedding immunogold labeling using the mouse monoclonal antibody CM1A10 alone (Fig. 8, C–E) or in combination with an anti-Pmp69p peptide antiserum (Fig. 8 F). Whereas the peroxisomal marker, Pmp69p (decorated with 6-nm gold particles), uniformly labels the organellar membrane, coatomer (decorated with 12-nm gold particles) is detected on some peroxisomes in cap-like patches (Fig. 8 C) as well as on budding and vesiculated structures (Fig. 8, D–F). This characteristic pattern was predominantly observed when peroxisomes were incubated in the presence of cytosol and GTP-γS, and not when GDP-βS replaced GTP-γS.
Figure 8.
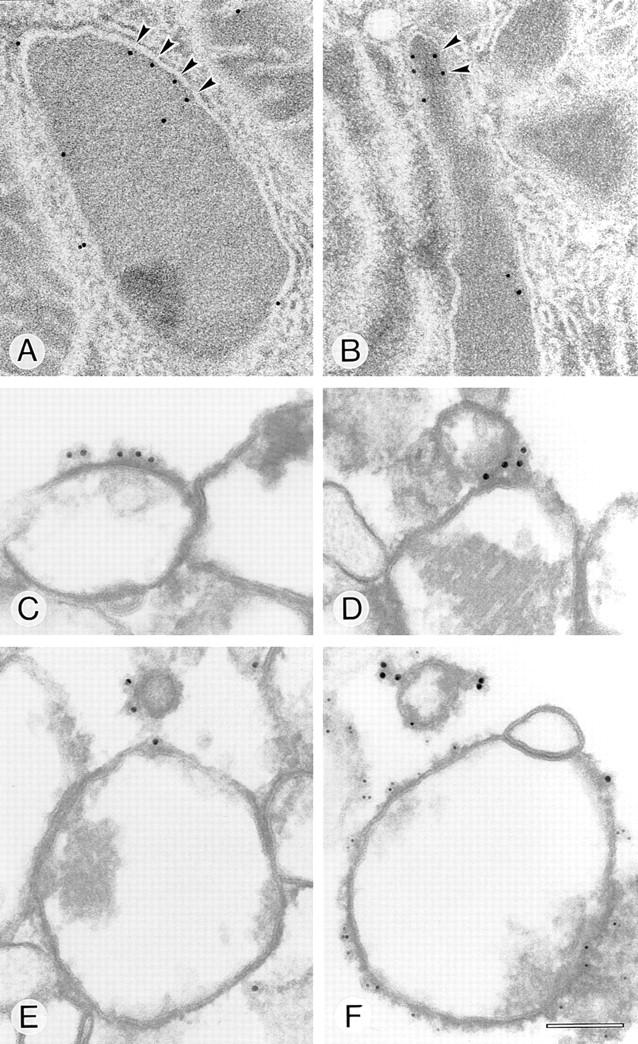
Peroxisomal localization of Pex11p (A and B) and coatomer binding (C–F) by immunogold electron microscopy (post- [A and B] and preembedding [C–F]). Clusters of Pex11p (arrowheads) are localized to the peroxisomal membrane in rat liver treated with clofibrate (A) or trifluoroacetate (B). Coatomer binding to clofibrate-induced isolated rat liver peroxisomes primed with bovine brain cytosol and GTP-γS was detected on peroxisomes, buds, and small vesicular structures. Note the nucleoid core in D and E that clearly distinguishes these organelles as peroxisomes. The mouse monoclonal antibody CM1A10 decorated with 12-nm gold-labeled second antibody was used to detect coatomer (C–F). A polyclonal anti-Pex11p tail peptide antiserum decorated with 14-nm protein A–gold (A and B) and anti-Pmp69p peptide antiserum decorated with 6-nm gold-labeled second antibody were used to detect the peroxisomal membrane (F). Bar, 100 nm.
Phenotypic Changes of Peroxisome Morphology by Overexpression of Pex11p or Functional Inactivation of Coatomer
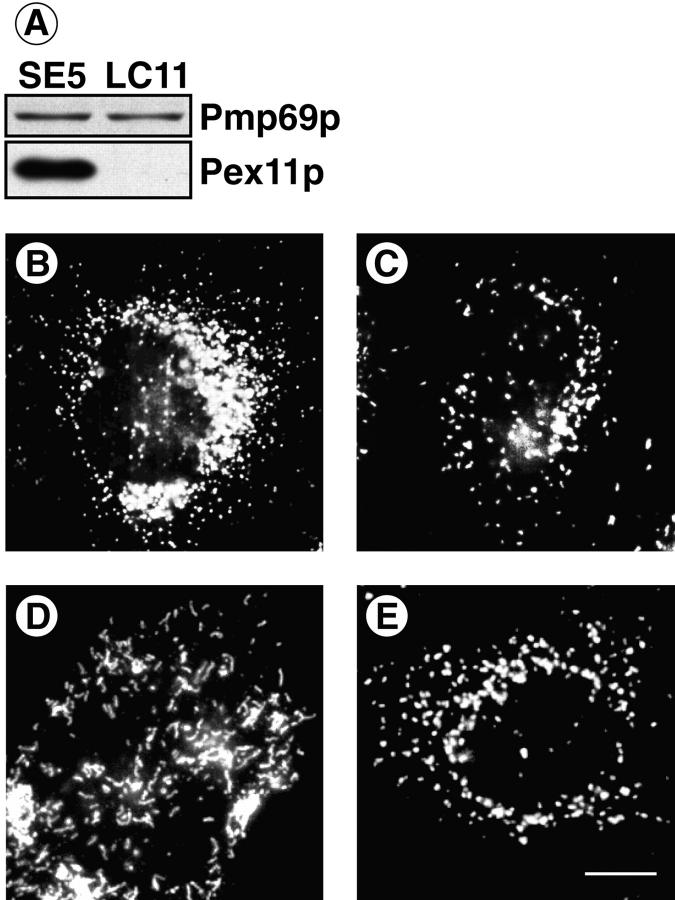
The presence of a KXKXX motif in Pex11p and the ability of the corresponding peptide to bind coatomer in vitro led us to assume an interaction of coatomer with Pex11p in vivo. Therefore, we transfected CHO cells with Pex11p-cDNA and isolated several stably transfected cell clones. Using immunological techniques, we then analyzed control cells transfected with vector containing no insert (clone LC11) and Pex11p-cDNA transfectants (clone SE5) for their content of Pex11p as well as their peroxisomal phenotype. Overexpression of Pex11p was demonstrated by immunoblotting, as shown in Fig. 9 A. However, the degree of overexpression was hardly determined with accuracy, since the amount of Pex11p in the control clone LC11 was extremely low. Comparative visualization of the peroxisomal compartment of LC11 and SE5 cells by immunofluorescence and electron microscopy showed an increase in number and a decrease in size of SE5 peroxisomes (Fig. 9, B and C). Quantitative evaluation revealed twice the number of peroxisomes in SE5 cells, demonstrating that proliferation had occurred.
Figure 9.
Phenotypic change in peroxisomal morphology by overexpression of Pex11p (A–C) as well as functional deficiency of coatomer (D, E). CHO cells were stably transfected with either Pex11p-cDNA cloned into pcDNA3 (SE5) or pcDNA3 lacking an insert (LC11). Carbonate membranes of postnuclear supernates (10 μg) were analyzed for their content of Pmp69p and Pex11p by SDS-PAGE and Western blotting (A). The peroxisomal compartment of SE5 (B) and LC11 (C) was visualized by immunofluorescence using the polyclonal anti-Pmp69p antiserum. ldlF cells expressing a ts mutant of ε-COP were kept for 24 h at nonpermissive (39.5°C, D) or permissive (34°C, E) temperature before immunofluorescence staining of peroxisomes using the anti-Pmp69p antiserum. Note numerical increase in small spherical peroxisomes (B) and clustering of tubular peroxisomes (D). Bar, 3 μm.
If coatomer is involved in peroxisomal vesiculation, as suggested by the phenotypic change of peroxisomes upon overexpression of Pex11p, a defect in coatomer function would be expected to block vesiculation, causing an enlargement of individual peroxisomes. The temperature-sensitive ldlF CHO mutant, recently established by Guo et al. (21, 22), contains a point mutation in ε-COP (Glu251 to Lys251) resulting in an instability of this coatomer subunit at the nonpermissive temperature of 39.5°C. As visualized by immunofluorescence, ldlF cells, exposed for 24 h to this temperature, exhibited a dramatic change in the morphology of the Golgi apparatus and peroxisomes. The Golgi apparatus disappeared (not shown) and dissociated into vesicular structures (see Fig. 10 B). Peroxisomes instead of being spherical and highly dispersed were heavily clustered (Fig. 9, D and E) and appeared as strikingly elongated tubules consistent with impaired peroxisomal vesiculation. The peroxisomal morphology of wild-type CHO cells kept at the nonpermissive temperature for the same time span was indistinguishable from normal controls (not shown).
Figure 10.
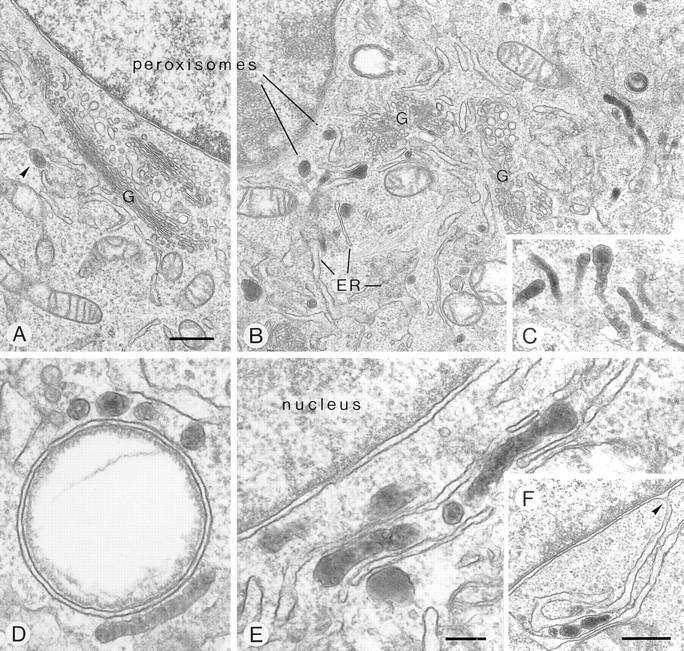
Fine structure of peroxisomes in the perinuclear region of mutant ldlF cells at permissive (34°C, A, arrowhead) and nonpermissive (39.5°C, B–F) temperature. Phenotypic peroxisome clustering and formation of peroxisome-ER aggregates is revealed, due to functional deficiency of ε-COP. At nonpermissive temperature, the intensely DAB-stained peroxisomes frequently exhibit a highly tortuous, tubular shape (B–E). They cluster at the periphery of lipid vacuoles (D) and form rows alternating with ER cisternae (E), the latter of which are often in continuity with the perinuclear membrane (F, arrowhead). G, Golgi complex. Bars: (A–C and F) 500 nm; (D and E) 150 nm.
Fine structural analysis largely confirmed the data obtained by immunofluorescence and in addition provided insight into details of the peroxisomal compartment in this mutant cell line. At permissive temperature, peroxisomes appear as small round or rod-shaped particles (∼150 nm in mean diameter) that are scattered throughout the cytoplasm, though a few are always present near the Golgi apparatus (Fig. 10 A, arrowhead). At nonpermissive temperature, peroxisomes are tubular and tortuous, frequently exceeding 1 μm in length (Fig. 10, B–E). They occur isolated or in clusters (Fig. 10, B and C), the majority intimately associated to smooth ER cisternae. Occasionally, they are arranged in parallel forming rows or stacks (Fig. 10, C–F) that are located at the periphery of lipid vacuoles (Fig. 10 D) or near the nucleus (Fig. 10, E and F). Continuities between the perinuclear cisternae and ER (Fig. 10 F) are frequently found developing into characteristic ER– peroxisome clusters that are also easily identified at the light microscopic level (Fig. 9 D).
Discussion
Membrane Topology and Intracellular Localization of Pex11p
We have characterized RnPex11p, an integral peroxisomal membrane protein with an NH2-terminal PTS2 and a COOH-terminal dilysine motif both oriented to the cytoplasm, and a putative lumenal mPTS sequence. At present it is not clear which signal is responsible for correct peroxisomal targeting of Pex11p. The PTS2 exactly matches the consensus sequence (67) and is located 15 amino acids distant from the NH2-terminal methionine, quite similar as in rat thiolase B (46). Its presence in a peroxisomal membrane protein is highly unusual and thus, its functional significance has to be proved. Our data on membrane integration and intracellular location of Pex11p indicate that it is directly inserted into the peroxisomal membrane because (a) sequence comparison with typical ER targeting signal peptides did not reveal an ER signal sequence in the Pex11p cDNA-derived structure (69), (b) the in vitro translation product of Pex11p on SDS gels has the same molecular mass as mature Pex11p, and (c) is not inserted into ER membranes neither co- nor posttranslationally (results not shown), but is inserted posttranslationally into peroxisomal membranes.
Specificity of Coatomer Binding to Peroxisomes
The following observations demonstrate specificity of coatomer recruitment to peroxisomes.
First, coatomer binding is strictly dependent on GTP-γS and does not occur with GDP-βS that is assumed to render ARF in its inactive state. This not only holds for isolated peroxisomes but also for peroxisomes in a permeabilized cell system. After binding, both coatomer and ARF cosedimented in a Nycodenz gradient with peroxisomes, suggesting their close physical association. In mammalian tissues at least six distinct ARFs have been identified so far that are highly conserved and function at distinct intracellular locations (27, 42, 45). Our observation that peroxisomes recruit more coatomer from liver cytosol than from brain cytosol might indicate at least one peroxisome-specific cytosolic factor, because liver contains far more peroxisomes than brain. Moreover, cytosol prepared from livers of rats treated with peroxisome proliferators that strongly increase the number of hepatic peroxisomes, is about twice as active in delivering coatomer to peroxisomes as normal rat liver cytosol (result not shown). The cytosolic factor might be a peroxisome-specific subtype of ARF that cross-reacts with the polyclonal antibody against ARF1 used in this study. This ARF subtype remains to be identified. However, at present we cannot exclude the possibility that additional GTP-binding proteins are involved that might be activated by GTP-γS.
Second, protease-pretreated peroxisomes have completely lost their coatomer binding capacity suggesting that binding is mediated by at least one peroxisomal membrane factor (Fig. 7 A), most likely the cytosolic COOH-terminal domain of Pex11p that in vitro binds coatomer efficiently (Fig. 7 B).
Third, mitochondria and ER-derived microsomal vesicles are unable to bind coatomer or bind it with much lower efficiency than peroxisomes, respectively (Fig. 5, A and B). Since the microsomal preparations used for these assays still contained Golgi membranes, the observed residual microsomal coatomer binding activity might well be due to the presence of this contamination.
Based on protein content Golgi membranes are about five times more efficient in recruiting coatomer from the cytosol than peroxisomes. This may be explained by different amounts of organelles used: whereas the protein to phospholipid ratio of highly purified Golgi fractions on a mg scale was analyzed to be 1.4, that of peroxisomes was 9 (Brügger, B., personal communication). If the amount of peroxisomes and Golgi used for the assays is based on phospholipid rather than protein content (which may reflect more accurately comparable amounts of membranes) then one equivalent of peroxisomal membrane binds approximately the same amount of coatomer as one equivalent of Golgi membrane.
Peroxisomal binding of coatomer does not seem to involve subspecies of the coat complex other than those that mediate biogenesis of Golgi-derived COP I–coated vesicles. The complete set of COPs is recruited to the peroxisomes, different from the binding of subcomplexes as Aniento et al. (1) have suggested on endosomal vesicles.
Peroxisome Biogenesis
There is general agreement that biological membranes cannot be synthesized de novo (47), but are derived from existing membranes. As mentioned, cells are equipped with various protein coats that mediate membrane budding and vesiculation (50, 56, 57, 60). The way peroxisomes are formed is being a matter of continuous discussion during the last 20 years. A variety of models have been forwarded, and most recently, the ER has again been invoked as a peroxisomal precursor organelle (4, 14). Our finding of an inducible membrane protein in peroxisomes that binds to coatomer, together with a GTP-γS–dependent recruitment to the peroxisomal membrane of ARF and the coatomer complex strongly indicates that coatomer is involved in peroxisomal biogenesis. Coatomer may either function in the formation of vesicles that would retrieve ER resident membrane proteins escaped from the ER (provided that the ER is a donor organelle, indeed), and/or in fission of peroxisomes to generate nucleating centers for the growth of new organelles. Consistent with this view are (a) the phenotypic changes of the peroxisomal compartment after overexpression of Pex11p or functional inactivation of coatomer, as described here, and (b) previous observations of a peroxisome population of lower equilibrium density but with protein import competence strikingly exceeding that of mature peroxisomes (26, 28, 31).
Acknowledgments
The authors thank Drs. B. Helms and W. Nickel (Biochemie-Zentrum, University of Heidelberg) for sharing reagents and for their stimulating discussions throughout the work. We also thank Dr. M. Eilers (ZMBH, University of Heidelberg) for samples of the anti-myc antibody and Dr. S. Bhakdi (University of Mainz) for the generous gift of SLO and are particularly grateful to Dr. M. Krieger (Massachusetts Institute of Technology, Cambridge) for providing us with the ldlF mutant CHO cell line and to Drs. Gottfried Schatz (Biozentrum Basel), Graham Warren (Imperial Cancer Research Fund, London), Mark Stamnes (Sloan Kettering Cancer Research Center, New York), and Mathew Hanah (Department Neurobiology, University of Heidelberg) for their critical comments on the manuscript. The expert technical assistance of Jutta Worsch, Ingrid Kuhn-Krause, and Ursula Wald is gratefully acknowledged.
Abbreviations used in this paper
- ARF
ADP-ribosylation factor
- COP
coat protein
- GDP-βS
guanosine 5′-O-(2-thiodiphosphate)
- GTP-γS
guanosine 5′-O-(3-thiotriphosphate)
- Pmp
peroxisomal membrane protein
- PTS
peroxisomal targeting signal
- PVDF
polyvinylidene difluoride
- RT-PCR
reverse transcriptase PCR
- SLO
streptolysin O
- ERGIC
ER Golgi intermediate compartment
Note Added in Proof
Higher eukaryotes contain a second constitutively expressed Pex11p distinct from the inducible one described by us (Gould, S.J., personal communication). In order to discriminate between these two proteins, we suggest to name the inducible protein Pex11pα and the constitutively expressed one Pex11pβ. We also would like to indicate a recent paper (Titorenko, V.I., and R.A. Rachubinski. 1998. Mutants of the yeast Yarrowia lipolyrica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol. Cell. Biol. In press).
Footnotes
The work was supported by the Deutsche Forschungsgemeinschaft (SFB 352).
References
- 1.Aniento F, Gu F, Parton RG, Gruenberg J. An endosomal β-COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol. 1996;133:29–41. doi: 10.1083/jcb.133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine B, Levrat F, Vallet V, Berbar T, Cartier N, Dubois N, Briand P, Kahn A. Gene expression in hepatocyte-like lines established by targeted carcinogenesis in transgenic mice. Exp Cell Res. 1992;200:175–185. doi: 10.1016/s0014-4827(05)80086-2. [DOI] [PubMed] [Google Scholar]
- 3.Aridor M, Bannykh SI, Rowe T, Balch WE. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. JCell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baerends RJ, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PT, Kiel JA, Cregg JM, van der Klei IJ, Veenhuis M. The Hansenula polymorphaPER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- 5.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–908. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 6.Bhakdi S, Bayley H, Valeva A, Walev I, Walker B, Kehoe M, Palmer M. Staphylococcal α-toxin, streptolysin-O, and Escherichia colihemolysin: prototypes of pore-forming bacterial cytolysins. Arch Microbiol. 1996;165:73–79. doi: 10.1007/s002030050300. [DOI] [PubMed] [Google Scholar]
- 7.Cosson P, Démollière C, Hennecke S, Duden R, Letourneur F. δ- and ζ-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO (Eur Mol Biol Organ) J. 1996;15:1792–1798. [PMC free article] [PubMed] [Google Scholar]
- 8.Cosson P, Letourneur F. Coatomer interaction with dilysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 9.Cosson P, Letourneur F. Coatomer (COPI)-coated vesicles: role in intracellular transport and protein sorting. Curr Opin Cell Biol. 1997;9:484–487. doi: 10.1016/s0955-0674(97)80023-3. [DOI] [PubMed] [Google Scholar]
- 10.Diestelkotter P, Just WW. In vitro insertion of the 22-kD peroxisomal membrane protein into isolated rat liver peroxisomes. J Cell Biol. 1993;123:1717–1725. doi: 10.1083/jcb.123.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW, et al. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duden R, Griffiths G, Frank R, Argos P, Kreis TE. β-COP, a 110 kd protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to β-adaptin. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- 13.Dyer JM, McNew JA, Goodman JM. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO (Eur Mol Biol Organ) J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erdmann R, Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiki Y, Rachubinski RA, Lazarow PB. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc Natl Acad Sci USA. 1984;81:7127–7131. doi: 10.1073/pnas.81.22.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galas MC, Helms JB, Vitale N, Thierse D, Aunis D, Bader MF. Regulated exocytosis in chromaffin cells: a potential role for a secretory granule associated ARF6 protein. J Biol Chem. 1997;272:2788–2793. doi: 10.1074/jbc.272.5.2788. [DOI] [PubMed] [Google Scholar]
- 18.Gerich B, Orci L, Tschochner H, Lottspeich F, Ravazzola M, Amherdt M, Wieland F, Harter C. Non-clathrin-coat protein α is a conserved subunit of coatomer and in Saccharomyces cerevisiaeis essential for growth. Proc Natl Acad Sci USA. 1995;92:3229–3233. doi: 10.1073/pnas.92.8.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman BM, Blobel G. Biogenesis of peroxisomes: intracellular site of synthesis of catalase and uricase. Proc Natl Acad Sci USA. 1978;75:5066–5070. doi: 10.1073/pnas.75.10.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorgas K. Serial section analysis of mouse hepatic peroxisomes. Anat Embryol Berl. 1985;172:21–32. doi: 10.1007/BF00318940. [DOI] [PubMed] [Google Scholar]
- 21.Guo Q, Penman M, Trigatti BL, Krieger M. A single point mutation in epsilon-COP results in temperature-sensitive, lethal defects in membrane transport in a Chinese hamster ovary cell mutant. J Biol Chem. 1996;271:11191–11196. doi: 10.1074/jbc.271.19.11191. [DOI] [PubMed] [Google Scholar]
- 22.Guo Q, Vasile E, Krieger M. Disruptions in Golgi structure and membrane traffic in a conditional lethal mammalian cell mutant are corrected by ε-COP. J Cell Biol. 1994;125:1213–1224. doi: 10.1083/jcb.125.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harter C, Pavel J, Coccia F, Draken E, Wegehingel S, Tschochner H, Wieland F. Nonclathrin coat protein gamma, a subunit of coatomer, binds to the cytoplasmic dilysine motif of membrane proteins of the early secretory pathway. Proc Natl Acad Sci USA. 1996;93:1902–1906. doi: 10.1073/pnas.93.5.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartl FU, Just WW. Integral membrane polypeptides of rat liver peroxisomes: topology and response to different metabolic states. Arch Biochem Biophys. 1987;255:109–119. doi: 10.1016/0003-9861(87)90300-6. [DOI] [PubMed] [Google Scholar]
- 25.Hartl FU, Just WW, Koster A, Schimassek H. Improved isolation and purification of rat liver peroxisomes by combined rate zonal and equilibrium density centrifugation. Arch Biochem Biophys. 1985;237:124–134. doi: 10.1016/0003-9861(85)90261-9. [DOI] [PubMed] [Google Scholar]
- 26.Heinemann P, Just WW. Peroxisomal protein import. In vivo evidence for a novel translocation competent compartment. FEBS Lett. 1992;300:179–182. doi: 10.1016/0014-5793(92)80191-i. [DOI] [PubMed] [Google Scholar]
- 27.Hosaka M, Toda K, Takatsu H, Torii S, Murakami K, Nakayama K. Structure and intracellular localization of mouse ADP- ribosylation factors type 1 to type 6 (ARF1-ARF6) J Biochem Tokyo. 1996;120:813–819. doi: 10.1093/oxfordjournals.jbchem.a021484. [DOI] [PubMed] [Google Scholar]
- 28.Just, W.W., M. Anton, P. Diestelkotter-Bachert, S. Rapp, U. Soto. 1995. Peroxisomal protein import. In Functions and Biogenesis of Peroxisomes in Relation to Human Disease. R.J.A. Wanders, R.B.H. Schutgens, H.F. Tabak, editors. North-Holland, Amsterdam. 107–124.
- 29.Just WW, Diestelkotter P. Protein insertion into the peroxisomal membrane. Ann NY Acad Sci. 1996;804:60–75. doi: 10.1111/j.1749-6632.1996.tb18608.x. [DOI] [PubMed] [Google Scholar]
- 30.Just WW, Hartl F-U, Schimassek H. Rat liver peroxisomes. I. New peroxisome population induced by thyroid hormones in the liver of male rats. Eur J Cell Biol. 1982;26:249–254. [PubMed] [Google Scholar]
- 31.Just WW, Soto U. Biogenesis of peroxisomes in mammals. Cell Biochem Funct. 1992;10:159–165. doi: 10.1002/cbf.290100305. [DOI] [PubMed] [Google Scholar]
- 32.Kaldi K, Diestelkotter P, Stenbeck G, Auerbach S, Jakle U, Magert HJ, Wieland FT, Just WW. Membrane topology of the 22 kDa integral peroxisomal membrane protein. FEBS Lett. 1993;315:217–222. doi: 10.1016/0014-5793(93)81167-x. [DOI] [PubMed] [Google Scholar]
- 33.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 34.Lamaze C, Schmid SL. The emergence of clathrin-independent pinocytic pathways. Curr Opin Cell Biol. 1995;7:573–580. doi: 10.1016/0955-0674(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 35.Lannert H, Bünning C, Jeckel D, Wieland FT. Lactosylceramide is synthesized in the lumen of the Golgi apparatus. FEBS Lett. 1994;342:91–96. doi: 10.1016/0014-5793(94)80591-1. [DOI] [PubMed] [Google Scholar]
- 36.Lazarow PB, de Duve C. The synthesis and turnover of rat liver peroxisomes. V. Intracellular pathway of catalase synthesis. J Cell Biol. 1973;59:507–524. doi: 10.1083/jcb.59.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 38.Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 39.Malhotra V, Serafini T, Orci L, Shepherd JC, Rothman JE. Purification of a novel class of coated vesicles mediating biosynthetic protein transport through the Golgi stack. Cell. 1989;58:329–336. doi: 10.1016/0092-8674(89)90847-7. [DOI] [PubMed] [Google Scholar]
- 40.Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM. Pmp27 promotes peroxisomal proliferation. J Cell Biol. 1995;129:345–355. doi: 10.1083/jcb.129.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monaghan, P., and D. Robertson. 1993. The postembedding techniques. In Immunocytochemistry. J.E. Beesley, editor. IRL Press, Oxford. 43–68.
- 42.Moss J, Vaughan M. Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson T, Jackson M, Peterson PA. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 44.Novikoff AB, Novikoff PM. Microperoxisomes and peroxisomes in relation to lipid metabolism. Ann NY Acad Sci. 1982;386:138–152. doi: 10.1111/j.1749-6632.1982.tb21412.x. [DOI] [PubMed] [Google Scholar]
- 45.Ostrom Liang, J., and S. Kornfeld. Comparative activity of ADP- ribosylation factor family members in the early steps of coated vesicle formation on rat liver Golgi membranes. J Biol Chem. 1997;272:4141–4148. doi: 10.1074/jbc.272.7.4141. [DOI] [PubMed] [Google Scholar]
- 46.Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- 47.Palade, G.E. 1983. Membrane biogenesis: an overview. Methods Enzymol. 96:xxix–iv. [DOI] [PubMed]
- 48.Palmer DJ, Helms JB, Beckers CJ, Orci L, Rothman JE. Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J Biol Chem. 1993;268:12083–12089. [PubMed] [Google Scholar]
- 49.Pause B, Diestelkotter P, Heid H, Just WW. Cytosolic factors mediate protein insertion into the peroxisomal membrane. FEBS Lett. 1997;414:95–98. doi: 10.1016/s0014-5793(97)00975-7. [DOI] [PubMed] [Google Scholar]
- 50.Pearse BM, Robinson MS. Clathrin, adaptors, and sorting. Annu Rev Cell Biol. 1990;6:151–171. doi: 10.1146/annurev.cb.06.110190.001055. [DOI] [PubMed] [Google Scholar]
- 51.Poole B, Higashi T, de Duve C. The synthesis and turnover of rat liver peroxisomes. 3. The size distribution of peroxisomes and the incorporation of new catalase. J Cell Biol. 1970;45:408–415. doi: 10.1083/jcb.45.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rachubinski RA, Fujiki Y, Mortensen RM, Lazarow PB. Acyl-CoA oxidase and hydratase-dehydrogenase, two enzymes of the peroxisomal β-oxidation system, are synthesized on free polysomes of clofibrate-treated rat liver. J Cell Biol. 1984;99:2241–2246. doi: 10.1083/jcb.99.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rapp S, Soto U, Just WW. Import of firefly luciferase into peroxisomes of permeabilized Chinese hamster ovary cells: a model system to study peroxisomal protein import in vitro. Exp Cell Res. 1993;205:59–65. doi: 10.1006/excr.1993.1058. [DOI] [PubMed] [Google Scholar]
- 54.Robbi M, Lazarow PB. Synthesis of catalase in two cell-free protein-synthesizing systems and in rat liver. Proc Natl Acad Sci USA. 1978;75:4344–4348. doi: 10.1073/pnas.75.9.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 57.Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 58.Sakai Y, Marshall PA, Saignaji A, Takabe K, Sakai H, Kato N, Goodman JM. The Candida boidinii peroxisomal membrane protein Pmp30 has a role in peroxisomal proliferation and is functionally homologous to Pmp27 from Saccharomyces cerevisiae. . J Bacteriol. 1995;177:6773–6781. doi: 10.1128/jb.177.23.6773-6781.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scales SJ, Pepperkok R, Kreis TE. Visualization of ER- to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- 60.Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 61.Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothman JE. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- 62.Simionescu N, Simionescu M. Galloylglucoses of low molecular weight as mordant in electron microscopy. II. The moiety and functional groups possibly involved in the mordanting effect. J Cell Biol. 1976;70:622–633. doi: 10.1083/jcb.70.3.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sohn K, Orci L, Ravazzola M, Amherdt M, Bremser M, Lottspeich F, Fiedler K, Helms JB, Wieland FT. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J Cell Biol. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soto U, Pepperkok R, Ansorge W, Just WW. Import of firefly luciferase into mammalian peroxisomes in vivo requires nucleoside triphosphates. Exp Cell Res. 1993;205:66–75. doi: 10.1006/excr.1993.1059. [DOI] [PubMed] [Google Scholar]
- 65.Stenbeck G, Harter C, Brecht A, Herrmann D, Lottspeich F, Wieland FT. β-COP, a novel subunit of coatomer with homology to the β-subunits of trimeric G proteins. EMBO (Eur Mol Biol Organ) J. 1992;12:2841–2845. doi: 10.1002/j.1460-2075.1993.tb05945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stenbeck G, Schreiner R, Herrmann D, Auerbach S, Lottspeich F, Rothman JE, Wieland FT. γ-COP, a coat subunit of non-clathrin-coated vesicles with homology to Sec21p. FEBS Lett. 1992;314:195–198. doi: 10.1016/0014-5793(92)80973-k. [DOI] [PubMed] [Google Scholar]
- 67.Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- 68.Tabas I, Kornfeld S. Purification and characterization of a rat liver Golgi alpha-mannosidase capable of processing asparagine-linked oligosaccharides. J Biol Chem. 1979;254:11655–11663. [PubMed] [Google Scholar]
- 69.von Heijne G. Signal sequences the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 70.Wattenberg BW, Rothman JE. Multiple cytosolic components promote intra-Golgi protein transport. Resolution of a protein acting at a late stage, before membrane fusion. J Biol Chem. 1986;261:2208–2213. [PubMed] [Google Scholar]