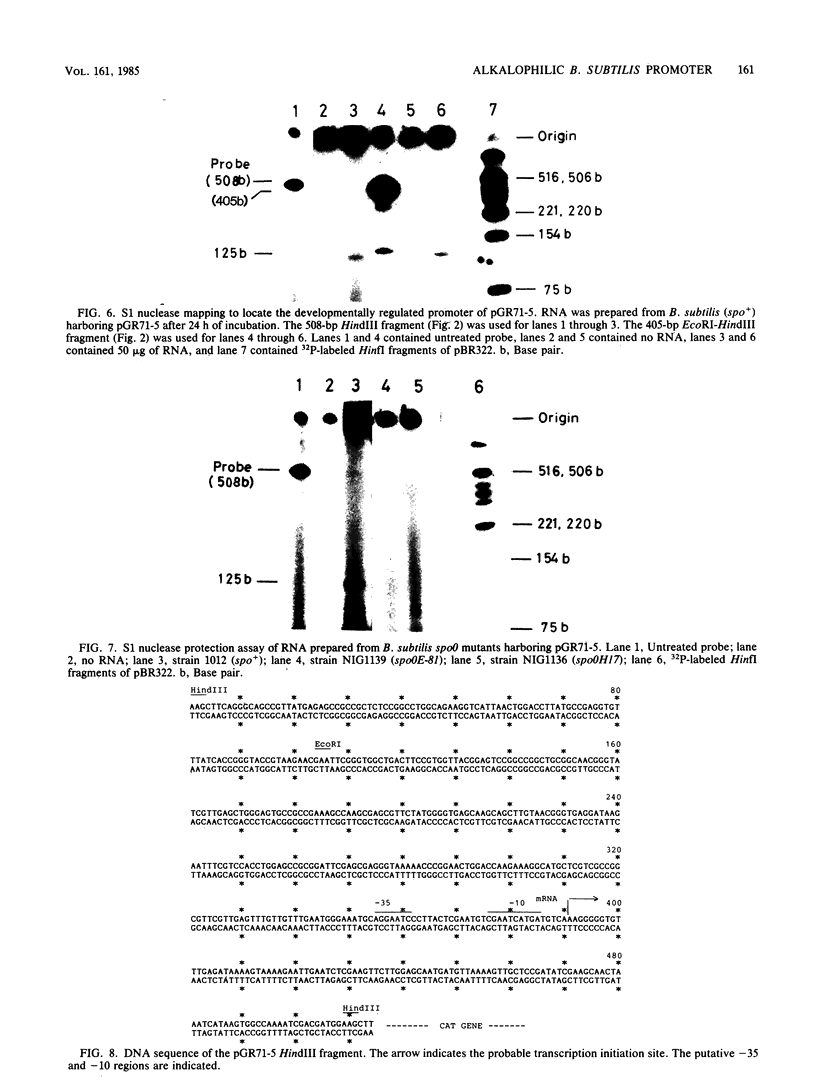
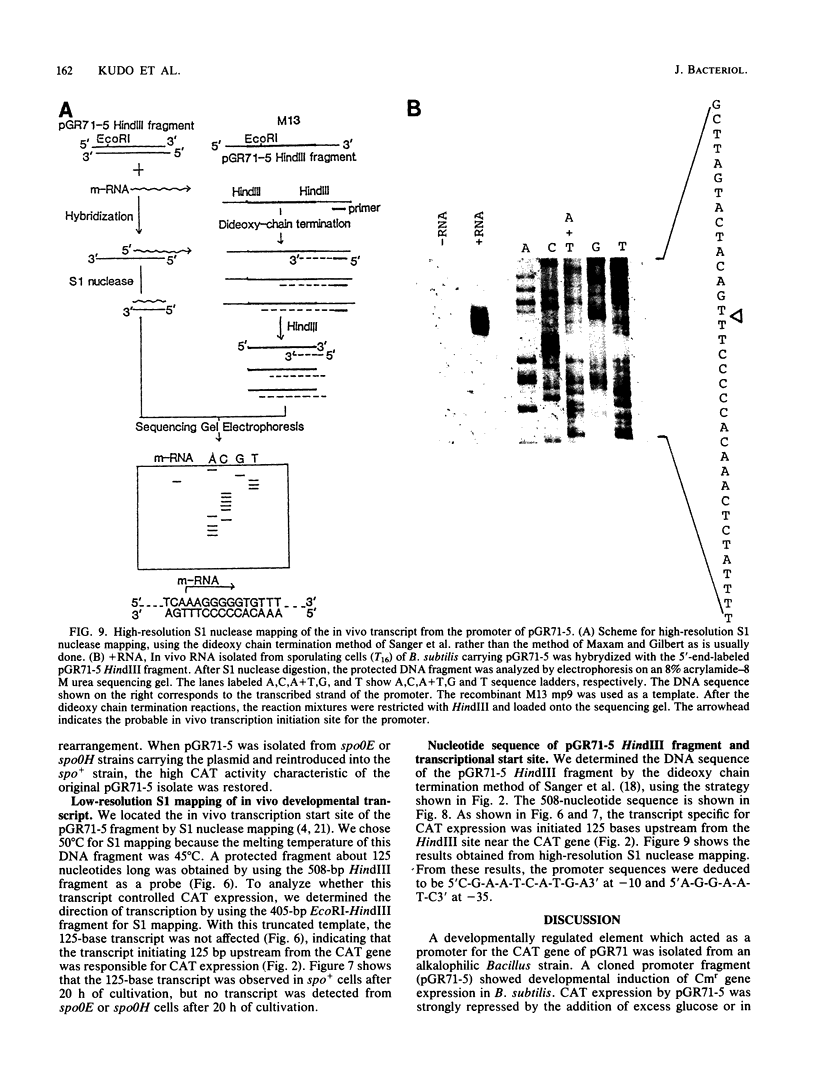
Abstract
An alkalophilic Bacillus DNA bank cloned in an expression probe plasmid, pGR71, was screened for the presence of developmentally regulated genetic elements. A 508-base pair HindIII fragment isolated from this bank in plasmid pGR71-5 expressed plasmid-encoded chloramphenicol resistance only during the sporulation phase of a Bacillus subtilis host grown on Schaeffer medium. This developmentally regulated expression was altered in spo0E and spo0H mutants which had very low levels of chloramphenicol acetyltransferase activity relative to the wild type or other spo0 mutants. We determined the nucleotide sequence of the entire 508-base pair fragment and located the site of regulated transcription initiation by high-resolution S1 nuclease mapping of the in vivo transcript. The deduced promoter sequences upstream from this start site were 5'C-G-A-A-T-C-A-T-G-A3' at -10 and 5' A-G-G-A-A-T-C3' at -35. This transcript was not detected in spo0E or spo0H mutants, indicating that the products of these genes control developmentally regulated chloramphenicol acetyltransferase expression at the level of transcription.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Doi R. H. Multiple RNA polymerase holoenzymes exert transcriptional specificity in Bacillus subtilis. Arch Biochem Biophys. 1982 Apr 1;214(2):772–781. doi: 10.1016/0003-9861(82)90084-4. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Goldfarb D. S., Doi R. H., Rodriguez R. L. Expression of Tn9-derived chloramphenicol resistance in Bacillus subtilis. Nature. 1981 Sep 24;293(5830):309–311. doi: 10.1038/293309a0. [DOI] [PubMed] [Google Scholar]
- Goldfarb D. S., Rodriguez R. L., Doi R. H. Translational block to expression of the Escherichia coli Tn9-derived chloramphenicol-resistance gene in Bacillus subtilis. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5886–5890. doi: 10.1073/pnas.79.19.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb D. S., Wong S. L., Kudo T., Doi R. H. A temporally regulated promoter from Bacillus subtilis is transcribed only by an RNA polymerase with a 37,000 dalton sigma factor. Mol Gen Genet. 1983;191(2):319–325. doi: 10.1007/BF00334833. [DOI] [PubMed] [Google Scholar]
- Honda H., Kato C., Kudo T., Horikoshi K. Cloning of leucine genes of alkalophilic Bacillus No. 221 in E. coli and B. subtilis. J Biochem. 1984 May;95(5):1485–1490. doi: 10.1093/oxfordjournals.jbchem.a134756. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S., Lovett P. S. Selective expression of a plasmid cat gene at a late stage of Bacillus subtilis sporulation. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3457–3460. doi: 10.1073/pnas.81.11.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollington J. F., Haldenwang W. G., Huynh T. V., Losick R. Developmentally regulated transcription in a cloned segment of the Bacillus subtilis chromosome. J Bacteriol. 1981 Aug;147(2):432–442. doi: 10.1128/jb.147.2.432-442.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sadaie Y., Kada T. Formation of competent Bacillus subtilis cells. J Bacteriol. 1983 Feb;153(2):813–821. doi: 10.1128/jb.153.2.813-821.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashihara N., Kudo T., Horikoshi K. Molecular cloning and expression of cellulase genes of alkalophilic Bacillus sp. strain N-4 in Escherichia coli. J Bacteriol. 1984 May;158(2):503–506. doi: 10.1128/jb.158.2.503-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Wong S. L., Price C. W., Goldfarb D. S., Doi R. H. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1184–1188. doi: 10.1073/pnas.81.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]