Abstract
Several families of evolutionarily conserved axon guidance cues orchestrate the precise wiring of the nervous system during embryonic development. The remarkable plasticity of freshwater planarians provides the opportunity to study these molecules in the context of neural regeneration and maintenance. Here we characterize a homologue of the Slit family of guidance cues from the planarian Schmidtea mediterranea. Smed-slit is expressed along the planarian midline, in both dorsal and ventral domains. RNA interference targeting Smed-slit results in the collapse of many newly regenerated tissues at the midline; these include the cephalic ganglia, ventral nerve cords, photoreceptors, and the posterior digestive system. Surprisingly, Smed-slit RNAi knockdown animals also develop morphologically distinguishable, ectopic neural structures near the midline in uninjured regions of intact and regenerating planarians. These results suggest that Smed-slit acts not only as a repulsive cue required for proper midline formation during regeneration but that it may also act to regulate the behavior of neural precursors at the midline in intact planarians.
Keywords: planarian, Schmidtea mediterranea, Slit, axon guidance, midline, regeneration, neural differentiation
Introduction
In contrast to most animals, freshwater planarians can regenerate a complete, functional central nervous system (CNS) (Reuter et al., 1996; Umesono et al., 1997; Cebrià et al., 2002a; Inoue, et al., 2004). The planarian CNS consists of two cephalic ganglia situated above two ventral nerve cords that run the length of the animal and are connected by transverse commissures (Reuter and Gustafsson, 1995; Agata et al., 1998; Tazaki et al., 1999; Umesono et al., 1999; Cebrià et al. 2002a; Okamoto et al., 2005). Regeneration in planarians relies upon a population of stem cells, called neoblasts, that proliferate in response to injury and differentiate to form the missing tissues (reviewed in Newmark and Sánchez Alvarado, 2002; Agata 2003; Reddien and Sánchez Alvarado, 2004; Sánchez Alvarado, 2006). Regeneration of the planarian CNS requires the neoblasts to adopt neuronal identities during the initial stages of regeneration. These nascent neurons must then elaborate appropriate axonal and dendritic projections, reconstructing the synaptic pathways required for nervous system function. In order to unravel how the newly regenerated CNS is re-wired, we have started to characterize the roles of planarian homologues of conserved axon guidance cues (Cebrià and Newmark, 2005; 2007). We have shown that planarian homologues of netrin and netrin receptor are required for the normal regeneration of the CNS and to maintain the architecture of the mature nervous system (Cebrià and Newmark, 2005). Fusaoka et al. (2006) have also reported that homologues of NCAM and DSCAM, members of the immunoglobulin superfamily of adhesion molecules, play roles in regeneration of the planarian CNS.
Slit proteins represent another family of conserved axon guidance cues that are critical for nervous system development. Slits are large, extracellular glycoproteins of ∼200 kDa that contain four leucine-rich repeats and seven-to-nine EGF repeats; these proteins have been conserved from Drosophila and C. elegans to vertebrates (Brose and Tessier-Lavigne, 2000; Wong et al., 2002; Dickson and Gilestro, 2006). The slit gene was initially identified in Drosophila (Rothberg et al. 1988); slit mutants are characterized by the collapse of commissural and longitudinal axonal tracts at the midline (Rothberg et al., 1990; Kidd et al., 1999). This repulsive role for slit in midline commissural axon guidance has been conserved in vertebrates (Long et al., 2004). In addition to midline-related defects, slit genes function in establishing proper dorso-ventral axonal tracts in C. elegans (Hao et al., 2001) and the mammalian forebrain (Bagri et al., 2002). Slit genes are also required for migration of both neuronal (Wu et al., 1999; Hu, 1999) and non-neuronal cell types (Kidd et al., 1999; Kramer et al., 2001; Wu et al., 2001; Kolesnikov and Beckendorf, 2005; Qian et al., 2005; Santiago-Martínez et al., 2006).
Here we report the characterization of a slit orthologue from the planarian Schmidtea mediterranea. Smed-slit is expressed in both dorsal and ventral domains along the planarian midline. Functional analyses using RNA interference (RNAi) reveal that Smed-slit functions as a repulsive midline cue: after knockdown of this gene, the nervous system regenerates at the midline, instead of forming the well-defined, bilateral CNS normally observed in these animals. In addition to defects in the nervous system, the regenerated posterior gut branches connect abnormally at the midline after Smed-slit RNAi. Surprisingly, in intact planarians Smed-slit knockdown results in the development of ectopic photoreceptors, cephalic ganglia, and ventral nerve cords at or near the midline. Our results suggest that Smed-slit functions not only as a midline repellent during nervous system regeneration, but that it may also play a role in regulating the behavior of neural precursors at the midline of intact planarians.
Materials and methods
Organisms
Animals from a clonal line (Sánchez Alvarado et al., 2002) of the diploid, asexual strain of Schmidtea mediterranea (Benazzi et al., 1972) were used. Planarians were maintained as described (Cebrià and Newmark, 2005) and 4-6 mm long animals were starved for at least one week before use in experiments.
Isolation of Smed-slit
In order to identify planarian slit homologues, Slit proteins from different organisms were used in tblastn searches of S. mediterranea genomic sequences (generated by the Washington University Genome Sequencing Center, St. Louis, and available from the NCBI Trace Archives). Genomic sequences encoding predicted ORFs similar to Slit were assembled using Sequencher 4.2.2 (Gene Codes Corp.). Sets of specific primers were designed to amplify Smed-slit from a planarian cDNA library (Zayas et al., 2005), then 5' and 3' RACE were used to obtain additional cDNA sequences. We isolated ∼4.6 kb of Smed-slit cDNA sequence (accession number DQ336176), encoding a predicted product with all of the structural features of Slit proteins, but still lacking a start codon and N-terminal signal sequence (Suppl. Fig. 1). In the genomic sequence, we identified a possible start codon and signal peptide 105 base pairs upstream of our longest RACE fragment; these predicted sequences aligned well with the amino termini of other Slit proteins (Suppl. Fig. 1). We then performed PCR on S. mediterranea cDNA using primers corresponding to sequences upstream of the putative start codon and downstream of the first intron (to rule out the possibility of contamination from genomic DNA); these experiments confirmed that we identified the full-length coding sequence of Smed-slit. The combined cDNA sequence consists of ∼4.8 kb (not including the poly(A) tail), in good agreement with the single ∼5 kb transcript observed in Northern blots (data not shown).
Whole-mount in situ hybridization
Planarians were fixed and then processed in an Insitu Pro hybridization robot (Abimed/Intavis) as previously described (Umesono et al., 1997; Sánchez Alvarado et al., 2002). Images of nonfluorescence in situ hybridizations for genes Smed-slit, H.10.2f (novel), Smed-netrin2, and SmedroboA were captured using a MicroFire digital camera (Optronics) attached to a Leica MZ125 stereomicroscope or a Nikon Eclipse TE200 inverted microscope. For Smed-slit fluorescent in situ hybridizations, after post-hybridization washes and blocking in 1% BSA in MABT buffer (100mM maleic acid, 150 mM MaCl, 0.1% tween-20; pH 7.5), the samples were incubated overnight at 4°C with an anti-digoxigenin-POD antibody (Roche) diluted 1:100 in 1%BSA in MABT and then washed at RT in PBS-0.1% tween-20 for 6 hours with several changes. Finally, samples were developed using Tyramide Signal Amplification (Molecular Probes) as recommended by the manufacturer. The samples were incubated in the tyramide solution for 5 minutes in the dark, then washed in PBS-0.1% tween for several hours, mounted in Vectashield (Vector Laboratories), and observed through a Nikon TE 2000-S inverted microscope (objectives used: 20x/0.75 plan fluor and 40x/1.0 plan apo) and a CARV spinning disk confocal (AttoBiosciences). Images were captured with a CoolSnap HQ CCD camera (Photometrics) and Metamorph software v6.1, and deconvolved using AutoDeblur 9.3 (AutoQuant Imaging, Inc).
Whole-mount immunostaining
Immunostaining was carried out essentially as described (Sánchez Alvarado and Newmark, 1999; Cebrià and Newmark, 2005). The following monoclonal antibodies were used: anti-tubulin Ab-4 (NeoMarkers, used at 1:200), an anti-phospho-tyrosine P-Tyr-100 (Cell Signaling technology, used at 1:500) to visualize the brain and the ganglia of the VNCs, the gut and the pharynx, and VC-1, specific for the photosensitive cells (Sakai et al., 2000). Highly cross-absorbed Alexa Fluor 488 or 568 goat anti-mouse IgG secondary antibodies (Molecular Probes) were used at 1:400 and 1:1000, respectively. Nuclei were stained with 0.5 μg/ml Hoechst overnight at 4°C. Images were taken as described above for fluorescent in situ hybridization.
RNAi analyses
Double-stranded RNA (dsRNA) of Smed-slit was synthesized using the MegaScript in vitro transcription system (Ambion) and injected into the animals as described previously (Sánchez Alvarado and Newmark, 1999). Injected planarians were amputated, allowed to regenerate, and then processed for in situ hybridization or immunostaining. For long-term experiments on intact animals, they were re-injected with Smed-slit dsRNA seventeen and thirty-eight days after the first round of injections. Animals were fixed seventeen days, thirty-eight days, and eight weeks after the first round of injections. Control animals were injected with water.
Results
Smed-slit is expressed along the midline of intact planarians, in distinct dorsal and ventral cell populations
Sequences encoding predicted proteins with significant similarity to slit genes from other organisms were identified from whole-genome shotgun sequence from the planarian, Schmidtea mediterranea and then isolated from a planarian cDNA library (Zayas et al., 2005; see Materials and methods). The predicted Smed-Slit protein shares the domain organization observed in other Slits: it contains four leucine-rich repeats (LRRs), seven EGF-like repeats, with a laminin G domain inserted between the sixth and seventh EGF repeats, and a C-terminal cysteine-rich domain (Suppl. Fig. 1).
Whole-mount in situ hybridization revealed that Smed-slit (for brevity, referred to as slit) was expressed along the midline of intact planarians, in distinct cells in dorsal (Fig. 1A) and ventral (Fig. 1B) domains. To visualize slit-expressing cells in relation to other structures in the body, intact planarians were double labeled to detect slit mRNA by fluorescence in situ hybridization (FISH) and anti-phospho-tyrosine (P-tyr) immunofluorescence to label the nervous system, the digestive system lumen, and cell-cell junctions of the epithelia. Confocal analysis of these specimens showed two discrete, dorsal populations of slit-positive cells (Figs. 1C, D) in the anterior half of the animal (from the anterior tip to the pharyngeal region): a more superficial, narrow layer of slit-positive cells (Fig. 1C, magenta; arrowheads) at the level of the submuscular nerve plexus (Fig. 1C, green) and a deeper layer of slit-positive cells (Fig. 1D, magenta; arrowheads) dorsal to the gut. In the posterior half of the planarians (the post-pharyngeal region), the more superficial population of slit-positive cells was not detected by in situ hybridization. However, slit-positive cells were detected along the midline, deeper in the mesenchyme (Fig. 1E, magenta), between the two posterior gut branches (Fig. 1E, green). Anti-Tubulin immunostaining labels a population of ciliated epithelial cells along the dorsal midline (Fig. 1F, arrowheads; Robb and Sánchez Alvarado, 2002). Cells positive for slit mRNA (Fig. 1G, arrowheads) were located beneath those epithelial cells, and in the same midline-restricted domain (Fig. 1H). Ventrally, slit-positive cells were also detected along the midline (Figs. 1B, I, J, magenta); however, they were distributed in a wider region of the midline than those observed in the more-restricted dorsal populations. These ventral, slit-positive cells were found between the submuscular plexus and the ventral nerve cords (Figs. 1I, J, green) along the length of the animal, both pre-pharyngeally (Fig. 1I) and post-pharyngeally (Fig. 1J).
Fig. 1.
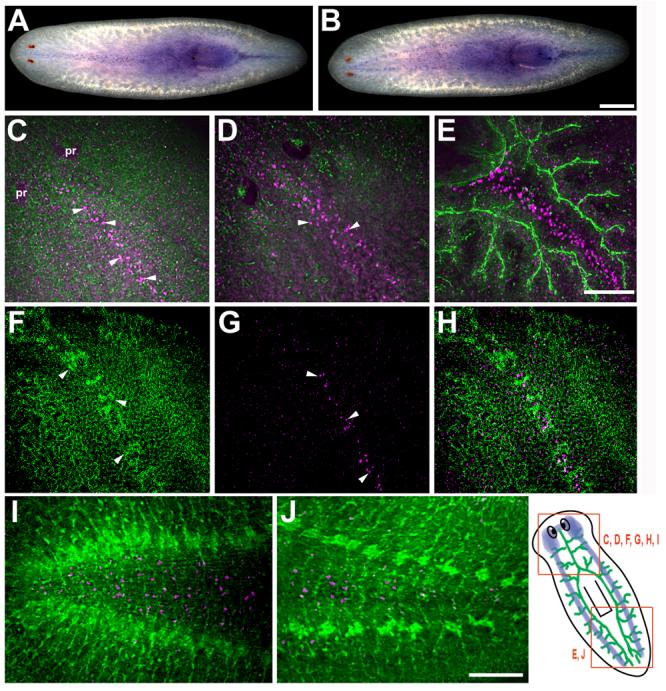
Expression pattern of Smed-slit mRNA in intact planarians. (A-B) Whole-mount in situ hybridization showing distinct populations of Smed-slit-positive cells at the midline observed from either dorsal (A) or ventral (B) views. (C-J) Double labeling to detect Smed-slit mRNA (in magenta) by whole-mount fluorescent in situ hybridization and immunostaining with either anti-phospho-tyrosine (green in C-E; I-J) or anti-Tubulin (green in F,H). (C) Smed-slit-positive cells (arrowheads) along the midline in the submuscular plexus (in green). (D) Smed-slit-positive cells (arrowheads) deeper in the mesenchyme. (C) and (D) are confocal projections from different focal planes of the same sample. (E) Smed-slit positive cells between the two posterior gut branches (in green). (F) Midline dorsal stripe of ciliated epithelial cells labeled with anti-Tubulin (arrowheads). (G) Smed-slit-positive cells (arrowheads) in the submuscular plexus, beneath the epithelium. (F) and (G) are confocal projections from different focal planes of the same sample. (H) is a merged image of (F) and (G). (I-J) Ventral views of Smed-slit mRNA expression between the cephalic ganglia (in green) in the head (I) and between the ventral nerve cords (in green) in the post-pharyngeal region (J). The drawing at the lower right shows the regions of the animal represented in the indicated panels. The CNS is depicted in blue and the digestive system is shown in green; the digestive system is located dorsally with respect to the CNS. Note that (C-H) are oriented anterior to the top left corner, whereas, (I, J) are oriented anterior to the left. (C-H) dorsal views, (I, J) ventral views. Abbreviation: pr, photoreceptors. Scale bars: 500 μm in A, B; 100 μm in C-H; 100 μm in I,J.
slit is expressed in the vicinity of the anterior commissure of the regenerating cephalic ganglia and in the regenerating pharynx
During anterior regeneration (animals regenerating a new head), slit expression was detected within the regeneration blastema in the initial stages after amputation (Figs. 2A-C), becoming apparent in the proximal blastema at day two of regeneration (arrowhead in Fig. 2B). Around day three of regeneration, thin processes (Fig. 2D, magenta; indicated by an arrow) that appeared to extend from the newly forming cephalic ganglia crossed the midline to connect the two ganglia. slit-positive cells (Fig. 2D, green; arrowhead) were observed at the midline in the same region as those initial processes. At day five slit-expressing cells (Fig. 2E, green; arrowheads) appeared close to the forming anterior commissure (Fig. 2E, magenta) of the planarian brain; confocal analysis revealed that the slit-expressing cells were located beneath the developing commissure. As regeneration proceeded the brain anterior commissure (Fig. 2F, magenta) thickened and slit-positive cells continued to be detected below it. In head pieces regenerating a new pharynx and tail, slit-positive cells (Fig. 2G, green) were observed in the new pharynx primordium (Fig. 2G, arrowheads). In the succeeding days slit-positive cells continued to be detected at lower levels in the growing pharynx and they also appeared between the regenerating posterior gut branches (Figs. 2H, I, magenta, labeled “g”), restoring the pattern observed in intact planarians (Fig. 1E).
Fig. 2.
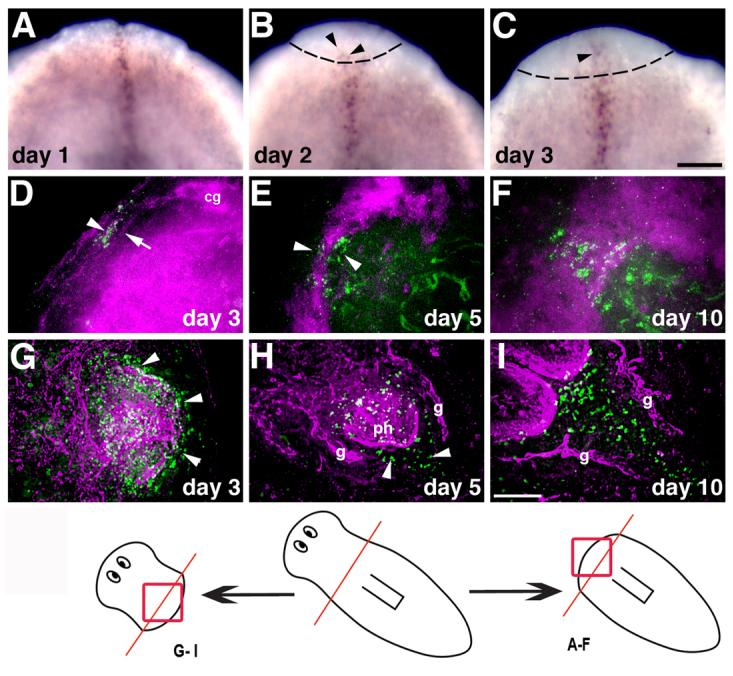
Expression of Smed-slit during regeneration. (A-C) Whole-mount in situ hybridization to detect Smed-slit mRNA during anterior regeneration; days after amputation (prepharyngeal) are indicated. Arrowheads in (B) and (C) point to Smed-slit-positive cells within the blastema, which is delimited by dotted lines. (D-I) Double labeling to detect Smed-slit mRNA (in green) by fluorescent in situ hybridization and anti-phospho-tyrosine immunofluorescence (in magenta) during anterior regeneration (D-F) and posterior regeneration (G-I). Arrowheads in (D) and (E) indicate Smed-slit-positive cells; arrow in (D) points to thin processes that connect the left and right halves of the new cephalic ganglia. Arrowheads in (G) indicate Smed-slit expression in the pharynx primordium. Arrowheads in (H) point to Smed-slit positive cells in the new post-pharyngeal region. The drawing at the bottom of the figure indicates the regions shown in the indicated panels; the prepharyngeal amputation site is shown in red. Abbreviations: cg, cephalic ganglia; g, gut; ph, pharynx. (A-C) anterior to the top; (D-I) anterior to the top left corner. Scale bars: 100 μm in A-C ; 50 μm in D-I.
slit RNAi knockdown results in cyclopic anterior regenerates
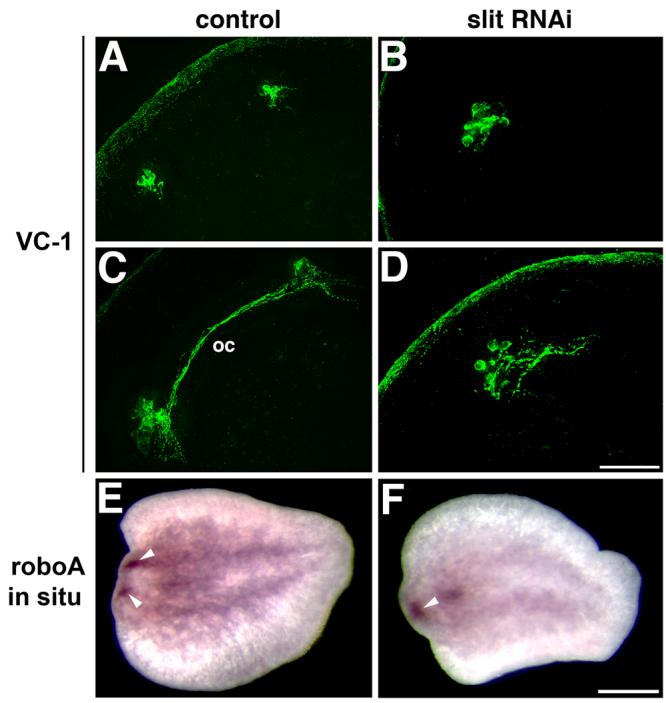
We analyzed the function of slit by RNA interference (RNAi), a technique used routinely to study the function of planarian genes (Sánchez Alvarado and Newmark, 1999; Pineda et al., 2000; Cebrià et al., 2002b; Newmark et al., 2003; Inoue et al. 2004; Mannini et al., 2004; Cebrià and Newmark, 2005; Reddien et al., 2005a,b; Salvetti et al., 2005; Fusaoka et al., 2006; Guo et al., 2006; Cebrià and Newmark, 2007). After delivery of double-stranded RNA, animals were amputated and the effects on regeneration were scored. At the gross morphological level, the most obvious phenotype after slit RNAi knockdown was cyclopia, in which the newly regenerated photoreceptors appeared fused at the midline. Most species of freshwater planarians have two anterior photoreceptors that consist of two cell types: pigmented cup cells and photosensitive cells (Hyman, 1951). Control anterior regenerates developed two normal photoreceptors (58/60; 2/60 did not regenerate a new head; Figs. 3A-C); these new photoreceptors (Fig. 3C, arrowheads) formed bilaterally, with one on either side of the dorsal stripe of ciliated epithelial cells at the midline (Fig. 3C, green). In contrast, after slit RNAi (Figs. 3D-F) the newly regenerated photoreceptors formed at the midline, as defined by the stripe of dorsal epithelial cells (Fig. 3F, arrowheads; 70/72; 2/72 did not regenerate a new head). In slit RNAi knockdowns both the pigment cups (Fig. 3D) and the photosensitive cells detected with the monoclonal antibody VC-1 (Sakai et al., 2000; Fig. 3E) or with anti-Tubulin (Fig. 3F) regenerated at the midline.
Fig. 3.
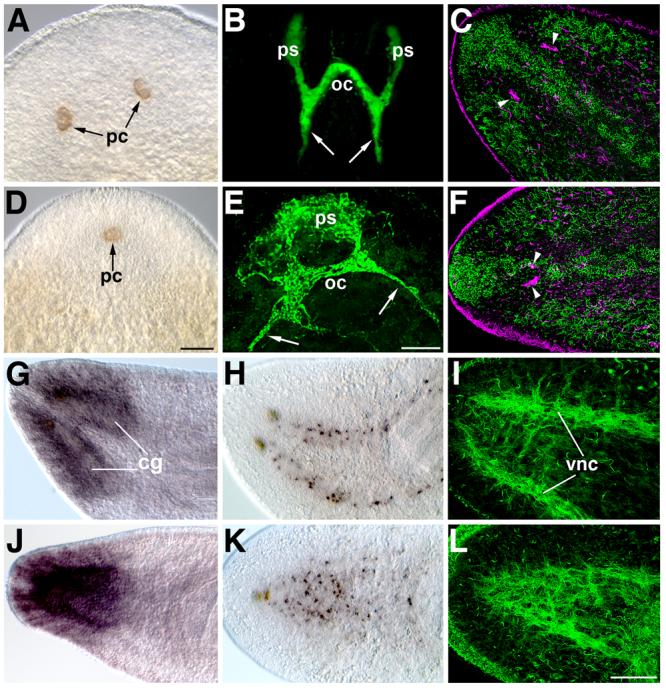
Midline defects in the regenerated nervous system after Smed-slit RNAi. (A-F) Photoreceptor regeneration. (A-C) Control planarians; (D-F) Smed-slit RNAi planarians. (A, D) DIC images showing the regenerated pigment cups (pc) of the photoreceptors. (B, E) VC-1 immunofluorescence to label the optic chiasm (oc) formed by axons of the photosensitive cells (ps); projections to the cephalic ganglia are indicated by arrows. Both the pigment cups (D) and the photosensitive cells (E) regenerate at the midline after Smed-slit RNAi. (C, F) Confocal projections of anti-Tubulin immunofluorescence, pseudocolored according to the depth of the focal plane: dorsal-most sections are shown in green; deeper sections in magenta. (C) In controls, the photoreceptors (arrowheads, magenta) flank the ciliated strip of dorsal epithelium at the midline (in green). (F) After Smed-slit RNAi the regenerated photoreceptors (arrowheads, magenta) are found beneath the dorsal midline epithelial strip. (G-L) Cephalic ganglia and ventral nerve cord markers. (G-I) Control planarians; (J-L) Smed-slit RNAi planarians. (G, J) Expression of H.10.2f in cephalic ganglia (cg). (H, K) Whole-mount in situ hybridization for Smed-netrin2 (Cebrià and Newmark, 2005). (I, L) Confocal projections of anti-Tubulin immunofluorescent labeling of the regenerated ventral nerve cords (vnc). All samples are 17-day regenerates. (A-B) and (D-E) anterior to the top. (C), (F) and (G-L) anterior to the left. Scale bars: 100 μm in A, D; 50 μm in B, E; 100 μm in C, F, G-L.
The regenerating CNS collapses at the midline after slit RNAi
slit homologues show conserved roles in the development of the CNS; therefore, we analyzed nervous system regeneration after slit RNAi. The planarian CNS consists of two anterior cephalic ganglia and two ventral nerve cords (VNCs) that extend the length of the animal (Agata et al., 1998; Cebrià et al., 2002a). Control planarians regenerated a normal CNS: the newly formed cephalic ganglia were well-defined and situated bilaterally about the midline (Figs. 3G, H), as were the newly formed VNCs (Fig. 3H, I) that were connected by transverse commissures (Fig. 3I). After slit RNAi, however, the two new cephalic ganglia collapsed at the midline (Fig. 3J). Smed-netrin2-positive cells (Cebrià and Newmark, 2005) appeared co-mingled along the midline (Fig. 3K), in contrast to their normal position around the cephalic ganglia and along the VNCs (Fig. 3H). Moreover the VNCs also appeared fused along the midline, generating a disorganized meshwork of axonal projections (Fig. 3L).
The midline defects observed after slit RNAi appear at early stages of regeneration
The brain primordia (Cebrià et al., 2002a) and photoreceptors (Inoue et al., 2004) begin to develop within the blastema at very early stages of anterior regeneration. After slit RNAi the new photoreceptors and brain primordia appeared at the midline during the initial stages of regeneration (Fig. 4). In controls, two distinct bilateral clusters of photosensitive cells were detected around day 3 of regeneration (Fig. 4A). Around day 4, axons from these photosensitive cells crossed the midline and projected contralaterally to form an optic chiasm (Fig. 4C). In the following days, visual axons also projected to specific posterior brain regions, restoring the stereotypical pattern of the visual system (Inoue et al., 2004; Cebrià and Newmark, 2005; Okamoto et al., 2005). In contrast, after slit RNAi only a single cluster of VC-1 positive cells appeared at the midline (Fig. 4B); axons from these cells projected posteriorly in the following days, making chiasm-like structures (Fig. 4D).
Fig. 4.
Midline defects appear at early stages of regeneration after Smed-slit RNAi. (A-D) Confocal projections of VC-1 immunofluorescence showing regeneration of the photosensitive cells in control (A,C) and Smed-slit RNAi animals (B,D). (A) In control planarians, two bilateral clusters of photosensitive cells are detected between days 2 and 3 of regeneration; this image corresponds to a 3-day blastema. (C) Between days 3 and 4, axons project contralaterally to form the optic chiasm (oc); this image corresponds to a 4-day blastema. (B) After Smed-slit RNAi a single cluster of photosensitive cells is observed within the blastema. (D) In Smed-slit RNAi animals axons project from the photosensitive cells, forming a greatly reduced chiasm at the midline. (B) and (D) show 3-day blastemas. (E-F) Regenerating cephalic ganglia visualized by whole-mount in situ hybridization to detect Smed-roboA (Cebrià and Newmark, 2007) in regenerating tail pieces 2 days after amputation. (E) Two bilateral clusters of cells (arrowheads) constitute the brain primordia that develop within the regeneration blastema between days 1 and 2. (F) The brain primordia collapse at the midline after Smed-slit RNAi: a single cluster (arrowhead) of new brain cells is observed within the blastema. (A-D) anterior to the top left corner. (E-F) anterior to the left. Scale bars: 50 μm in A-D; 250 μm in E, F.
In order to visualize early defects in the regeneration of the brain primordia we performed in situ hybridizations with Smed-roboA (Cebrià and Newmark, 2007). In control planarians two bilateral clusters of cells expressing Smed-roboA were clearly visible after 1-2 days of regeneration (Fig. 4E, arrowheads). However, in slit RNAi knockdown animals only one cluster of Smed-roboA-positive cells was detected at the midline of animals regenerating a new head (Fig. 4F, arrowhead).
Ectopic neural tissues form at the midline of uninjured tissues in regenerating head pieces after slit RNAi
We also analyzed CNS regeneration in head pieces that were regenerating new central and posterior regions. In both control and slit RNAi head pieces, new pharynges developed normally (Figs. 5A, B, asterisks). In control animals the VNCs grew as two distinct cords that converged at the tip of the tail (Fig. 5A). However, the regenerated VNCs in the new tail of slit RNAi knockdown animals connected at the midline (Fig. 5B, arrow, and see below), similar to the midline collapse observed during anterior regeneration. Surprisingly, around one week after amputation a small, ectopic photoreceptor (Fig. 5C, arrow) developed at the midline between the two pre-existing photoreceptors of slit RNAi knockdown animals. These supernumerary photoreceptors were observed in ∼70% (32/46) of slit RNAi-treated head pieces (0/44 controls developed ectopic photoreceptors); they consisted of both pigment cup (Fig. 5C) and photosensitive cells (Fig. 5D, arrowhead), the latter of which appeared to send axonal projections towards the pre-existing optic chiasm (Fig. 5D, arrow).
Fig. 5.

Ectopic neural tissues develop near the midline of regenerating head pieces after Smed-slit RNAi. (A-B) Montages of confocal projections showing anti-phospho-tyrosine (green) and VC1 (bright green in A) immunofluorescence in regenerating head pieces (17 days of regeneration). (A) Control animal showing a properly regenerated pharynx (asterisk) and the two parallel ventral nerve cords (vnc) that grow into the regenerated tail. (B) Smed-slit RNAi animal: the pharynx appears to regenerate normally (asterisk), but the regenerated ventral nerve cords collapse at the midline of the regenerated tail (arrow). Ectopic neural tissues are found near the midline (arrowhead), throughout the uninjured region of the animal. (C-D) A small, ectopic photoreceptor develops at the midline of regenerating head pieces, between the two pre-existing photoreceptors. These ectopic photoreceptors consist of both pigment cup cells (arrow in (C); DIC image) and photosensitive cells (arrowhead in (D); VC-1 immunofluorescence). Arrow in (D) points to axonal projections connecting the ectopic photoreceptor with the pre-existing optic chiasm. (E-G) Higher magnification views of the animal shown in (B); arrowheads in E indicate ectopic neural tissue that develops medial to the pre-existing ventral nerve cords. (F) Hoechst staining of the same projection as (E). (G) Merged view of (E) and (F). (H-J) Ectopic structures resembling small cephalic ganglia (arrowheads) develop medial to the pre-existing cephalic ganglia (magenta asterisks). (I) Nuclear staining with Hoechst shows that these ectopic cephalic ganglia-like structures are organized similarly to the pre-existing cephalic ganglia with peripheral cell bodies and a central neuropil (white asterisks). Magenta asterisks label the pre-existing cephalic ganglia. (J) Merged view of (H) and (I); white asterisks label the ectopic cephalic ganglia-like tissue. (E-G) and (H-J) correspond to different focal planes of the same sample. All the samples correspond to 17-day regenerates. (A-B, E-J) anterior to the left. (C) anterior to the top. (D) anterior to the upper left. Scale bars: 150 μm in A; 100 μm in B; 50 μm in C-D; 50 μm in E-J.
In addition to the photoreceptors, other ectopic neural tissues developed in uninjured portions of slit RNAi knockdown animals (Figs. 5B, E-J). Confocal sections at the level of the VNCs revealed that ectopic clusters of cells morphologically similar to the ganglia of the VNCs (Figs. 5B, E, arrowheads) developed along the length of the ventral midline. Confocal sections at the level of the cephalic ganglia showed that ectopic structures resembling small, cephalic ganglia (Fig. 5H, arrowheads) developed medially with respect to the pre-existing cephalic ganglia (Fig. 5H, asterisks). These ectopic structures were organized similarly to the cephalic ganglia, with a central neuropil and peripheral cell bodies (Figs. 5I, J, white asterisks).
Abnormal gut regeneration after slit RNAi
Triclad planarians are characterized by a digestive system consisting of three primary intestinal branches connected to a central pharynx. One primary branch extends anteriorly along the midline, ending between the cephalic ganglia; the other two primary branches extend posteriorly toward the tail and are situated dorsally to the ventral nerve cords (see control tail regenerate in Figs. 6A, C). slit RNAi fragments regenerating a new tail showed defects in the pattern of the newly formed gut branches: like the newly regenerated VNCs, (Fig. 6B, green), the gut branches also connected inappropriately at the midline (Fig. 6D, arrow). Although the posterior gut branches occasionally appear connected in a small percentage of control animals (4/26 vs. 22/26 with normal parallel branches), such anastomoses were observed in almost all slit RNAi knockdown animals (29/30 vs. 1/30 with a normal pattern). In control lateral regenerates (animals cut longitudinally), a new posterior gut branch regenerates parallel to the pre-exisitng one (Fig. 6E); in contrast, a single posterior gut branch was observed in slit RNAi lateral regenerates (Fig. 6F).
Fig. 6.
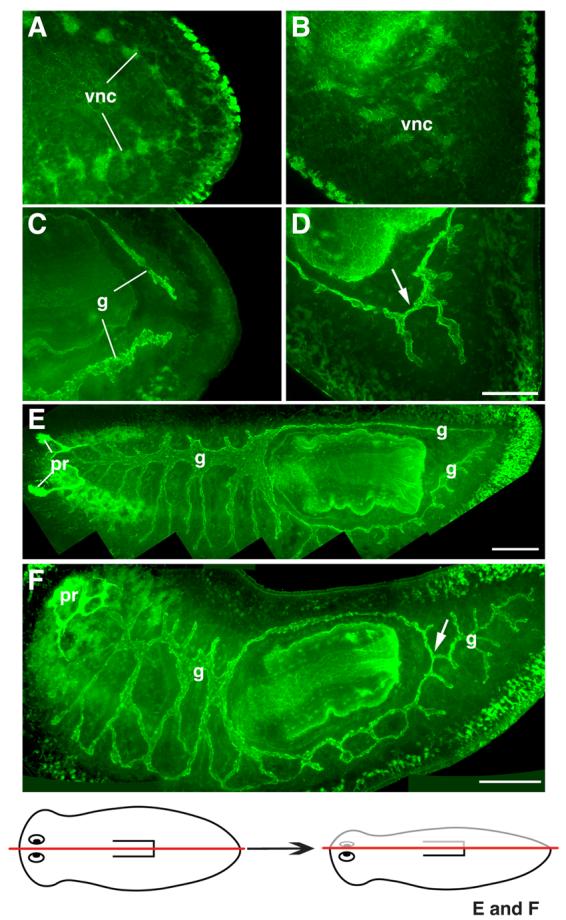
Defects in gut patterning after Smed-slit RNAi. (A-D) Confocal projections of posterior regenerates visualized using anti-phospho-tyrosine immunofluorescence. (A, C) Control planarian imaged at different focal planes: (A) at level of the ventral nerve cords (vnc); (C) dorsal to those shown in (A), at the level of the gut (g). Note separation between regenerated ventral verve cords (in A) and regenerated gut branches (in C). (B, D) Different focal planes from a Smed-slit RNAi planarian: (B) at the level of ventral nerve cords; (D) dorsal to those shown in (B), at the level of gut. Note midline collapse of ventral nerve cords (in B) and improper connection between the left and right posterior gut branches (arrow in D). (E,F) Confocal projections of lateral regenerates visualized using anti-phospho-tyrosine (green) and VC-1 immunofluorescence (bright green labeling of the photoreceptors, pr). (E) Control planarian: note two posterior gut branches. (F) Smed-slit RNAi planarian: only a single main gut branch is observed posterior to the pharynx (arrow). The drawing at the bottom depicts the longitudinal amputation site for the samples shown in (E) and (F). (A, C, E, F) Anterior is to the left; (B, D) anterior is to the upper left. Scale bars: 50 μm in A-D; 200 μm in E; 200 μm in F.
Ectopic neural development at the midline of intact planarians after slit RNAi
Previous studies have shown the plasticity of the nervous system in intact planarians during growth and degrowth (Oviedo et al., 2003; Cebrià and Newmark, 2005). In order to analyze the function of slit in intact planarians, we performed long-term RNAi experiments (see Materials and methods). Similarly to what was observed in regenerating head pieces, an ectopic eye developed at the midline between the two pre-existing photoreceptors, seventeen days after starting the slit RNAi treatement (10/13 animals vs. 0/14 controls; data not shown) Throughout the duration of the experiment, control animals showed a normal brain with peripheral cell bodies, a central neuropil, and the visual axons projecting along the medial border of the cephalic ganglia (Figs. 7A, B depict an 8-week control). By contrast, in slit RNAi knockdown animals, structures resembling small cephalic ganglia developed medially to the pre-existing brain within 17 days of the first RNAi treatment (arrowheads in Figs. 7C,D). These ectopic ganglia were strikingly similar to those that developed in the same region of regenerating head pieces (Figs. 5H-J). In intact planarians examined after longer RNAi treatments (Figs. 7E-H) these ectopic cephalic ganglia became bigger (arrowheads in Figs. 7E,F) and in the most extreme case, three ectopic ganglia were observed (arrowheads in Figs. 7G,H). In this last case, visual axons projected to those ectopic brain-like structures (arrow in Figs. 7G,H), providing evidence that they represent ectopic cephalic ganglia.
Fig. 7.
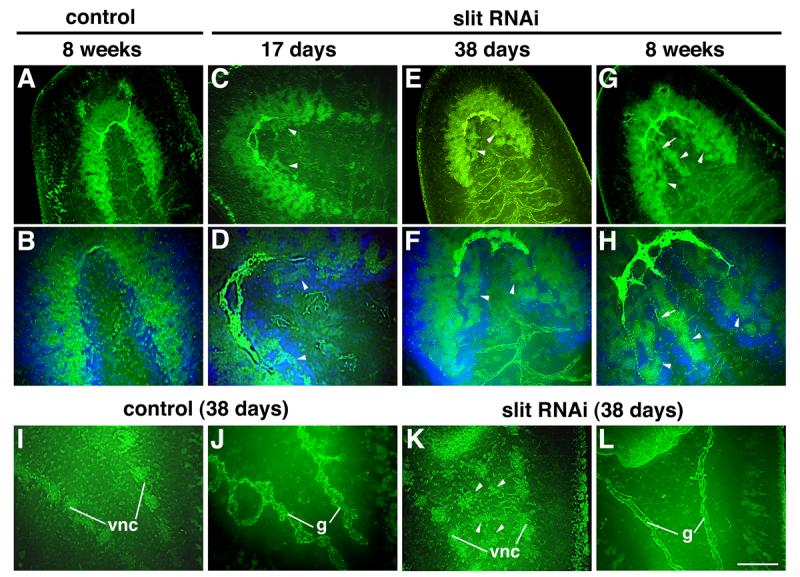
Development of ectopic neural tissues in intact animals after Smed-slit RNAi. (A-H) Confocal projections of intact heads visualized using anti-phospho-tyrosine (green) and VC-1 immunofluorescence (bright green). (A, B) Control planarian 8 weeks after control injections. (C-H) Smed-slit RNAi animals fixed at the indicated times after initiating dsRNA injections. (B), (D), (F) and (H) are counterstained with Hoechst (in blue) and correspond to higher magnification views of (A), (C), (E) and (G), respectively. Arrowheads point to ectopic neural tissues; arrows in (G,H) label photoreceptor projections to the ectopic ganglia. (I-L) Confocal projections of the post-pharyngeal regions of intact planarians visualized by anti-phosphotyrosine immunofluorescence. (I-J) Control and (K-L) Smed-slit RNAi knockdown planarians fixed 38 days after the first injection. (I) and (K) show planes at the level of the ventral nerve cords; (J) and (L) show planes dorsal to those shown in (I) and (K), respectively, at the level of the gut branches. Ectopic neural tissue is indicated by the arrowheads in (K). Anterior to the upper left. Abbreviations: g, gut; vnc, ventral nerve cords. Scale bar (shown in L): 100 μm in A, C, E, G, I-L; 50 μm in B, D, F, H.
Development of ectopic neural structures was not restricted to the cephalic region, but was observed all along the antero-posterior axis between the pre-existing VNCs. Structures resembling ganglia of the VNCs developed in the medial region of the tail (Fig. 7K, arrowheads). These ectopic structures were never observed in control animals (Fig. 7I). In contrast to this ectopic neural development along the midline of intact slit RNAi planarians, abnormalities in the digestive system were not observed. Even in those animals in which ectopic neural structures were observed, the gut pattern was unaffected (compare Figs. 7J and L); neither transverse connections between the two posterior gut branches nor ectopic gut branches at the midline were detected after slit RNAi.
Discussion
We identified a slit gene from the planarian Schmidtea mediterranea based upon the similarity of its predicted product to Slit proteins from other organisms. Given its conserved expression pattern, repulsive function at the midline, and domain structure of its product, this gene appears to be a planarian orthologue of slit. Like slit genes from other organisms, Smed-slit mRNA is expressed along the midline of the planarian body, in both dorsal and ventral domains. We observed different populations of Smed-slit-expressing cells along the midline: some were located at the level of the submuscular plexus, whereas others were observed deeper in the mesenchyme. In Drosophila and mouse slit genes are expressed by glial cells at the midline. Glia-like, neuroaccessory cells in planarians have been observed by electron microscopy (Morita and Best, 1966), but we currently lack specific markers for such cells. Recent work has shown that DjPiwi-1, a piwi homologue, is expressed in a sub-population of neoblasts at the dorsal midline in the planarian Dugesia japonica (Rossi et al., 2006). This population is sensitive to gamma-irradiation, whereas, the Smed-slit expressing cells are radiation-insensitive (data not shown). Radiation sensitivity is a hallmark of the planarian neoblasts (Reddien et al., 2005a,b; Salvetti et al., 2005; Guo et al., 2006); thus, Smed-slit does not appear to be expressed in the DjPiwi-1-expressing neoblasts and the exact nature of the cells that express Smed-slit remains to be determined.
Slit proteins play critical roles in axon guidance in both vertebrates and invertebrates, acting as repulsive cues for commissural axons at the midline (reviewed in Brose and TessierLavigne, 2000; Wong et al., 2002; Dickson and Gilestro, 2006) and for cell migration (Hu, 1999; Wu et al. 1999; Kramer et al., 2001; Santiago-Martínez et al., 2006). Our results suggest that such a repulsive role for slit at the midline has been conserved in planarians. In Smed-slit RNAi knockdown animals, the regenerating axon tracts of the ventral nerve cords (Figs. 3L and 5B) and the cell bodies of the regenerated cephalic ganglia (Figs. 3J, K) collapse at the midline, instead of forming the well-defined, bilateral nerve cords and cephalic ganglia that are normally observed (Figs. 3G, I). Similarly, the photoreceptors regenerate at the midline after Smed-slit RNAi (Figs. 3D, 4B), rather than forming bilaterally (Figs. 3A, 4A). The appearance of photoreceptor precursors and cephalic ganglia primordia (Fig. 4F) at the midline of Smed-slit RNAi knockdown planarians during the initial stages of regeneration suggests that slit cues may normally serve to guide these cells away from the midline, thereby generating the bilaterally symmetric photoreceptors and CNS.
Analysis of slit mRNA expression reveals that slit-expressing cells are in close proximity to the axons that project during the initial stages of anterior regeneration (Fig. 2D). As regeneration proceeds, slit-positive cells appear to underlie the growing anterior commissure (Fig. 2E). Thus, slit may be acting to “channel” the growing anterior commissure during regeneration, in a manner similar to Slit1 and Slit2 function in thalamocortical and corticothalamic axons (Bagri et al., 2002) and retinal ganglion cell axons (Plump et al., 2002) in mice, as well as Slit2 and Slit3 function in the postoptic commissure in zebrafish (Barresi et al., 2005).
Smed-slit does not appear to be required for the proper projection and positioning of all neurons. For example, the lateral branches of the cephalic ganglia seem to differentiate normally and project towards the head periphery after Smed-slit RNAi (Figures 3G and 3J). Likewise, the distribution of cintillo-positive cells, putative sensory cells distributed in the head periphery around the cephalic ganglia (Oviedo et al., 2003), appears unaltered in Smed-slit RNAi knockdowns animals (data not shown). Thus, the position and projection of these neurons are unlikely to be regulated by Smed-slit function.
In addition to defects in the architecture of the regenerated nervous system, Smed-slit RNAi knockdown planarians also show defects in the patterning of the regenerated posterior gut. The regenerated posterior gut branches connect aberrantly at the midline, instead of forming two independent, longitudinally running tracts (Fig. 6D). This improper development of the posterior gut is most dramatic in lateral regenerates after longitudinal amputation, in which a single gut branch is formed along the midline, posterior to the pharynx (Fig. 6F, arrow). Smed-slit mRNA is expressed in the mesenchymal space between the two gut branches in intact animals (Fig. 1C) and during posterior regeneration (Figs. 2H,I). These expression data, combined with our functional analysis, suggest that Smed-slit acts as a repulsive cue on the posterior gut; Smed-slit expression is likely to guide the posteriorly directed, regenerating gut branches away from the midline. Slit function in non-neuronal tissues is well documented, guiding heart cell and salivary gland migration in Drosophila (Kolesnikov and Beckendorf, 2005; Qian et al., 2005; MacMullin and Jacobs, 2006; Santiago-Martínez et al., 2006), as well as playing roles in angiogenesis, kidney, and mammary gland development (Piper et al., 2000; Wang et al., 2003; Klagsbrun and Eichmann, 2005; Strickland et al., 2006).
Slit proteins serve as ligands for the Roundabo ut (Robo) family of receptors (Brose et al., 1999; Kidd et at. 1999). We recently identified two planarian robo homologues (Cebrià and Newmark, 2007). However, RNAi experiments suggested that neither of these two robo genes is likely to encode the receptor for Smed-slit: the phenotypes obtained from robo RNAi knockdowns are distinct from those reported here in both neural and non-neural tissues (Cebriàand Newmark, 2007). These results suggest that additional robo gene(s) likely exist in the planarian genome; it is also possible that some function(s) of Smed-slit may be mediated independently of robo. For example, in the Drosophila heart, a slit mutant lacking the leucine-rich repeats required for Robo binding can restore normal heart morphology (MacMullin and Jacobs, 2006). Furthermore, mutations affecting the function of Integrins and Integrin ligands act as dominant enhancers of the heart assembly phenotype in slit transheterozygotes, while such interactions are not observed in the heart with mutations affecting robo and its downstream signaling components (MacMullin and Jacobs, 2006).
Our results suggest that Smed-slit encodes an important component of midline signaling in planarians that is required for restoring the proper pattern of the nervous system and posterior gut. However, other factors are likely to be involved in production of the midline signal(s) during planarian regeneration. The phenotypic analysis reported here indicated that the dorsal strip of ciliated epithelial cells at the midline (Robb and Sánchez Alvarado, 2002) appeared to regenerate normally after Smed-slit RNAi (Fig. 3F). This result suggests that the initial specification of the midline occurs in Smed-slit knockdown animals and that Smed-slit may function downstream of other midline cues. One possible candidate for such a cue is a BMP family member: in the planarian Dugesia japonica a BMP homologue is expressed along the dorsal midline (Orii et al., 1998). Although no RNAi phenotype has yet been described for this gene, the large-scale RNAi screen of Reddien et al. (2005a) reported that RNAi knockdown of either bmp-1 or smad4 homologues produced indented blastemas, indicative of possible defects in midline signaling.
Perhaps the most dramatic and unexpected aspect of the Smed-slit RNAi phenotype is the development of ectopic neuronal tissues at the midline of uninjured portions of regenerating (Fig. 5) and intact (Fig. 7) planarians. Because we do not observe obvious defects in the pre-existing neural structures, it seems unlikely that the ectopic neurons are derived from these differentiated tissues. Instead, we suggest that the most likely source of the ectopic neurons is neoblasts that have adopted neuronal identities and differentiated at the midline. As the only proliferating somatic cells in the planarian, neoblasts give rise to missing cells during regeneration and to new differentiated cells during the course of growth and tissue maintenance (Baguñà et al., 1989; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005b). In Smed-slit knockdown planarians, newly born neurons in uninjured regions or in intact animals no longer integrate properly into the bilateral nerve cords, cephalic ganglia, and photoreceptors; instead, they form these structures at or near the midline. Further analyses will be required to determine if these ectopic neurons arise from: (i) neural precursors generated near the midline that would normally migrate away from midline-derived, slit-mediated repulsive cues; and/or (ii) more widely distributed neural precursors that would normally be prevented from migrating toward the midline by slit-mediated repulsive cues. Whether the ectopic neurons at the midline represent the typical amount of cell turnover observed in the planarian nervous system or are due to neuronal hyperplasia also remains to be resolved. Whatever the origin of these ectopic neural tissues, they clearly implicate Smed-slit function in the maintenance of proper nervous system architecture in intact planarians, in addition to its role as a midline cue during regeneration.
Supplementary Material
Acknowledgements
We would like to thank: David Forsthoefel and Ricardo Zayas for comments on the manuscript; Alejandro Sánchez Alvarado for providing EST clone H.10.2f from the asexual strain; and Kiyokazu Agata for providing the VC-1 mAb. Planarian genomic sequence data were generated by the Washington University Genome Sequencing Center in St. Louis. F.C. was supported by a Long-Term Fellowship from EMBO and the programme Beatriu de Pinós of the Generalitat de Catalunya. This work was supported by NIH R01 HD43403 and NSF CAREER Award IBN-0237825 to P.A.N. P.A.N. was a Damon Runyon Scholar supported by the Damon Runyon Cancer Research Foundation (DRS 33-03).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agata K. Regeneration and gene regulation in planarians. Curr. Op. Gen. Dev. 2003;13:492–496. doi: 10.1016/j.gde.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Agata K, Soejima Y, Kato K, Kobayashi C, Umesono Y, Watanabe K. Structure of the planarian central nervous system (CNS) revealed by neuronal cell markers. Zool. Sci. 1998;15:433–440. doi: 10.2108/zsj.15.433. [DOI] [PubMed] [Google Scholar]
- Bagri A, Marin O, Plump AS, Mak J, Pleasure SJ, Rubenstein JL, Tessier-Lavigne M. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron. 2002;33:233–248. doi: 10.1016/s0896-6273(02)00561-5. [DOI] [PubMed] [Google Scholar]
- Baguñà J, Saló E, Auladell C. Regeneration and pattern formation in planarians. III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development. 1989;107:77–86. [Google Scholar]
- Barresi MJ, Hutson LD, Chien CB, Karlstrom RO. Hedgehog regulated Slit expression determines commissure and glial cell position in the zebrafish forebrain. Development. 2005;132:3643–3656. doi: 10.1242/dev.01929. [DOI] [PubMed] [Google Scholar]
- Benazzi M, Ballester R, Baguñà J, Puccinelli I. The fissiparous race of the planarian Dugesia lugubris S.L. found in Barcelona (Spain) belongs to the biotype G: comparative analysis of the karyotypes. Caryologia. 1972;25:59–68. [Google Scholar]
- Brose K, Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Cur. Op. Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionary conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Newmark PA. Morphogenesis defects are associated with abnormal nervous system regeneration after roboA RNAi in planarians. Development. 2007;134:833–837. doi: 10.1242/dev.02794. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Agata K. Dissecting planarian CNS regeneration by the expression of neural-specific genes. Dev. Growth Differ. 2002a;44:135–146. doi: 10.1046/j.1440-169x.2002.00629.x. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sánchez Alvarado A, Agata K. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002b;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- Dickson BJ, Gilestro GF. Regulation of commissural pathfinding by slit and its Robo receptors. Annu. Rev. Cell Dev. Biol. 2006;22:651–675. doi: 10.1146/annurev.cellbio.21.090704.151234. [DOI] [PubMed] [Google Scholar]
- Fusaoka E, Inoue T, Minetas K, Agata K, Takeuchi K. Structure and function of primitive immunoglobulin superfamily neural cell adhesion molecules: a lesson from studies on planarian. Genes to Cells. 2006;11:541–555. doi: 10.1111/j.1365-2443.2006.00962.x. [DOI] [PubMed] [Google Scholar]
- Guo T, Peters AHFM, Newmark PA. A bruno-like gene is required for stem cell maintenance in planarians. Developmental Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Hao JC, Yu TW, Fujisawa K, Culotti JG, Gengyo-Ando K, Mitani S, Moulder G, Barstead R, Tessier-Lavigne M, Bargmann CI. C. elegans slit acts in midline, dorso-ventral, and antero-posterior guidance via the sax-3/robo receptor. Neuron. 2001;32:25–38. doi: 10.1016/s0896-6273(01)00448-2. [DOI] [PubMed] [Google Scholar]
- Hu H. Chemorepulsion of neuronal migration by slit2 in the developing mammalian forebrain. Neuron. 1999;23:703–711. doi: 10.1016/s0896-6273(01)80029-5. [DOI] [PubMed] [Google Scholar]
- Hyman LH. The Invertebrates: Platyhelminthes and Rhynchocoela. McGraw-Hill Book Company; New York: 1951. [Google Scholar]
- Inoue T, Kumamoto H, Okamoto K, Umesono Y, Sakai M, Sánchez Alvarado A, Agata K. Morphological and functional recovery of the planarian photosensing system during head regeneration. Zool. Sci. 2004;21:275–283. doi: 10.2108/zsj.21.275. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the Robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Klagsbrun M, Eichmann A. A role for axon guidance receptors and ligands in blood vessel development and tumor angiogenesis. Cytokine Growth Factor Rev. 2005;16:535–548. doi: 10.1016/j.cytogfr.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Kolesnikov T, Beckendorf SK. NETRIN and SLIT guide salivary gland migration. Dev. Biol. 2005;282:102–111. doi: 10.1016/j.ydbio.2005.04.037. [DOI] [PubMed] [Google Scholar]
- Kramer SG, Kidd T, Simpson JH, Goodman CS. Switching repulsion to attraction: changing response to Slit during transition in mesoderm migration. Science. 2001;292:737–740. doi: 10.1126/science.1058766. [DOI] [PubMed] [Google Scholar]
- Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M. Conserved roles for slit and robo proteins in midline commissural axon guidance. Neuron. 2004;42:213–223. doi: 10.1016/s0896-6273(04)00179-5. [DOI] [PubMed] [Google Scholar]
- MacMullin A, Jacobs JR. Slit coordinates cardiac morphogenesis in Drosophila. Dev. Biol. 2006;293:154–164. doi: 10.1016/j.ydbio.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Mannini L, Rossi L, Deri P, Gremigni V, Salvetti A, Saló E, Batistoni R. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev. Biol. 2004;269:346–359. doi: 10.1016/j.ydbio.2004.01.042. [DOI] [PubMed] [Google Scholar]
- Morita M, Best JB. Electron microscopic studies of Planaria. 3. Some observations on the fine structure of planarian nervous tissue. J. Exp. Zool. 1966;161:391–411. doi: 10.1002/jez.1401610307. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 2000;220:142–53. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nat. Rev. Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Reddien PW, Cebrià F, Sánchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl1):11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Takeuchi K, Agata K. Neural projections in planarian brain revealed by fluorescent dye tracing. Zool. Sci. 2005;22:535–546. doi: 10.2108/zsj.22.535. [DOI] [PubMed] [Google Scholar]
- Orii H, Kato K, Agata K, Watanabe K. Molecular cloning of bone morphogenetic protein (BMP) gene from the planarian Dugesia japonica. Zool. Sci. 1998;15:871–877. [Google Scholar]
- Oviedo NJ, Newmark PA, Sánchez Alvarado A. Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev. Dyn. 2003;226:326–333. doi: 10.1002/dvdy.10228. [DOI] [PubMed] [Google Scholar]
- Pineda D, González J, Callaerts P, Ikeo K, Ghering WJ, Saló E. Searching for the prototypic eye genetic network: sine oculis is essential for eye regeneration in planarians. Proc. Natl. Acad. Sci. USA. 2000;97:4525–4529. doi: 10.1073/pnas.97.9.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Georgas K, Yamada T, Little M. Expression of the vertebrate Slit gene family and their putative receptors, the Robo genes, in the developing murine kidney. Mech. Dev. 2000;94:213–217. doi: 10.1016/s0925-4773(00)00313-0. [DOI] [PubMed] [Google Scholar]
- Plump AS, Erskine L, Sabatier C, Brose K, Epstein CJ, Goodman CS, Mason CA, Tessier-Lavigne M. Slit1 and slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron. 2002;33:219–232. doi: 10.1016/s0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Slit and Robo control cardiac cell polarity and morphogenesis. Curr. Biol. 2005;15:2271–2278. doi: 10.1016/j.cub.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell. 2005a;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005b;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reuter M, Gustafsson MK. The flatworm nervous system: pattern and phylogeny. In: Breibach O, Kutsch W, editors. The nervous system of invertebrates: an evolutionary and comparative approach. Birkäuser Verlag; Basel: 1995. pp. 25–59. 1995. [Google Scholar]
- Reuter M, Sheiman IM, Gustafsson MK, Halton DW, Maule AG, Shaw C. Development of the nervous system in Dugesia tigrina during regeneration after fission and decapitation. Invert. Reprod. Dev. 1996;29:199–211. [Google Scholar]
- Robb SMC, Sánchez Alvarado A. Identification of immunological reagents for use in the study of freshwater planarians by jeans of whole-mount immunofluorescence and confocal microscopy. Genesis. 2002;32:293–298. doi: 10.1002/gene.10087. [DOI] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Lena A, Batistoni R, Deri P, Pugliesi C, Loreti E, Gremigni V. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev. Genes Evol. 2006;216:335–346. doi: 10.1007/s00427-006-0060-0. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Hartley DA, Walther Z, Artavanis-Tsakonas S. Slit: an EGFhomologous locus of D. melanogaster involved in the development of the embryonic central nervous system. Cell. 1988;55:1047–1059. doi: 10.1016/0092-8674(88)90249-8. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. Slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- Sakai F, Agata K, Orii H, Watanabe K. Organization and regeneration ability of spontaneous supernumerary eyes in planarians-Eye regeneration field and pathway selection by optic nerves. Zool. Sci. 2000;17:375–381. doi: 10.2108/jsz.17.375. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–184. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A. Planarian regeneration: its end is its beginning. Cell. 2006;124:241–245. doi: 10.1016/j.cell.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl. Acad. Sci. USA. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA, Robb SMC, Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Santiago-Martínez E, Soplop NH, Kramer SG. Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proc. Natl. Acad. Sci. USA. 2006;103:12441–12446. doi: 10.1073/pnas.0605284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland P, Shin GC, Plump A, Tessier-Lavigne M, Hinck L. Slit2 and netrin1 act synergistically as adhesive cues to generate tubular bi-layers during ductal morphogenesis. Development. 2006;133:823–832. doi: 10.1242/dev.02261. [DOI] [PubMed] [Google Scholar]
- Tazaki A, Gaudieri S, Ikeo K, Gojobori T, Watanabe K, Agata K. Neural network in planarian revealed by an antibody against planarian synaptotagmin homologue. Bioch. Biophys. Res. Comm. 1999;260:426–432. doi: 10.1006/bbrc.1999.0933. [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K. A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev. Growth Differ. 1997;39:723–727. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K. Distinct structural domains in the planarian brain defined by the expression of evolutionary conserved homeobox genes. Dev. Genes Evol. 1999;209:31–39. doi: 10.1007/s004270050224. [DOI] [PubMed] [Google Scholar]
- Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4:19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Wong K, Park HT, Wu JY, Rao Y. Slit proteins: molecular guidance cues for cells ranging from neurons to leukocytes. Curr. Op. Gen. Dev. 2002;12:583–591. doi: 10.1016/s0959-437x(02)00343-x. [DOI] [PubMed] [Google Scholar]
- Wu JY, Feng L, Park HT, Havlioglu N, Wen L, Tang H, Bacon KB, Jiang ZH, Zhang XC, Rao Y. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Wong K, Chen J, Jiang Z, Dupuis S, Wu JY, Rao Y. Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature. 1999;400:331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas RM, Hernández A, Habermann B, Wang Y, Stary J, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: Analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. USA. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


