Abstract
CENP-B is a constitutive centromere DNA-binding protein that is conserved in a number of mammalian species and in yeast. Despite this conservation, earlier cytological and indirect experimental studies have provided conflicting evidence concerning the role of this protein in mitosis. The requirement of this protein in meiosis has also not previously been described. To resolve these uncertainties, we used targeted disruption of the Cenpb gene in mouse to study the functional significance of this protein in mitosis and meiosis. Male and female Cenpb null mice have normal body weights at birth and at weaning, but these subsequently lag behind those of the heterozygous and wild-type animals. The weight and sperm content of the testes of Cenpb null mice are also significantly decreased. Otherwise, the animals appear developmentally and reproductively normal. Cytogenetic fluorescence-activated cell sorting and histological analyses of somatic and germline tissues revealed no abnormality. These results indicate that Cenpb is not essential for mitosis or meiosis, although the observed weight reduction raises the possibility that Cenpb deficiency may subtly affect some aspects of centromere assembly and function, and result in reduced rate of cell cycle progression, efficiency of microtubule capture, and/or chromosome movement. A model for a functional redundancy of this protein is presented.
The protein components of the mammalian centromere can be broadly classified into two groups. Proteins from the first group are constitutively present on the centromere throughout the cell cycle, and include CENP-A, CENP-B, and CENP-C. The second group of proteins has been referred to as passenger proteins, since these proteins undergo complex relocations to other cellular organelles during the cell cycle, appearing on the centromere only during specific stages of the cycle (Brinkley et al., 1992; Earnshaw and Mackay, 1994). Examples of passenger proteins are INCENPs, MCAK, CENP-E, CENP-F, 3F3/2 antigens, and cytoplasmic dynein (reviewed by Earnshaw and Mackay, 1994; Pluta et al., 1995; Choo, 1997a ). The proposed biological roles for these passenger proteins have included centromere formation and maturation, motor movement of chromosomes, sister chromatid cohesion and release, modulation of spindle dynamics, nuclear organization, intercellular bridge structure and function, and cytokinesis (reviewed by Choo, 1997a).
Amongst the constitutive centromere proteins, CENP-A has been localized to the outer kinetochore domain, and is a member of a growing class of proteins referred to as histone H3-like proteins whose members also include the S. cerevisiae homologue of CENP-A, CSE4p (Sullivan et al., 1994; Wilson et al., 1994; Stoler et al., 1995). Since CENP-A is found in association with histone H4 and the other core histones in particles that copurify with nucleosome core particles (Palmer and Margolis, 1985; Palmer et al., 1987), the protein is thought to act as a histone H3 homologue, replacing one or both copies of histone H3 in a certain set of centromeric nucleosomes, and is thought to serve to differentiate the centromere from the rest of the chromosome at the most fundamental level of chromatin structure: the nucleosome (Sullivan et al., 1994). CENP-C is located at the inner kinetochore plate, and has been shown to have an essential although yet undetermined centromere function as seen from its association with the active, but not the inactive centromeres of human dicentric chromosomes (Earnshaw et al., 1989; Page et al., 1995; Sullivan and Schwartz, 1995), arrest of mitotic progression after microinjection of anti-CENP-C antibodies into cultured mammalian cells (Bernat et al., 1990; Tomkiel et al., 1994) or gene knockout (Fukagawa and Brown, 1997; Kalitsis et al., 1998), and the significant sequence homology it shares with Mif2, a protein involved in budding yeast chromosome segregation and believed to have a role in kinetochore function (Brown et al., 1993; Brown, 1995; Meluh and Koshland, 1995).
Human CENP-B is an 80-kD polypeptide that has been localized throughout the heterochromatin or central domain of the centromere (Earnshaw and Rothfield, 1985; Earnshaw et al., 1987; Cooke et al., 1990; Sullivan and Glass, 1991; Saitoh et al., 1992). The protein is encoded by an intronless gene present in a single copy within the genome (Sugimoto et al., 1993; Seki et al., 1994). The number of CENP-B protein molecules has been estimated to be ∼20,000 per diploid genome in HeLa cells (Cooke et al., 1990; Muro et al., 1992). On different human chromosomes, variable but generally detectable levels of the protein have been observed (Earnshaw et al., 1987). A notable exception is the Y chromosome, which has been shown to consistently lack this protein (Earnshaw et al., 1987). Through the recognition of a 17-bp PyTTCGTTGGAAPuCGGGA sequence known as the CENP-B box motif, CENP-B protein has been demonstrated to bind human centromeric α-satellite DNA directly (Masumoto et al., 1989; Muro et al., 1992; Pluta et al., 1992; Yoda et al., 1992).
Comparison of cloned human and mouse CENP-B gene sequences (Earnshaw et al., 1987; Sullivan and Glass, 1991) reveals a high degree of homology between the two species, with the coding regions showing an overall 96% sequence similarity and substantial stretches demonstrating 100% nucleotide identity between the two species (Sullivan and Glass, 1991). Of particular importance, both the NH2-terminal DNA-binding and COOH-terminal dimerization domains are totally conserved. Surprisingly, even the 5′ and 3′ untranslated sequences demonstrate an unusually high level (95% and 83%, respectively) of homology that is suggestive of possible posttranscriptional regulatory mechanisms (Mullner and Kühn, 1988; Caput et al., 1986). Like its human counterpart, the mouse gene is single-copy and intronless. Although the mouse genome does not contain recognizable α-satellite DNA, CENP-B binding occurs through the 17-bp consensus CENP-B box motif that is found in the mouse centromeric minor satellite DNA (Pietras et al., 1983; Rattner, 1991). In addition to mouse and humans, the CENP-B gene is conserved in hamster, African green monkey, great ape, tupaias, calf, Indian muntjac, and sheep (Sullivan and Glass, 1991; Haaf and Ward, 1995; Yoda et al., 1996; Bejarano and Valdivia, 1996; EMBL accession no. U35655). Significant homology is also found between CENP-B and two S. pombe centromere DNA-binding proteins Cbh+ and Abp1p, where cbh+ has been shown to be an essential gene (Lee et al., 1997), while abp1-deleted strains exhibit slower growth and a pronounced meiotic defect (Halverson et al., 1997). The CENP-B box motif has been found in the centromeric satellite DNA of species as diverse as primates, Mus musculus, Mus caroli, tree shrews, giant panda, gerbils, and ferrets (Pietras et al., 1983; Masumoto et al., 1989; Rattner, 1991; Muro et al., 1992; Pluta et al., 1992; Yoda et al., 1992; Haaf and Ward, 1995; Kipling et al., 1995; Kipling et al., 1994; Wu et al., 1990; Volobouev et al., 1995; Choo et al., 1991; Laursen et al., 1992; Haaf et al., 1995). Based on this observed conservation of CENP-B and its DNA-binding motif, it may be speculated that CENP-B is a functionally important component of the mammalian centromere.
Through its CENP-B box-binding and dimerization properties, the protein has the hallmark of a cross-linking protein that is involved in assembly of the large arrays of centromeric α-satellite or minor satellite DNA (Yoda et al., 1992; Muro et al., 1992). However, the absence of this protein on the Y chromosome in humans and mouse (Earnshaw et al., 1987), and on the centromeres of African green monkeys, which are known to be composed largely of α-satellite DNA containing little or no binding sites for CENP-B (Goldberg et al., 1996), suggests that this role may not be universal. In other studies, the protein has been shown to be present on both the active and inactive centromeres of mitotically stable pseudodicentric human chromosomes (Earnshaw et al., 1989; Page et al., 1995; Sullivan and Schwartz, 1995), suggesting that CENP-B binding does not immediately translate into centromere activity. Furthermore, an increasing number of stable human neocentromeric marker chromosomes (Voullaire et al., 1993; Ohashi et al., 1994; Choo, 1997b ; Depinet et al., 1997; du Sart et al., 1997) have now been described that are capable of normal mitotic division in the absence of CENP-B binding, indicating that CENP-B is nonessential for mitotic chromosome segregation, at least for these marker chromosomes. Earlier attempts at defining the role of CENP-B in mammals have yielded conflicting results. Microinjection of polyclonal anti-CENP-B antibodies into human and mouse cells resulted in disruption of centromere assembly during interphase, and led to inhibition of kinetochore morphogenesis and function in mitosis (Bernat et al., 1990; Simerly et al., 1990; Bernat et al., 1991). However, a different study has indicated that expression of truncated versions of CENP-B in HeLa cells does not lead to a mitotic or cell cycle arrest phenotype (Pluta et al., 1992). To date, the role of CENP-B in meiosis has not been investigated.
To better understand the role of CENP-B in centromere function, we used gene targeting in mouse embryonic stem cells to derive animals with a null mutation in the CENP-B gene. A major advantage of such mouse mutants is that it enables us to study CENP-B functions not only in mitosis, but also in meiosis. We report that CENP-B–deficient mice appear to be mitotically and meiotically normal, but develop lower body and testis weights. We discuss the implications of these results and propose a model in favor of a redundancy of CENP-B in centromere function.
Materials and Methods
Construction of Targeting Vectors
A hybridization probe spanning the coding region of Cenpb was prepared from genomic DNA of mouse embryonic stem (ES)1 cell line E14 by PCR using primers Bprot-1 (5′-GCGCAGATCTATGGGCCCCAAGCGGCGGCAGC-3′) and Bprot-3 (5′-TCAGAATTCAGCTTTGATGTCCAAGACCC-3′). Screening of mouse genomic phage libraries with this probe resulted in the identification of a positive clone (designated E1) from a 129/OLA library (gift of M. Kennedy) of E14 cells, and a second clone (designated D1) from a 129/SV library (Stratagene) of R1 mouse ES cells. An E1-derived fragment spanning nucleotides 920–2676 of mouse Cenpb gene (Sullivan and Glass, 1991, EMBL accession no. X55038) was ligated with a D1-derived fragment spanning 2676–5800 and cloned into a modified pSP72 vector (Promega Corp., Madison, WI). An oligonucleotide linker sequence designated D/TAA (5′-GTACCTAGGTATACTTTTAAACTGAC-3′) was inserted at position 1207, which is 72 amino acids downstream of the ATG start site of the 1.8 kb-coding sequence of Cenpb (Fig. 1 b). This linker introduced a DraI site, a frameshift mutation, and three stop codons in all three reading frames, of which TAA was in frame with Cenpb translation, disrupting not only the critical NH2-terminal 125–amino acid centromere DNA-binding domain (Yoda et al., 1992; Kitagawa at al., 1995), but also removing all remaining COOH-terminal regions including the dimerization domain (Yoda et al., 1992; Kitagawa et al., 1995). The IRES-neomycin (Mountford et al., 1994) and IRES-hygromycin (A. Smith, personal communication) markers were separately cloned into an AvrII site at position 3202 in the 3′ untranslated region before the polyadenylation signal to produce the targeting constructs IRES(neo) and IRES(hygro), respectively.
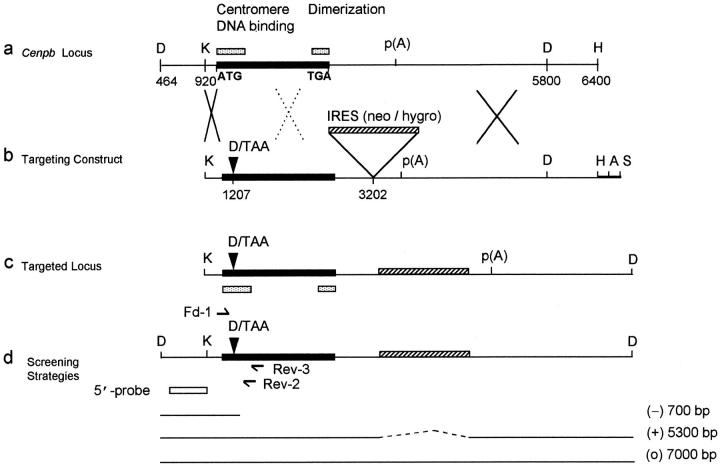
Figure 1.
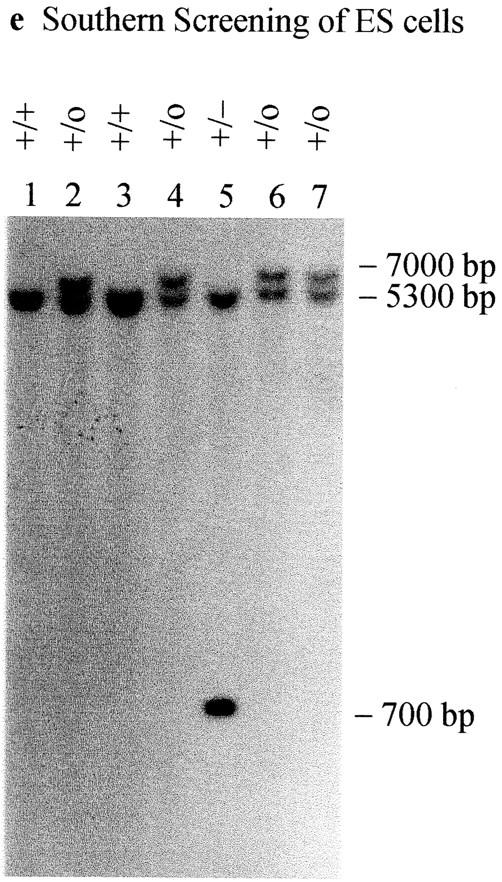
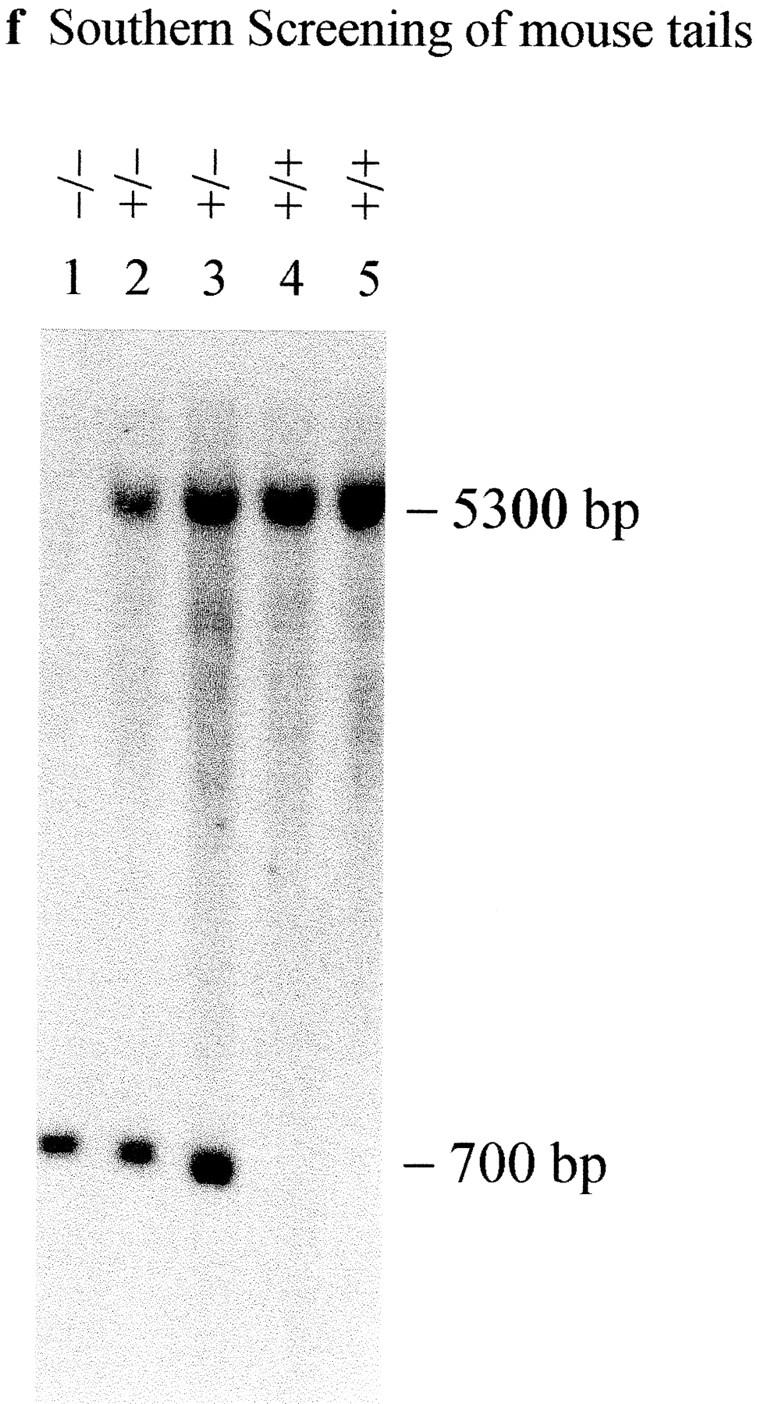
Cenpb targeting construct and screening strategies. (a) Intronless wild-type Cenpb locus showing the start and stop codons of the 1.8-kb coding region (solid box). The positions of the NH2-terminal 125–amino acid centromere DNA-binding domain and the COOH-terminal 59– amino acid dimerization domain are indicated (stippled boxes). Numbers below the line indicate nucleotide positions of the Cenpb sequence (EMBL accession no. X55038). (b) The IRES(neo) and IRES(hygro) targeting constructs, showing a 5480-bp contiguous KspI/HindIII Cenpb fragment that is interrupted by a frame shift-linker (D/TAA) containing stop codons and a new DraI site within the centromere DNA-binding domain, and an IRES(neo or hygro) cassette within the 3′ untranslated region. The H-A-S region on the right is vector DNA. Solid crosses denote the preferred regions of homologous recombination that result in the desired gene disruption event shown in c, whereas the broken cross indicates an undesired alternative region of recombination which, together with that at the solid-cross region to the right, results in incorporation of the IRES(neo or hygro) cassette, but not the D/TAA linker, generating a targeted event that is not accompanied by a gene disruption. p(A), polyadenylation signal; D, DraI; K, KspI; H, HindIII; A, AatII; S, SspI. (c) Correctly targeted Cenpb allele. (d) Screening strategies for targeted events. In Southern analysis of DraI digests, the 5′-probe (open box) detects a wild-type allele (+) of 5,300 bp, a desired targeted gene disruption allele (−) of 700 bp, and an undesired targeted but undisrupted allele (o) of 7,000 bp. Fd-1 and Rev-2/Rev-3 are PCR primers flanking the D/TAA linker region used for mouse tail DNA genotyping and RT-PCR. (e) Southern blot screening of transfected ES cells digested with DraI using the 5′-probe. Lane 5 shows a heterozygous colony with the desired gene disruption event. (f) Southern blot screening of tail DNA from mouse progeny of a +/− × +/− cross, digested with DraI and probed with the 5′-probe, showing the detection of homozygous Cenpb disruption (lane 1), heterozygous mice (lanes 2 and 3), and wild-type animals (lanes 4 and 5). (g) RT-PCR of total RNA from +/+ R1, +/− IRES(neo)-targeted R1-26, and −/− IRES(neo) and IRES(hygro) double-targeted R1-189N/H cells using the Fd-1/Rev-2 (lanes 1–3) and Fd-1/Rev-3 (lanes 4–7) primer sets. Incorporation of the D/TAA linker in the targeted allele increases the PCR products for both primer sets by 26 bp compared with the wild-type allele. Only the larger band containing the D/TAA linker was detected in the −/− cells (lanes 3 and 6). Lane 7, PCR control with no RNA.
Southern Blot and PCR Analyses of Targeting Events
Correct gene targeting in ES cells and mouse tail genomic DNA was determined by Southern analysis using a 5′ genomic probe generated from the E1 phage clone with NheI (position 564) and SacII (position 920) situated outside the targeting construct sequence (see Fig. 1 d). For PCR genotyping, the following primers flanking the D/TAA linker were used: Fd-1 (5′-ACCATCCTGAAGAGAACAACGG-3′) and Rev-2 (TGGAACCAAGCATGAGAGAAG), which gave 128-bp and 154-bp products for wild-type and targeted alleles, respectively; or Fd-1 and Rev-3 (3′-TGGAACCAAGCATGAGAAG-5′), which gave a 173-bp and 199-bp product for wild-type and targeted alleles, respectively (see Fig. 1, d and g). PCR conditions were as follows: 95°C for 30 s, 55°C for 1 min, and 72°C for 1.5 min for a total of 35 cycles using a 50-μl vol containing 50–200 ng genomic tail DNA, 1 U Taq polymerase, 200 μM dNTPs, and 300 ng of each primer in 1 X Taq PCR buffer (Perkin-Elmer Corp., Norwalk, CT).
Generation of Targeted ES Cells and Mouse Chimeras
For transfection, 50 μg of the IRES(neo) construct was linearized at the 3′ end with AatII or SspI, or at the 5′ end with SacII, and electroporated into approximately 108 ES cells in 800 μl vol using a single pulse from a Bio-Rad Gene Pulser at 800 V, 3 μFD, ∋⃒ Ω. The ES cell lines used in this study were R1 (Nagy et al., 1993), W9.5, and W9.8 (Buzin et al., 1994). Transfected cells were plated onto mitomycin C-treated, neomycin-resistant, STO-neoR (Robertson, 1987) plus 103U/ml LIF (Amrad-Pharamacia) and selected in G418 (Gibco-BRL) active at 300 μg/ml. One R1-derived G418-resistant colony, designated R1–26, demonstrated correct targeted disruption at the Cenpb allele, and was used for blastocyst injection to produce germline chimeric mice and for a second round of gene targeting to produce double-targeted, Cenpb-null cell lines in culture.
For chimeric mouse production, R1–26 cells were microinjected into host (C57 bla/6) blastocysts, followed by breeding of the resulting germ-line transmitting chimeras to generate heterozygous and homozygous Cenpb-null mice. For the second targeting event, the R1–26 cells were electroporated with 50 μg of the IRES(hygro) construct that has been linearized at the 3′ end with SspI. Transfected cells were grown in the absence of STO fibroblast feeder layer, and were selected in 300 μg/ml G418 and 110–140 μg/ml hygromycin. This resulted in a Cenpb-null cell line, designated R1–189N/H, in which both the Cenpb alleles were disrupted. This cell line was injected into C57 bla/6 blastocysts, and the resulting germline chimeras were used in a back-cross with C57 black mice to allow segregation of the two targeted alleles and the derivation of heterozygous and homozygous mouse strains carrying only the IRES(hygro)-targeted allele. In this way, mice with two independently targeted Cenpb alleles were generated.
Reverse Transcription (RT)-PCR Analysis and Sequencing
Total genomic RNA was extracted from wild-type R1 cells (+/+), the IRES(neo)-targeted cell line R1–26 (+/−), and the double-targeted cell line R1-189N/H (−/−). 0.5–1.0 μg total RNA was reverse-transcribed using a first-strand RT kit (Boehringer Mannheim Corp., Indianapolis, IN) and oligo-d(T) as primer. Annealing was carried out at room temperature for 10 min, followed by transcription at 42°C for 60 min and cooling at 4°C for 5 min. PCR was then performed on +/+, +/−, and −/− cell lines using conditions described for primers Fd-1 and Rev-2/Rev-3. For sequencing, the 199-bp Fd-1/Rev-3 PCR product from the −/− cell line was gel-isolated and cloned into the pGEM-T vector (Promega Corp., Madison, WI). Sequencing was performed on both strands in two separate clones using M13 Rev and T7 primers. Reactions were carried out using fluorescent dye terminator cycle sequencing (ABI PRISM™; Perkin-Elmer Corp., Norwalk, CT).
Immunohistochemistry
Autoimmune serum CREST no. 6 (gift of S. Wittingham and T. Kaye) was from a patient with calcinosis, Raynaud's phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasia (Fritzler et al., 1980; Moroi et al., 1981; Brenner et al., 1981) and detects CENP-A and CENP-B (du Sart et al., 1997). Anti-CENP-B monoclonal antibody 2D-7 (Earnshaw et al., 1987) was purchased as hybridoma cells from American Type Culture Collection (Rockville, MD) and prepared as ascites fluid in pristane primed mice. Anti-Cenpc polyclonal antibody, Am-C1, was produced in a rabbit against a mouse Cenpc/GST-fusion product expressed in Escherichia coli (du Sart et al., 1997). Antihuman CENP-E antibody, HX1, was a gift from T. Yen (Yen et al., 1991; Yen et al., 1992). Cultured cells were arrested in mitosis with 10 μg/ml colcemid for 2 h. Immunofluorescence staining was performed as previously described (Jeppeson et al.,1992; du Sart et al., 1997). After the antibody binding, the cells were postfixed in 10% formalin, washed, and counterstained with DAPI and DABCO mountant. Images were analyzed using an Axioskop fluorescence microscope equipped with a 100× objective (Carl Zeiss, Inc., Thornwood, NY) and a cooled CCD camera (Photometrics Image Point) linked to a PowerMac computer.
Histology, Advanced Sperm Count (ASC), and Stereology
The organs analyzed for histology were dissected from 10-wk-old and 6 mo-old mice using standard techniques. Testicular determination of homogenization-resistant ASC was performed as previously described (Robb et al., 1978) on 10-wk-old animals. Stereological analysis was performed on testicular materials from 10-wk-old mice using the optical dissector (sic) technique (Wreford et al., 1995) to investigate the efficiency of meiosis by determining the ratio of pachytene spermatocytes to round spermatids associated with stages I–VIII of spermatogenesis. This ratio has an expected value of 4:1 if the efficiency of division through meiosis I and II is 100% and there is no loss of round spermatids after meiosis.
Cytogenetics and Flow Sorting
For karyotyping, −/− R1-189 N/H cell line as well as spleen and bone marrow cell cultures (with and without phytohaemagglutinin) were isolated from −/− mice and compared with the +/+ cell line and +/+ animal. Cells were treated with colcemid and GTL-stained using standard cytogenetic techniques. For flow sorting, 50,000 cells from spleen, bone marrow, or testis were isolated from 30-wk-old mice. These were analyzed by two-parameter analysis of DNA content vs. cell diameter using a FACScan™ (Becton Dickinson & Co., Sparks, MD) cell sorter equipped with an argon laser at 488 nm. Signals were collected by a FL2 detector with a 585-nm band pass filter. Testis cells were profiled into five main regions corresponding to the different steps in maturation from elongated and round haploid spermatids to diploid, S-phase, and tetraploid cells.
Results
Generation of Cenpb-null ES Cells and Mice
For disruption of the mouse Cenpb gene in ES cells, a promoterless targeting vector was constructed that incorporated a translation frame-shift linker, designated D/TAA, containing stop codons in all three reading frames, and the IRES-neomycin or IRES-hygromycin marker (Fig. 1 b and Methods). The D/TAA linker inserted 72 amino acids downstream of the translation start site not only disrupted the critical NH2-terminal 125–amino acid centromere DNA-binding domain (Kitagawa et al., 1995; Yoda et al., 1992), but also removed all remaining COOH-terminal regions including the dimerization domain (Kitagawa et al., 1995; Yoda et al., 1992). The IRES-neo and IRES-hygro selectable markers were placed in the 3′ untranslated region before the polyadenylation signal. For correct targeting and gene disruption, two homologous recombination events external to the linker and IRES(neo/hygro) regions were required (Fig. 1 b, solid-cross regions). When the IRES(neo) construct was linearized at the 3′ end with AatII or SspI and transfected into R1 and W9.5 cells, 2% (or 2 out of 103 neomycin resistant colonies) of R1 cells, 1.3% (1 out of 72 colonies) of W9.8, and 3.5% (12 out of 344 colonies) of W9.5 cells gave the desired targeted gene disruption (Fig. 1 e, lane 5). Interestingly, a significantly higher frequency (90% for R1, 67% for W9.8, and 92% for W9.5) of an undesired targeting event was detected (Fig. 1 e, lanes 2, 4, 6, and 7) where incorporation of the IRES-neo element at the Cenpb locus had not been accompanied by the D/TAA linker. This result was due to recombinations occurring within the region between the IRES-neo cassette and the D/TAA linker (Fig. 1 b, broken-cross region) instead of in the region 5′ of the linker. The observation of a higher recombination frequency in this region was perhaps not surprising in view of the fact that 1995 bp of homologous DNA was present in this region compared with only 287 bp of homologous DNA between the D/TAA linker and the 5′ end of the construct. In subsequent experiments, it was further demonstrated that use of construct DNA linearized at the 5′ end using KspI to expose the Cenpb DNA end, as distinct from the plasmid vector DNA end using 3′ AatII or SspI, gave a 2.5-fold increase in the frequency of the desired targeting in R1 cells and a 3.8-fold increase in W9.5 cells (data not shown).
From the above screening, 21 heterozygous ES cell colonies with a disrupted Cenpb allele were obtained from the R1, W9.5, and W9.8 cell lines. Two of these colonies, R1-26 from the R1 line and W-190 from the W9.8 cell line, were retransfected with the IRES(hygro) construct to obtain a Cenpb null cell line. Selection of the transfected cells in neomycin and hygromycin gave rise to one double-targeted colony (out of five resistant colonies screened) designated R1-189N/H from R1-26, and two double-targeted colonies (out of 76 colonies screened) from W-190. All three colonies showed normal cell morphology and apparently normal growth rates. No desired double-targeted event (0 out of 81 colonies) were seen for both W9.8 and R1 when the transfected cells were selected in hygromycin alone, due presumably to a direct replacement of the IRES(neo)-targeted allele with the IRES(hygro) cassette. Furthermore, as with the IRES(neo) construct, a much higher frequency (three out of five colonies for R1-26, and 57 out of 76 colonies for W-190) of the undesired targeting event involving the loss of the D/TAA linker was observed.
The heterozygous R1-26 cell line was injected into C57 bla/6 blastocysts to produce germline chimeras, from which heterozygous (+/− neo) and homozygous (−/− neo) mice carrying the IRES(neo)-targeted allele were produced (Fig. 1 f). The double-targeted R1-189N/H cell line was similarly injected into C57 bla/6 blastocysts and, through selective breeding, heterozygous (+/− hygro) and homozygous (−/− hygro) mice carrying the IRES (hygro) allele were generated (data not shown). These mice, together with the various cell lines created above, were subjected to further detailed studies.
Abolition of Cenpb Gene Expression in the Targeted Cell Lines
Cenpb gene disruption was determined by RNA analysis using PCR performed with primers designed across the D/TAA linker region. The results (Fig. 1 g) indicated the presence of normal Cenpb transcripts in the wild-type (Fig. 1 g; lanes 1 and 4) and heterozygous cell lines (lanes 2 and 5), but not in the double-targeted R1-189N/H cell line (lanes 3 and 6). This result suggested that transcription of both copies of the wild-type Cenpb alleles in the −/− cell line had been abolished and replaced by that of the targeted alleles. In addition, we wished to determine whether the D/TAA linker had incorporated correctly into the NH2-terminal centromere DNA-binding domain of the targeted Cenpb gene, and that no unforeseen sequence rearrangement undetected by the Southern or RT-PCR analyses had occurred. This was done by purifying and cloning the 199-bp fragment corresponding to the targeted allele (Fig. 1 g, lane 6) and direct sequencing analysis. The results (not shown) confirmed the correct insertion of the D/TAA linker and therefore the stop codons.
Absence of Cenpb Binding on Centromeres by Direct Immunofluorescence Staining
Immunofluorescence staining of metaphase chromosomes was used to detect specific centromere-binding proteins. Fig. 2 shows results obtained with the +/+ R1 and −/− R1-189N/H cell lines; the results for the +/− R1-26 cell line were similar to those of the +/+ R1 cell line, and are not shown. The anti-Cenpb monoclonal antibody clearly demonstrated the presence of Cenpb on the centromeres of the +/+ cell line, but not on those of the −/− cell line. The intensity of the Cenpb signals in the +/+ cells varied considerably on different chromosomes, reflecting the intrinsic quantitative variation in the amount of Cenpb boxes and Cenpb binding on different centromeres (Earnshaw et al., 1987). When these cell lines were tested with a CREST antibody and antibodies for Cenpc and CENP-E, uniform staining of the centromere was observed (Fig. 2). Similar results (not shown) were obtained with cells established from the +/+, +/−, and −/− IRES(neo) and IRES(hygro) mice. These data therefore provided direct evidence that the Cenpb gene has been disrupted in the −/− neo and −/− hygro knockout mice. They also demonstrated that Cenpb is not essential for centromeric binding of Cenpc and Cenpe and for CREST antibody binding on active centromeres.
Figure 2.
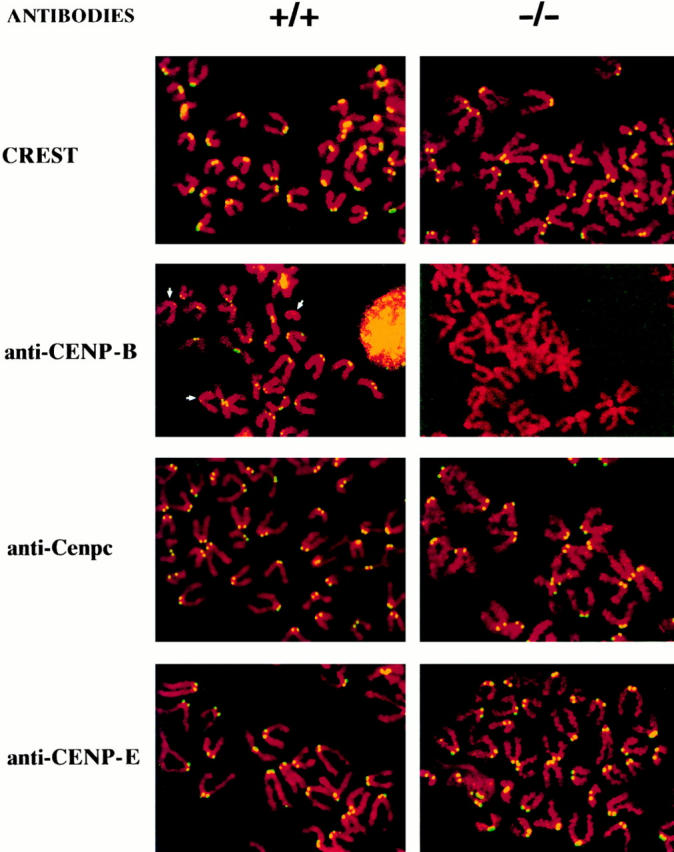
Immunostaining of centromere proteins (yellow signals) in +/+ R1 and −/− R1-189N/H cell lines using anticentromere antibodies. Results for the +/− R1-26 cell line were similar to those for the +/+ cell line and are not shown. Uniform signals were observed in all the centromeres in both cell lines when stained with CREST, anti-Cenpc, and anti-CENP-E antibodies. Note differences in the intensity of anti-CENP-B staining on different chromosomes in the +/+ cell line, with some centromeres (arrows) showing little or no detectable signals. No Cenpb signal was seen on the centromeres of the −/− cell line, even after maximal enhancement of fluorescence signal (thus the paler background) using computer imaging facility.
Cenpb Null Mice Have Lower Body Weight and Testis Size, but Are Otherwise Developmentally Normal
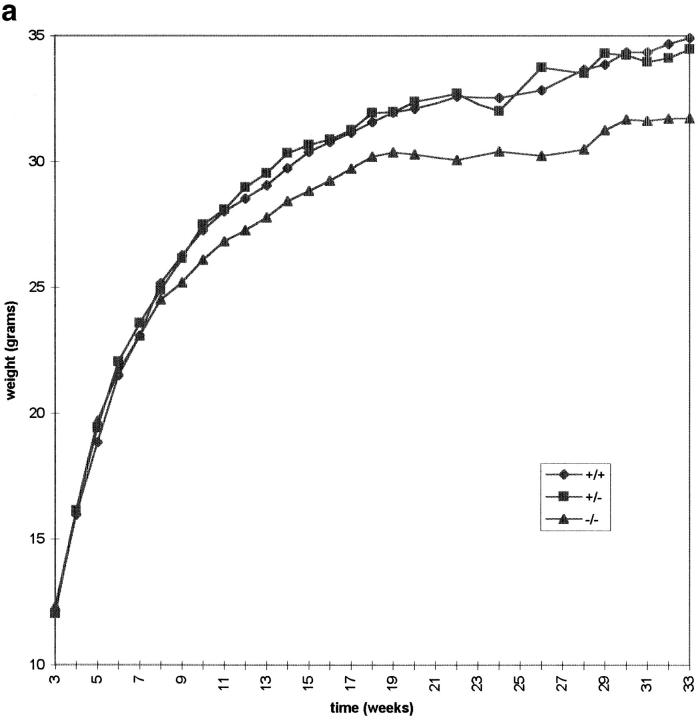
Cenpb null mice appeared phenotypically normal, and routinely gave normal litter size and the expected Mendelian ratios of offspring, suggesting that Cenpb deficiency did not drastically affect cell division, development, and reproduction of the mice. This phenotype is in stark contrast to that of another mouse model we have recently created with a disruption of Cenpc, where null mutants display severe mitotic disarray and die during early embryogenesis (Kalitsis et al., 1998). To determine if Cenpb gene disruption has a more subtle effect on growth, we measured the body weight of the IRES(neo) animals over an 8-mo period (Fig. 3). The −/− mice as a group appeared uniform in size at birth, and presented with normal weights at weaning (3 wk), but subsequent weight gain in this group lagged behind those of sex-matched +/+ and +/− animals, with the difference reaching a level of significance (P < 0.05) after 22 wk in males and 12 wk in females (see Table I for representative weight data for 26-wk-old animals). When the weight data were collected from the IRES(hygro) animals, similar trends as those obtained for the IRES(neo) mice were seen for the different genotypes in the two sexes (data not shown).
Figure 3.
Total body weight of male (a) and female (b) IRES(neo) mice. The male data were collected from an average of 33, 46, and 19 animals, whereas the female data were from an average of 24, 39, and 17 animals for each time point for the +/+, +/−, and −/− genotypes, respectively. Individual weight value, measured weekly or fortnightly, represents the means of the weights for the total number of animals used at that time point.
Table I.
Total Body Weight of IRES(neo) Mice at 26 wk.(b) Total Testis Weight of IRES(neo) Mice at 10 wk
| No. | Weight ± SD | t test | ||||
|---|---|---|---|---|---|---|
| g | ||||||
| (a) Male (body) | ||||||
| +/+ | 34 | 32.86 ± 3.58 | +/+ vs. +/− (P = 0.2469) | |||
| +/− | 54 | 33.75 ± 3.45 | +/− vs. −/− (P = 0.0002) | |||
| −/− | 19 | 30.25 ± 3.29 | +/+ vs. −/− (P = 0.0114) | |||
| Female (body) | ||||||
| +/+ | 22 | 27.36 ± 3.79 | +/+ vs. +/− (P = 0.7786) | |||
| +/− | 37 | 27.62 ± 3.16 | +/− vs. −/− (P = 0.0005) | |||
| −/− | 17 | 24.45 ± 2.06 | +/+ vs. −/− (P = 0.0079) | |||
| (b) Male (testis) | ||||||
| +/+ | 15 | 0.208 ± 0.029 | +/+ vs. +/− (P = 0.27743) | |||
| +/− | 18 | 0.195 ± 0.036 | +/− vs. −/− (P = 0.00255) | |||
| −/− | 16 | 0.154 ± 0.038 | +/+ vs. −/− (P = 0.00012) |
Histological Analysis Reveals No Gross Abnormality
To investigate the reasons for the observed weight difference, various organs from the IRES(neo)-targeted animals were subjected to histological examination. The organs analyzed were stomach, duodenum, descending colon, liver, hairy skin, ear flap, salivary gland, spleen, pancreas, kidney, thymus, brain, lung, adrenal, seminal vesicle, ovary, uterus, and pituitary. When organs from 10-wk- and 6-mo-old male and female −/− mice were directly compared with those derived from age- and sex-matched +/+ and +/− animals, the results indicated no obvious abnormality in any of these organs. During this analysis, the testes of −/− mice were found to be markedly smaller (29%; P < 0.01) than those of the wild-type mice (Table I). Follicle-stimulating hormone (FSH) and luteinizing hormone (LH) levels were measured and found to be normal. When the testes of 10-wk-old animals were assessed for sperm content (ASC), a 39.5% reduction (P = 0.0007) was seen in the −/− animals (N = 9) compared with the +/+ animals (N = 12). When the efficiency of meiosis was determined using the more comprehensive stereological analysis on sectioned testicular materials from +/+ (N = 5), +/− (N = 6), and −/− (N = 4) mice, values of 3.9 ± 0.2 (mean ± SEM), 3.8 ± 0.1, and 3.6 ± 0.2, respectively, were obtained, which were not significantly different from the expected 4:1 ratio. Thus, although it appears that reduction in germ cell content is correlated with testicular weight reduction, results of the stereological analysis have revealed no substantial difference in the efficiency of either mitotic or meiotic division.
Karyotyping and Flow Sorting of Cenpb−/− Cells Indicate Normal Meiosis and Mitosis
The chromosomes of the Cenpb-disrupted cells were investigated by cytogenetic analysis and flow sorting. For cytogenetic analysis, ES-derived R1-189 N/H −/− cells in culture and spleen and bone marrow cells from 30-wk-old −/− mice (N = 4) were analyzed and compared with the wild-type ES cells and animal. The results indicated a normal karyotype in each case. For flow sorting, 50,000 cells from the spleen, bone marrow, or mitotically and meiotically dividing testis cells were isolated from +/+ (N = 8), +/− (N = 13), and −/− (N = 7) 10-wk-old mice, and +/+ (N = 3), +/− (N = 1), and −/− (N = 8) 30-wk-old mice. Again, no detectable aberration was observed. These results, together with those obtained using the stereological techniques, provide further evidence that Cenpb is not essential for mitosis or meiosis.
Discussion
The question of whether CENP-B is essential for chromosome segregation has been intensely debated in recent years. Conservation of the protein in different mammalian species and in lower eukaryotes and its demonstrated centromere DNA-binding property attest to a significant functional role. However, various cytological observations have hinted at CENP-B not being critical for mitosis, although such evidence are often indirect and open to interpretation. For example, detection of CENP-B on both the active and inactive centromeres of mitotically stable pseudodicentric human chromosomes (Earnshaw et al., 1989; Page et al., 1995; Sullivan and Schwartz, 1995), rather than indicating a lack of functional importance for CENP-B, can be interpreted to mean that additional centromere proteins are necessary to make the inactive centromere fully active. Similarly, the finding that the protein is absent on the Y chromosome in both humans and mouse (Earnshaw et al., 1987) is intriguing but needs to be interpreted in light of the unresolved peculiarity that this observed absence is associated with the only centromere in these genomes that does not undergo sister centromere pairing in meiosis. The absence of CENP-B on various analphoid neocentromeres (Choo, 1997b ) also does not per se exclude functions for CENP-B (or α-satellite DNA) on normal centromeres since these neocentromeres may have gained centromere function through some epigenetic modifications (Karpen and Allshire, 1997). Finally, the observation that the α-satellite DNA-containing centromeres of African green monkey lacks binding sites for CENP-B (Goldberg et al., 1996) could be because monkeys have evolved a different and functionally equally important way to compensate for their CENP-B deficiency.
In addition to the uncertainty on mitotic functions, the requirement of CENP-B in meiosis has also not previously been investigated. Such an investigation is especially important in light of recent evidence indicating a specific role of centromeric heterochromatin in meiotic chromosome segregation in Drosophila (Dernburg et al., 1996; Karpen et al., 1996), and in view of the fact that CENP-B binds directly to centromeric heterochromatic DNA and is thought to be involved in the higher order organization of this DNA (Yoda et al., 1992; Muro et al., 1992). The production and characterization of Cenpb null mice allows several conclusions regarding the functional significance of this protein in mitosis and meiosis to be drawn. The apparently normal growth and reproductive characteristics of these mice indicate that Cenpb is not essential for either of these cell division processes. This result is confirmed by direct cytogenetic and FACS analyses of chromosomes, which have not detected any karyotypic abnormality in the −/− mice. A closer look at the different stages of male meiosis in these animals has similarly not revealed any obvious defect. The protein also appears not to be required for the structural integrity of the centromere–kinetochore complex since the centromeres of Cenpb-deficient cells continue to show clear association with at least two of the functionally important centromere proteins: Cenpc and Cenpe. The observation that Cenpb is not essential for mitosis or meiosis therefore drastically contrasts the severe phenotypes previously reported for the cbh+ and abp1 null yeast strains (Halverson et al., 1997; Lee et al., 1997), suggesting that the functions of these homologues have diverged significantly.
Despite the lack of any detectable mitotic and meiotic phenotype, adult Cenpb null mice are significantly smaller in body weight compared with age- and sex-matched wild-type or heterozygous mice. In addition, the −/− male testes show a pronounced reduction both in weight (by 30%) and in total sperm count (by 39.5%) compared with wild-type animals. Extensive histological analysis of many different organs and direct measurement of FSH and LH hormones have not revealed any abnormality. It is possible that the absence of CENP-B may have a subtle effect on centromere assembly and function, and result in a slight reduction in the rate of progression through one or more phases of the cell cycle, in the efficiency of chromosome capture by microtubules, and/or chromosome movement. Alternatively, a small number of cells beyond our detection ability may not enter mitosis at all, or carry severe chromosomal abnormality, resulting in loss of valuable cells from the cycling cell population sufficiently to cause a significant weight reduction over time. The possibility that the weight phenotype is caused by some as yet unidentified hormonal or metabolic factors cannot be discounted at present.
In formulating any model on the role of CENP-B, the following observations need to be taken into consideration: (a) the protein is highly conserved in divergent mammals; (b) the protein binds centromeric repetitive DNA via the CENP-B-box motif and possesses dimerization properties that allow the protein to cross-link centromeric repeats (Yoda et al., 1992; Muro et al., 1992); (c) CENP-B box and CENP-B protein are not detected on human and mouse Y chromosomes, and are poorly represented on centromeric subdomains of certain human chromosomes (e.g. α13-II, α14-II, and α21-II domains of chromosomes 13, 14, and 21; Trowell et al., 1993; Ikeno et al., 1994; Choo, 1997a); (d) the centromeres of African green monkey are composed largely of α-satellite DNA containing few if any binding sites for CENP-B (Goldberg et al., 1996); (e) the protein is found on both active and inactive centromeres of dicentric chromosomes (Earnshaw et al., 1989); (f) despite the lack of CENP-B binding, human neocentromeres derived from noncentromeric chromosomal regions display full mitotic functions (Voullaire et al., 1993; Depinet et al., 1997; du Sart et al., 1997; Choo, 1997b ); and (g) the protein is neither essential for mitosis nor meiosis in Cenpb knockout mice.
Based on the sequence similarity between CENP-B and certain transposases, Kipling and Warburton (1997) suggested that CENP-B may share the DNA strand cleavage function of transposases and promote nicks adjacent to CENP-B boxes to facilitate the evolution and maintenance of satellite DNA. This model does not, however, take into consideration the dimerization property of CENP-B, offers no direct evidence for the proposed strand cleavage function, and cannot explain the absence of CENP-B on human and mouse Y chromosomes or the paucity of this protein on the α-satellite–containing centromeres of African green monkey and centromeric subdomains of at least some human chromosomes. Here we present a different model that satisfies all the reported observations. Simply stated, we propose that the role of CENP-B is to organize structurally the great abundance of repetitive DNA found in the centromere, with this role neither being exclusive to CENP-B nor directly essential or sufficient for centromere function. Our model further implicates the existence of a functionally related but perhaps lower-affinity protein, arbitrarily designated CENP-Z, that can perform a similar function to CENP-B in its absence (Fig. 4). The proposal of a role for CENP-B in organizing centromeric repeats is based on the biochemical (observation b above), cytogenetic (observation e above), and, indirectly, evolutionary (observation a above) properties of the protein. The suggestion that this role is not exclusive to CENP-B is based on the fact that centromeric repetitive DNAs with little or no CENP-B binding (observations c, d and g above) are nonetheless organized in a way that is compatible with centromere function. The suggestion that CENP-B is neither essential nor sufficient for centromere function is evident from the fact that the protein can be totally absent on centromeres without detrimental effects on chromosome segregation (observations c, d, f, and g above), and that its mere presence on some centromeres does not immediately lead to centromere activity (observation e above).
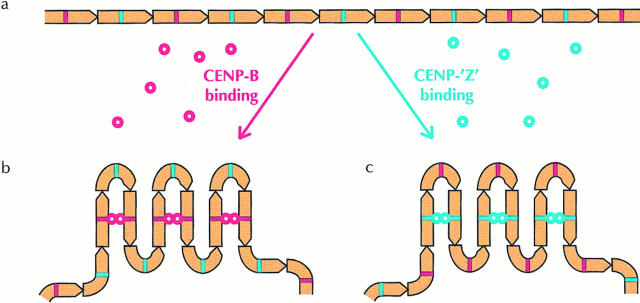
Figure 4.
Model depicting the role of CENP-B and its putative functional homolog CENP-Z. (a) A centromeric satellite DNA array, showing individual monomers containing either a CENP-B-box (red bar) or a CENP-Z-box motif (blue bar). (b) CENP-B proteins (red circles) bind to CENP-B-box motifs and undergo dimerization to cross-link the array into a more stable higher order configuration. This mode of organization is presumably found on chromosomes where CENP-B boxes are prevalent, including the human and mouse autosomes and X chromosomes. (c) In the absence of CENP-B-binding, a functionally related protein CENP-Z (blue circle) assumes the role of CENP-B to cross-link CENP-Z box–containing monomers. This mode of organization is suggested for the human and mouse Y chromosomes, various CENP-B box-poor centromeric subdomains, and the centromeres of African green monkey and Cenpb null mice.
A nuclear protein, pJα (Gaff et al., 1994), has previously been described that binds a 9-bp sequence motif, GTG(G/ A)AAAAG, that is present as an alternative nucleotide configuration to the CENP-B-box motif on a significant proportion (∼14%) of α-satellite monomers, including those that constitute the centromere of the human Y chromosome and the various CENP-B box-poor human centromeric subdomains (Tyler-Smith and Brown, 1987; Alexandrov et al., 1993; Vissel and Choo, 1992; Romanova et al., 1996). A recent study has further demonstrated that pJα box-containing α-satellite monomers are the primordial DNA from which the CENP-B box-containing monomers arose (Romanova et al., 1996). In preliminary studies, we have detected pJα proteins in the nuclear extracts of the +/+, +/−, and −/− Cenpb knockout mice, and in that of the African green monkey (D.F. Hudson and K.H.A. Choo, unpublished data). In addition, the consensus sequence for the CENP-B box-poor α-satellite DNA of African green monkey has been shown to contain a perfectly conserved pJα-box motif GTGAAAAAG (Yoda et al., 1996). These analyses therefore suggest that pJα may be a suitable candidate for the proposed CENP-Z protein. The availability of the Cenpb null mice should provide an amenable system to allow the further study of the role of Cenpb, as well as investigation of the proposed CENP-Z protein.
Acknowledgments
We thank L. Robertson, J. Mann, and S. Delaney for STO-NeoR, W9.5, W9.8, and R1 cell lines, T. Yen, S. Wittingham, and T. Kaye for antibodies, P. Mountford and A. Smith for IRES-neo and IRES-hygro markers, S. Gazeas and R. Breslin for mouse breeding, R. O'Dowd for technical assistance, C.W. Chow for histology, S. Bol and A. Fryga for flow sorting, A. O'Connor for DSP assay, L. Wilton and S. Pompolo for tissue dissection, and Qing Song for stereology. This work was supported by the National Health and Medical Research Council of Australia. KHA Choo is a Senior Associate of the Univeristy of Melbourne and a Principal Research Fellow of the National Health and Medical Research Council of Australia.
Abbreviations used in this paper
- ASC
advanced sperm count
- ES
embryonic stem
- RT
reverse transcription
Footnotes
Address all correspondence to Dr. Andy Choo, The Murdoch Institute for Research into Birth Defects, Royal Children's Hospital, Flemington Road, Parkville 3052, Australia. Phone: 61-3-9345-5045; FAX: 61-3-9348-1391; E-mail: choo@cryptic.rch.unimelb.edu.au
References
- Alexandrov IA, Medvedev LI, Mashkova TD, Kisselev LL, Romanova LY, Yurov YB. Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucl Acids Res. 1993;21:2209–2215. doi: 10.1093/nar/21.9.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano LA, Valdivia MM. Molecular cloning of an intronless gene for the hamster centromere antigen CENP-B. Biochim Biophys Acta. 1996;1307:21–25. doi: 10.1016/0167-4781(96)00039-5. [DOI] [PubMed] [Google Scholar]
- Bernat RL, Borisy GG, Rothfield NF, Earnshaw WC. Injection of anticentromere antibodies in interphase disrupts events required for chromosome movement in mitosis. J Cell Biol. 1990;111:1519–1533. doi: 10.1083/jcb.111.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat RL, Delannoy MR, Rothfield NF, Earnshaw WC. Disruption of centromere assembly during interphase inhibits kinetochore morphogenesis and function in mitosis. Cell. 1991;66:1229–1238. doi: 10.1016/0092-8674(91)90045-z. [DOI] [PubMed] [Google Scholar]
- Brenner S, Pepper D, Berns MW, Tan E, Brinkley BR. Kinetochore structure, duplication and distribution in mammalian cells: analysis by human autoantibodies from scleroderma patients. J Cell Biol. 1981;91:95–102. doi: 10.1083/jcb.91.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR, Ouspenski I, Zinkowski RP. Structure and molecular organization of the centromere-kinetochore complex. Trends Cell Biol. 1992;2:14–21. doi: 10.1016/0962-8924(92)90139-e. [DOI] [PubMed] [Google Scholar]
- Brown M. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- Brown MT, Goetsch L, Hartwell LH. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzin CH, Mann JR, Singer-Sam J. Quantitative RT-PCR assays show Xist RNA levels are low in mouse female adult tissue, embryos and embryoid bodies. Development. 1994;120:3529–3536. doi: 10.1242/dev.120.12.3529. [DOI] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H.A. 1997a. The Centromere. Oxford University Press, Oxford, United Kingdom. 304 pp.
- Choo KHA. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am J Hum Genet. 1997b;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KHA, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all human chromosomes and derivation of a new consensus sequence. Nucl Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Bernat RL, Earnshaw WC. CENP-B: a major human centromere protein located beneath the kinetochore. J Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, VanDyke DL, Willard HF, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role of heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao J, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KHA. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, MacKay AM. Role of nonhistone proteins in the chromosomal events of mitosis. FASEB J. 1994;8:947–956. doi: 10.1096/fasebj.8.12.8088460. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzler MJ, Kinsella TD. The CREST syndrome: a distinct serologic entity with anticentromere antibodies. Am J Med. 1980;69:520–526. doi: 10.1016/0002-9343(80)90462-3. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Brown WRA. Efficient conditional mutation of the vertebrate CENP-C gene. Hum Mol Genet. 1997;6:2301–2308. doi: 10.1093/hmg/6.13.2301. [DOI] [PubMed] [Google Scholar]
- Gaff C, du Sart D, Kalitsis P, Iannello R, Nagy A, Choo KHA. A novel nuclear protein binds centromeric alpha satellite DNA. Hum Mol Genet. 1994;3:711–716. doi: 10.1093/hmg/3.5.711. [DOI] [PubMed] [Google Scholar]
- Goldberg, I.G., A.F. Sawhney, A.F. Pluta, P.E. Warburton, and W.C. Earnshaw. 1996. Surprising deficiency of CENP-B binding sites in African green monkey alpha satellite DNA: implications for CENP-B function at centromeres. Mol. Cell. Biol 16:5156–5168. [DOI] [PMC free article] [PubMed]
- Haaf T, Mater AG, Wienberg J, Ward DC. Presence and abundance of CENP-B box sequences in great ape subsets of primate-specific alpha satellite DNA. J Mol Evol. 1995;41:487–491. doi: 10.1007/BF00160320. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Rabl orientation of CENP-B box sequences in Tupaia belangerifibroblasts. Cytogenet Cell Genet. 1995;70:258–262. doi: 10.1159/000134047. [DOI] [PubMed] [Google Scholar]
- Halverson D, Baum M, Stryker J, Carbon J, Clarke L. A centromere DNA-binding protein from fission yeast affects chromosome segregation and has homology to human CENP-B. J Cell Biol. 1997;136:487–500. doi: 10.1083/jcb.136.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Masumoto H, Okazaki T. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum Mol Genet. 1994;3:1245–1257. doi: 10.1093/hmg/3.8.1245. [DOI] [PubMed] [Google Scholar]
- Jeppesen P, Mitchell A, Turner B, Perry P. Antibodies to defined histone epitopes reveal variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Earle E, Hill J, Choo KHA. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc Natl Acad Sci USA. 1998;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Le M-H, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophilafemale meiosis. Science. 1996;273:118–273. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- Kipling D, Mitchell AR, Matsumoto H, Wilson HE, Nicol L, Cooke HJ. CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. . Mol Cell Biol. 1995;15:4009–4020. doi: 10.1128/mcb.15.8.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Warburton PE. Centromeres, CENP-B and Tiggertoo. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]
- Kipling D, Wilson HE, Mitchell AR, Taylor BA, Cooke HJ. Mouse centromere mapping using oligonucleotide probes that detects variants of the minor satellite. Chromosoma. 1994;103:46–55. doi: 10.1007/BF00364725. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Masumoto H, Ikeda M, Okazki T. Analysis of protein-DNA and protein-protein interactions of centromere protein B (CENP-B) and properties of the DNA-CENP-B complex in the cell cycle. Mol Cell Biol. 1995;15:1602–1612. doi: 10.1128/mcb.15.3.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen, H.B., A.L. Jargensen, C. Jones, and A.L. Bak. 1992. Higher rate of evolution of X chromosome alpha-repeat DNA in human than in the great apes. EMBO J 11:2367–2372. [DOI] [PMC free article] [PubMed]
- Lee J, Huberman JA, Hurwitz J. Purification and characterization of a CENP-B homologue protein that binds to the centromeric K-type repeat DNA of Schizosaccharomyces pombe. . Proc Natl Acad Sci USA. 1997;94:8427–8432. doi: 10.1073/pnas.94.16.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Sugimoto K, Okazaki T. Alphoid satellite DNA is tightly associated with centromere antigens in human chromosomes throughout the cell cycle. Exp Cell Res. 1989;181:181–196. doi: 10.1016/0014-4827(89)90192-4. [DOI] [PubMed] [Google Scholar]
- Meluh P, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiaeencodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Cell Biol. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi Y, Hartman AL, Nakane PK, Tan EM. Distribution of kinetochore (centromere) antigen in mammalian cell nuclei. J Cell Biol. 1981;90:254–259. doi: 10.1083/jcb.90.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner EW, Kühn LC. A stem-loop in the 3′ untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988;53:815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Dicistronic targeting constructs: Reporters and modifiers of mammalian gene expression. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi, H., K. Wakui, K. Ogawa, T. Okano, N. Niikawa, and Y. Fukushima. 1994. A stable acentric marker chromosome: possible existence of an intercalary ancient centromere at distal 8p. Am. J. Hum. Genet. 55:1202–1208. [PMC free article] [PubMed]
- Page SL, Earnshaw WC, Choo KHA, Shaffer LG. Further evidence that CENP-C is a necessary component of active centromeres: studies of a dic(X;15) with simultaneous immunofluorescence and FISH. Hum Mol Genet. 1995;4:289–294. doi: 10.1093/hmg/4.2.289. [DOI] [PubMed] [Google Scholar]
- Palmer DK, Margolis RL. Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol Cell Biol. 1985;5:173–186. doi: 10.1128/mcb.5.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras DF, Bennett KL, Siracusa LD, Woodworth-Gutai M, Chapman VM, Gross KW, Kane-Haas C, Hastie ND. Construction of a small Mus musculus repetitive DNA library: Identification of a new satellite sequence in Mus musculus. . Nucl Acids Res. 1983;11:6965–6983. doi: 10.1093/nar/11.20.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Pluta AF, Saitoh N, Goldberg I, Earnshaw WC. Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J Cell Biol. 1992;116:1081–1093. doi: 10.1083/jcb.116.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB. The structure of the mammalian centromere. Bioessays. 1991;13:51–56. doi: 10.1002/bies.950130202. [DOI] [PubMed] [Google Scholar]
- Robb GW, Amann RP, Killian GJ. Daily sperm production and epididymal sperm reserves of pubertal and adult rats. J Reprod Fertil. 1978;54:103–107. doi: 10.1530/jrf.0.0540103. [DOI] [PubMed] [Google Scholar]
- Robertson, E.J. 1987. Embryo-derived stem cell lines. In Teratocarcinomas and Embryo Stem Cells: A Practical Approach, E.J. Robertson, editor. IRL Press, Oxford, United Kingdom. 71–112.
- Romanova LY, Deriagin GV, Mashkova TD, Tumeneva IG, Mushegian AR, Kisselev LL, Alexandrov IA. Evidence for selection in evolution of alpha satellite DNA: the central role of CENP-B/pJα binding region. J Mol Biol. 1996;261:334–340. doi: 10.1006/jmbi.1996.0466. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Tomkiel J, Cooke CA, Ratrie H, III, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Seki N, Saito T, Kitagawa K, Masumoto H, Okazaki T, Horit TA. Mapping of the human centromere protein B gene (CENP-B) to chromosome 20p13 by fluorescence in situ hybridization. Genomics. 1994;24:187–188. doi: 10.1006/geno.1994.1600. [DOI] [PubMed] [Google Scholar]
- Simerly C, Balczon R, Brinkley BR, Schatten G. Microinjected kinetochore antibodies interfere with chromosome movement in meiotic and mitotic mouse oocytes. J Cell Biol. 1990;111:1491–1504. doi: 10.1083/jcb.111.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Glass CA. CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma. 1991;100:360–370. doi: 10.1007/BF00337514. [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Yata H, Himemo M. Mapping of the human CENP-B gene to chromosome 20 and the CENP-C gene to chromosome 12 by a rapid cycle DNA amplification procedure. Genomics. 1993;17:240–242. doi: 10.1006/geno.1993.1312. [DOI] [PubMed] [Google Scholar]
- Tomkiel J, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowell HE, Nagy A, Vissel B, Choo KHA. Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Hum Mol Genet. 1993;2:1639–1649. doi: 10.1093/hmg/2.10.1639. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Brown WR. Structure of the major block of alphoid satellite DNA on the human Y chromosome. J Mol Biol. 1987;195:457–470. doi: 10.1016/0022-2836(87)90175-6. [DOI] [PubMed] [Google Scholar]
- Vissel B, Choo KH. Evolutionary relationships of multiple alpha satellite subfamilies in the centromeres of human chromosomes 13, 14, and 21. J Mol Evol. 1992;35:137–146. doi: 10.1007/BF00183225. [DOI] [PubMed] [Google Scholar]
- Volobouev, V., N. Voght, E. Viegas-Pequignot, B. Malfoy, and B. Dutrillaux. 1995. Characterization and chromosomal location of two repeated DNAs in three Gerbillus species. Chromosoma. 104:252–259. [DOI] [PubMed]
- Voullaire LE, Slater HR, Petrovic V, Choo KHA. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? . Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Wreford NG. Theory and practice of stereological techniques applied to the estimation of cell number and nuclear volume in the testis. Microscopy Res Tech. 1995;32:423–436. doi: 10.1002/jemt.1070320505. [DOI] [PubMed] [Google Scholar]
- Wu ZA, Liu W-X, Murphy C, Gall J. Satellite 1 DNA sequence from genomic DNA of the giant panda Ailuropoda melanoleuca. . Nucl Acids Res. 1990;18:1054. doi: 10.1093/nar/18.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RR, Anscough K, Anderson C, Baynes M, Berks J, Bonfield J, Burton M, Connell T, Copsey J, Cooper A, et al. 2.2. Mb of contiguous nucleotide sequence from chromosome III of C. elegans. . Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol. 1992;119:1413–1427. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Nakamura T, Masumoto H, Suzuki N, Kitagawa K, Nakano M, Shinjo A, Okazaki T. Centromere protein B of African green monkey cells: Gene structure, cellular expression, and centromeric localization. Mol Cell Biol. 1996;16:5169–5177. doi: 10.1128/mcb.16.9.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]