Abstract
Cleavage of the hemagglutinin (HA) molecule by proteases is a prerequisite for the infectivity of influenza A viruses. Here, we describe a novel mechanism of HA cleavage for a descendant of the 1918 pandemic strain of human influenza virus. We demonstrate that neuraminidase, the second major protein on the virion surface, binds and sequesters plasminogen, leading to higher local concentrations of this ubiquitous protease precursor and thus to increased cleavage of the HA. The structural basis of this unusual function of the neuraminidase molecule appears to be the presence of a carboxyl-terminal lysine and the absence of an oligosaccharide side chain at position 146 (N2 numbering). These findings suggest a means by which influenza A viruses, and perhaps other viruses as well, could become highly pathogenic in humans.
Influenza A viruses possess two envelope glycoproteins: hemagglutinin (HA) and neuraminidase (NA). The HA, a type I membrane protein containing an N-terminal ectodomain and a C-proximal anchor, binds to cell surface sialylglycoconjugates and mediates virus attachment to target cells (1). It is synthesized as a single polypeptide and then is cleaved into HA1 and HA2 subunits by host proteases (2). The NA, a type II membrane protein containing an N-proximal anchor and a C-terminal ectodomain, cleaves the α-glycosidic linkage between sialic acid and an adjacent sugar residue, facilitating elution of virus progeny from infected cells and preventing self-aggregation of the virus (3, 4).
HA cleavage at the virion surface is essential for the infectivity of influenza viruses (5, 6); hence, both the susceptibility of the HA to cellular proteases and the distribution of these enzymes among host tissues are key determinants of viral pantropicity. Although well described in birds (2), the molecular mechanisms that give rise to highly pathogenic influenza viruses are poorly understood in human hosts. Among human influenza viruses, only the A/WSN/33 (H1N1)(WSN) strain can replicate in a variety of cultured cells without the addition of trypsin (7), suggesting a unique mechanism of HA cleavage. The WSN mutant, which was selected by mouse brain passage of the A/WS/33 (H1N1)(WS) virus, a descendant of the virus responsible for the 1918 pandemic, replicates in a range of murine tissues, including brain (8). Genetic studies indicate that the WSN NA molecule contributes critically to HA cleavage and thus to viral pantropicity (9). Moreover, Lazarowitz et al. (10) showed that serum plasminogen (upon activation by a cellular plasminogen activator) is the main source of protease responsible for WSN HA cleavage in cell cultures lacking exogenous trypsin. Here, we demonstrate that the WSN NA can sequester plasminogen, providing a mechanism for enhanced HA cleavage and greater virulence by this pantropic influenza A virus.
MATERIALS AND METHODS
Viruses and Cells.
All influenza viruses used in this study were obtained from the repository at St. Jude Children’s Research Hospital. A reassortant, WSN–HK, which possesses the NA gene from A/Hong Kong/1/68 (H3N2) and the remainder of its genes from the WSN strain, was obtained from M. Ueda (SRL, Tokyo, Japan). Madin–Darby bovine kidney (MDBK) cells were grown in MEM supplemented with 10% fetal calf serum (FCS), l-glutamine (GIBCO/BRL), vitamin and amino acid solutions (GIBCO), and penicillin–streptomycin solution (Sigma). 293T cells (constitutively expressing the simian virus 40 large T antigen) were cultured in high-glucose DMEM containing 10% FCS, l-glutamine, and antibiotics.
Plasmids.
Full-length cDNAs encoding the HA and NA genes of influenza viruses were cloned as described before (11) and then subcloned into a mammalian expression vector, pCAGGS, containing the chicken β-actin promoter (12).
Determination of HA Cleavage.
293T cells growing in 6-well tissue culture plates were transfected with 2 μg of expression plasmids using Lipofectamine reagent (GIBCO/BRL) and then were cultured in OPTI-MEM I medium (GIBCO/BRL) without FCS for 24 h. Transfected cells were starved in methionine and cysteine-deficient medium for 30 min and labeled with 100 μCi of Tran35S (ICN) for 10 min. After 1 h of chase, the cells were lysed in 50 mM Tris⋅HCl (pH7.2), 0.6 M KCl, and 0.5% Triton X-100 and clarified by centrifugation. Cell lysates were mixed with monoclonal antibodies to WSN HA and then protein A–Sepharose (Pharmacia). Precipitated proteins were analyzed by SDS/PAGE and fluorography.
Polykaryon Formation.
Transfected cells were incubated in OPTI-MEM I medium containing FCS, bovine plasminogen, or human plasminogen (Sigma) for 30 min at 37°C. Cells were then treated with PBS (pH 5.0) for 3 min and incubated in OPTI-MEM I for 6 h at 37°C. They were fixed and stained with Giemsa solution. For inhibition of polykaryon formation, we added an inhibitor specific for plasmin/plasminogen, 6-amino-n-hexanoic acid (Sigma), to the medium at 1 mg/ml.
Detection of Plasminogen Binding to the NA by Fluorescence-Activated Cell Sorting.
Transfected or infected cells were suspended in PBS and incubated with human plasminogen, 100 μg/ml or 10 μg/ml, respectively, for 30 min on ice. Bound plasminogen was detected by flow cytometry using rabbit anti-human plasminogen antibody (Boehringer Mannheim) and fluorescein isothiocyanate-labeled anti-rabbit antibody (Boehringer Mannheim).
RESULTS
Previous studies showed that plasmin cleaves the influenza virus HA into HA1 and HA2 (10). However, because plasmin is invariably associated with a plasmin inhibitor (α2-antiplasmin) in tissue fluids (13), we considered that plasminogen, whose conversion to plasmin would promote HA cleavage, might be a key factor in the virulence of the WSN virus. Indeed, plasminogen is involved in HA cleavage of the WSN strain in cell culture (10). To confirm whether the ability of WSN to undergo multiple cycles of replication in tissue culture without trypsin is due to HA cleavage by residual plasminogen derived from serum in the growth medium, we performed plaque assays after extensive washing of MDBK cells. Plaque formation was apparent only when 0.1% FCS was added to the agar medium. Moreover, plaque formation in the presence of FCS was inhibited by the addition of a lysine analog, 6-amino-n-hexanoic acid, which specifically inhibits plasminogen and plasmin.
To determine whether multiple cycles of replication in the presence of FCS is unique to the WSN virus, we tested replication of eight other influenza viruses in MDBK cells by titrating their egg-grown stocks (all had at least 1 × 106 50% egg infectious doses/ml) in the presence or absence of FCS or trypsin. Replication of these viruses was tested in liquid culture because some of them did not produce plaques in these cells, even though they underwent multiple cycles of replication in the presence of trypsin (Table 1). Results showed that whereas the WSN virus replicated 10-fold better in the presence of trypsin than FCS, all other viruses tested reached at least 1,000-fold higher titers in the presence of trypsin than FCS. These results suggested that plasminogen derived from residual FCS in culture medium was critical for multiple cycles of replication of the WSN virus in these cells. Because some bacteria (e.g., group A streptococci) contain plasminogen-binding proteins that can increase local concentrations of plasmin, leading to more efficient tissue invasion (14), we postulated that the NA surface protein mediates cleavage of the WSN HA through sequestration of plasminogen.
Table 1.
Dependency of WSN virus replication on FCS in MDBK cells
| Virus | Virus titers (log10/ml) in the presence of
|
||
|---|---|---|---|
| Trypsin (0.5 μg/ml) | 0.1% FCS | None | |
| A/WSN/33 (H1N1) | 6* | 5 | 1 |
| A/WS/33 (H1N1) | 5 | <1 | <1 |
| WSN-HK (H1N2) | 6 | 2 | 1 |
| A/duck/Ukraine/1/63 (H3N8) | 6 | 2 | 2 |
| A/Los Angeles/2/87 (H3N2) | 4 | 1 | 1 |
| A/duck/Hokkaido/8/80 (H3N8) | 4 | 1 | 1 |
| A/mallard/Alberta/22/76 (H3N6) | 4 | <1 | <1 |
| A/Udorn/307/72 (H3N2) | 6 | 2 | 2 |
| A/duck/Hong Kong/342/78 (H5N2) | 4 | <1 | <1 |
MDBK cells were washed five times with medium without FCS before virus infection. Cells were then infected with egg-grown stock viruses serially diluted 10-fold. After 1-h incubation, cells were then washed five times with medium without FCS and incubated with or without medium containing trypsin (0.5 μg/ml) or 0.1% FCS.
Reciprocal of the highest dilution of virus stock at which virus growth was detected by HA.
The WSN NA, but not Other NAs, Facilitates Plasminogen-Mediated HA Cleavage.
To verify involvement of the NA and plasminogen in cleavage of the WSN HA, we first expressed either or both surface proteins in the presence or absence of trypsin as well as 1% FCS or bovine or human plasminogen. When expressed alone, the HA was cleaved in the presence of trypsin, but not FCS or either of the two plasminogens (Fig. 1). However, when coexpressed with the WSN NA, it was cleaved into HA1 and HA2 subunits in the presence of FCS or either plasminogen. Under these conditions, HA cleavage occurred at the authentic site, as indicated by the massive polykaryon formation that occurred when cells coexpressing both the HA and NA were incubated at pH 5.0 (Fig. 2). Cleavage was inhibited by a plasmin-specific inhibitor, the lysine analog 6-amino-n-hexanoic acid, confirming the involvement of plasmin. The minimal concentration of human plasminogen required for HA cleavage (hence, polykaryon formation) was 5 μg/ml when the HA and NA were coexpressed, whereas even a 1,000-fold higher concentration of plasminogen did not result in cleavage when only the HA was expressed (Table 2).
Figure 1.
HA cleavage mediated by the WSN NA. 293T cells were transfected with a plasmid expressing the WSN HA alone, the WSN HA and WSN NA, or the WSN HA and the NA from A/Hong Kong/1/68 (HK) (H3N2). Cells were labeled with [35S]methionine at 24 h after transfection for 10 min and chased for 1 h. They were then incubated with trypsin (0.5 μg/ml), 1% FCS, or bovine (50 μg/ml) or human (5 μg/ml) plasminogen for 30 min. HAs were then immunoprecipitated from cell lysates by monoclonal antibodies to the WSN HA.
Figure 2.
Polykaryon formation by cells expressing HA or both the HA and NA. 293T cells were transfected with plasmids expressing the WSN HA (a–c), WSN HA and WSN NA (d–f) or WSN HA and HK NA (g–i). At 24-h posttransfection, the cells were treated with 0.5 μg/ml trypsin (a, d, and g), 1% FCS (b, e, and h), or 5 μg/ml human plasminogen (c, f, and i) for 30 min. After exposure to PBS (pH 5.0), the cells were then incubated for 6 h, fixed with 3% formaldehyde, and stained with Giemsa solution.
Table 2.
Minimal concentration of human plasminogen required for NA-mediated polykaryon formation by the HA
| Expressed protein* | Concentration of human plasminogen, μg/ml
|
Trypsin, μg/ml
|
||||
|---|---|---|---|---|---|---|
| 5000 | 500 | 50 | 5 | 0.5 | 0.5 | |
| HA/WSN NA | NT | NT | NT | + | − | + |
| HA/WSN NA− | NT | + | + | + | − | + |
| HA/WSN NA R146N | NT | + | + | − | − | + |
| HA only | − | − | NT | NT | NT | + |
Polykaryon formation was determined as described in the legend to Fig. 2. −, no polykaryon formation; +, 100% polykaryon formation as shown in Fig. 2; NT, not tested.
NA− denotes a mutant with loss of NA enzymatic activity due to Glu-to-Gln change at position 227. The R146N mutant possesses a potential glycosylation site at position 146.
We next established the specificity of plasminogen-mediated HA cleavage for the WSN virus by examining the NAs of human, swine, and avian viruses in experiments similar to those depicted in Figs. 1 and 2. None of the NAs from 10 different viruses, including the parent strain of the WSN strain A/WS/33 (H1N1), was able to promote HA cleavage, and thus polykaryon formation, in the presence of FCS or either bovine or human plasminogen [see, for example, A/Hong Kong/1/68 (H3N2) in Figs. 1 and 2]. Together, these results indicate that sequestration of plasminogen by the NA protein, leading to enhanced HA cleavage, is a unique property of the WSN influenza A virus.
Is the enzymatic activity of the NA a requirement for plasminogen-mediated HA cleavage? To address this question, we constructed a NA with a Glu-to-Gln substitution at position 227 (15), a change that abolishes detectable enzymatic activity (unpublished data). As shown in Table 2, the mutant functioned as well as the wild-type WSN NA in plasminogen-mediated HA cleavage.
To determine whether plasminogen-mediated cleavage occurs with other HAs, we coexpressed the A/Udorn/307/72 (H3N2) HA and the WSN NA in the presence of plasminogen and measured polykaryon formation upon exposure of cells to low pH. A/Udorn/307/72 (H3N2) induced an equivalent level of polykaryon formation as the WSN HA did under these conditions (data not shown), indicating that plasminogen-mediated cleavage in the presence of the WSN NA is not restricted to the WSN HA, but extends to other HAs.
Structural Basis of the WSN NA for Facilitating HA Cleavage.
What is the molecular basis for the unique activity of the WSN NA that promotes plasminogen-mediated HA cleavage? Some plasminogen-binding proteins possess a carboxyl-terminal lysine that is critical for plasminogen binding (16–18). Among all influenza A virus NAs studied to date, only those of the N1 subtype to which the WSN strain belongs contain this residue. We therefore tested the role of the carboxyl-terminal lysine at position 453 in plasminogen-mediated HA cleavage by mutating it to arginine (K453R) or leucine (K453L). Both mutants lost the ability to promote polykaryon formation (Table 3), even though they were transported to the cell surface and catalyzed removal of sialic acid from oligosaccharides as efficiently as the wild-type NA (data not shown). Additionally, the WSN NA lacks a glycosylation site at position 146 [according to the N2 numbering system (130 by N1 numbering), which is used throughout this report to relate this position to the three-dimensional structure of the subtype N2 NA molecule] on top of the NA head in the vicinity of the carboxyl-terminal residue (Fig. 3). This site appears to be critically important in view of its conservation in all other influenza A viruses and the inability of a reverse genetics-generated WSN mutant possessing this site to undergo multiple cycles of replication in cultured cells without the addition of trypsin (19). The NA mutant with the glycosylation site lost its ability to promote polykaryon formation in the presence of either bovine (50 μg/ml) or human (5 μg/ml) plasminogen (Table 3). However, at a 10-fold higher concentration, human plasminogen promoted polykaryon formation (Table 2), indicating that the presence of an oligosaccharide chain at position 146 inhibits the contribution of the NA to promote HA cleavage. Moreover, the NA of the WS virus (the parent of the WSN virus) possessing the glycosylation site did not promote polykaryon formation in the presence of human or bovine plasminogen, whereas its mutant (WS NA N146R) lacking this site did (Table 3). Together, these findings suggest that the presence of a carboxyl-terminal lysine and the absence of an oligosaccharide side chain at position 146 are important determinants of whether or not an NA molecule can participate in plasminogen-mediated HA cleavage.
Table 3.
Polykaryon formation by cells expressing the WSN HA and various other NAs
| Expressed protein* | Cell treatment
|
|||
|---|---|---|---|---|
| Trypsin (0.5 μg/ml) | FCS (1%) | Bovine plasminogen (50 μg/ml) | Human plasminogen (5 μg/ml) | |
| HA only | + | − | − | − |
| HA/WSN NA | + | + | + | + |
| HA/HK NA | + | − | − | − |
| HA/WSN NA− | + | + | + | + |
| HA/WSN NA K453R | + | − | − | − |
| HA/WSN NA K453L | + | − | − | − |
| HA/WSN NA R146N | + | − | − | − |
| HA/WS NA | + | − | − | − |
| HA/WS NA N146R | + | + | + | + |
Polykaryon formation was assayed as described in the legend to Fig. 2. −, no polykaryon formation; +, 100% polykaryon formation.
NA− denotes a mutant with loss of NA enzymatic activity due to a Glu-to-Gln change at position 227. The WSN NA K453R mutant is defined by conversion of the carboxyl-terminal Lys-to-Arg; K453L, by conversion of the carboxyl-terminal Lys to Leu; and R146N, by the presence of a potential glycosylation site at position 146. The WS NA N146R is defined by the loss of a potential glycosylation site at position 146.
Figure 3.
Three-dimensional structure of the NA monomer illustrating the relative positions of the carboxyl-terminal amino acid residue (space filled) and oligosaccharide chain at position 146 (space filled with dots). The sialic acid is shown in red in the substrate binding pocket (stick). The last seven carboxyl-terminal residues (490–496) of the NA are shown in purple. (A) Top-to-bottom view. The NA is a tetramer, and the upper and right-hand sides of the depicted molecule face with other monomers. (B) Side view. In a tetramer, the right-hand side is internal and the left-hand side external. The fourfold axes are indicated by a square with an arrow. This figure is based on the NA structure of an N2 virus, A/Tokyo/3/67, as determined by Varghese et al. (23), rather than that of an N1 virus, which is not available. However, because the basic folding of the NA is essentially identical between type A and type B NAs (24), the basic folding of the N1 NA should not differ appreciably from the one shown.
The WSN NA Is a Plasminogen-Binding Protein.
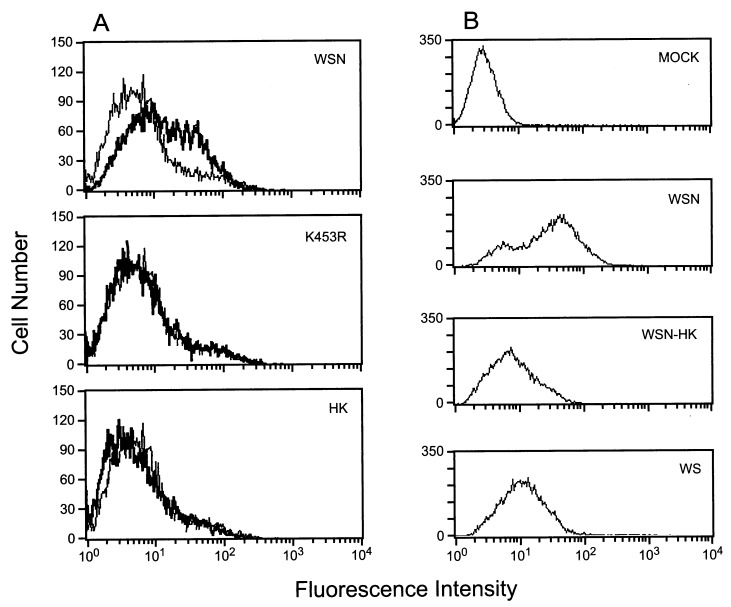
We next attempted to directly detect binding of the WSN NA to plasminogen by expressing the NA in 293T cells and incubating them with plasminogen. By fluorescence-activated cell-sorting analysis, plasminogen bound to cells expressing the WSN NA but not to those expressing either the NA of A/Hong Kong/1/68 (H3N2) or a mutant WSN NA with arginine substituted for the carboxyl-terminal lysine (K453R) (Fig. 4A). We also tested plasminogen binding in virus-infected cells. Among MDBK cells infected with either WSN or WSN–HK (a reassortant virus with the NA gene from A/Hong Kong/1/68 and all remaining genes from WSN) or the WS virus, plasminogen bound most efficiently to WSN-infected cells (Fig. 4B).
Figure 4.
Plasminogen binding to NA on the cell surface. (A) 293T cells were transfected with the WSN NA (WSN) gene, a mutant containing an alteration at the carboxyl-terminal residue, from lysine to arginine (K453R), or the HK NA gene and incubated for 24 h. Transfected cells were suspended in PBS containing 1% BSA and then incubated with 100 μg/ml human plasminogen. Bound plasminogen was detected by anti-human plasminogen antibody and fluorescein isothiocyanate-labeled anti-rabbit antibody. Cells were analyzed by flow cytometry. The thin line indicates control cells transfected with an expression plasmid with no insert, while the thick line refers to NA gene-transfected cells. (B) MDBK cells were infected with the WSN, WSN-HK, or WS virus. Twelve hours after infection, the cells were incubated with human plasminogen (10 μg/ml) and bound human plasminogen was detected as described above. MOCK, mock-infected cells.
DISCUSSION
Our results contrast sharply with the virulence mechanism established for avian influenza viruses, in which susceptibility of the HA to ubiquitous furin and PC6 proteases drives viral pantropicity (2). As we have demonstrated here, human influenza viruses can become pantropic by utilizing a ubiquitous protease, plasmin, for HA cleavage. Because plasmin is associated with an inhibitor, α2-antiplasmin, in tissue fluids (as a means of preventing tissue injury), we propose that plasminogen is converted to an active protease, plasmin, by cellular plasminogen activators. However, we cannot exclude a direct contribution to HA cleavage from plasmin, if its affinity for the NA exceeds that for α2-antiplasmin, resulting in its local accumulation in the vicinity of the HA substrate.
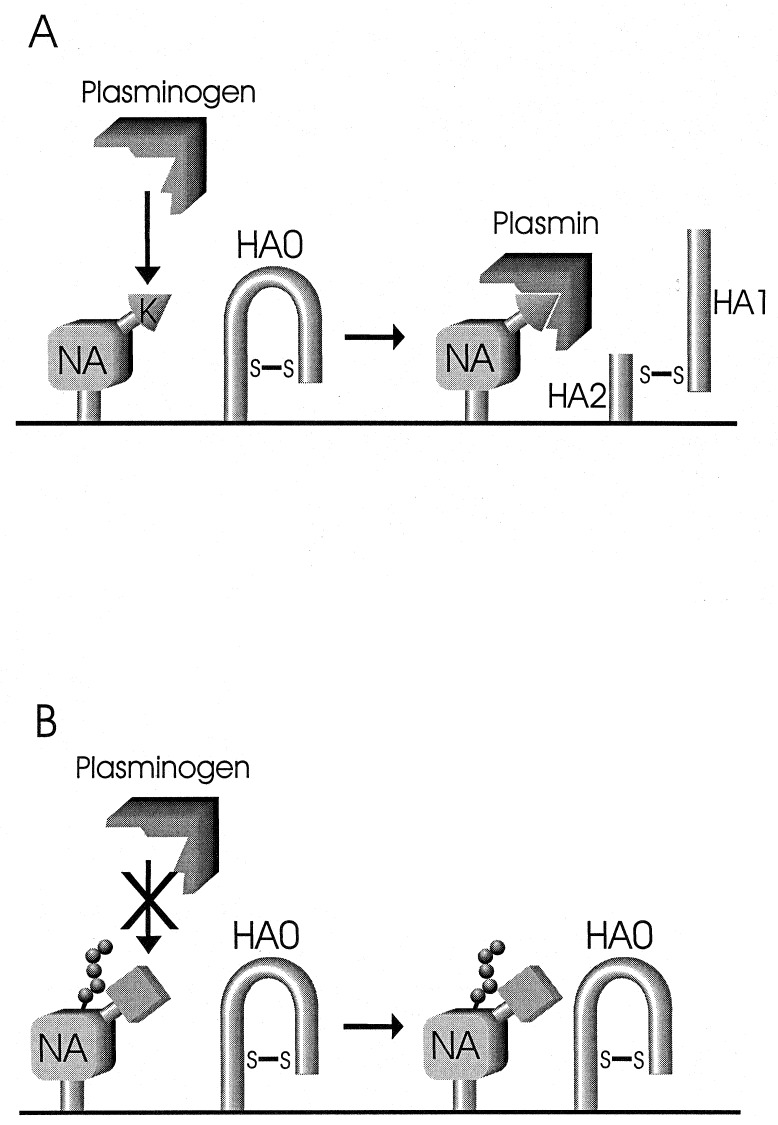
Fig. 5 summarizes our proposal of how the NA may promote HA cleavage in cells infected with the WSN strain of human influenza A viruses. Plasminogen binds to the NA because of its carboxyl-terminal lysine and lack of an oligosaccharide chain at position 146 and then is converted to plasmin, which cleaves the HA into HA1 and HA2 subunits that become incorporated into virions. If cells from which the virus buds lack a plasminogen activator or if the local concentration of plasminogen is low, the HA would remain uncleaved until the virus binds to cells expressing the appropriate plasminogen activator. Previous studies suggested that plasminogen is activated more efficiently at the cell surface than in culture medium (10). On the other hand, Boycott et al. (20) showed that the WSN HA is cleaved in endosomes by serine proteases. Considering that plasminogen activators are present on the cell surface (21), HA cleavage most likely occurs primarily at that site upon activation of plasminogen to plasmin; however, we cannot exclude a continuation of this process after the internalization of virions, as suggested by the apparent HA cleavage in endosomes (20).
Figure 5.
Schematic representation of plasminogen-mediated HA cleavage. (A) Plasminogen binds to a lysine at the carboxyl terminus of the WSN NA. Bound plasminogen is activated to plasmin by a plasminogen activator (most likely of cellular origin). Enzymatically active plasmin then cleaves HA0 into HA1 and HA2 subunits. (B) If NA does not contain a lysine at the carboxyl terminus or an oligosaccharide chain is present in the vicinity, plasminogen does not bind to the NA and HA0 remains uncleaved.
Although a single mutation that restored the NA glycosylation site at position 146 inhibited the plasminogen-binding activity of the protein, we do not conclude that a single mutation will convert nonplasminogen-binding NAs to efficient plasminogen binders, thus rendering the virus highly pathogenic. The WSN NA differs from its parent strain by 18 amino acids, including the change that abolished the glycosylation site; thus, additional mutations may be needed to promote efficient binding of plasminogen by the NA.
At least two structural features of the NA, the carboxyl-terminal lysine and the glycosylation site at position 146, should be considered when evaluating the health hazard posed by human influenza viruses. Any N1 strain whose NA possesses a carboxyl-terminal lysine and lacks a glycosylation site at position 146 should be regarded as potentially dangerous. Thus, although only the WSN strain has met these criteria in the modern era of virological research, it is tempting to speculate that the 1918 pandemic strain, an ancestor of A/WSN/33, may have acquired its unprecedented virulence from the mechanism we describe. In fact, Taubenberger et al. (22) reported that the 1918 pandemic strain does not possess multiple basic amino acids at the HA cleavage site, making it distinct from pathogenic avian influenza viruses, which use ubiquitous proteases furin and PC6 for their HA cleavage. Thus, the extreme virulence of the 1918 strain cannot be explained by the mechanism established for avian influenza. Although this group established that a short sequence of the NA obtained from a victim of the 1918 pandemic strain was identical to that of the WSN NA, they did not include sequence information on the NA at position 146 or the C terminus (22). Further sequencing of the NA from this strain is needed to address the issue of its unprecedented virulence in the 1918 pandemic.
Viral proteins other than the HA (e.g., some paramyxovirus F proteins) must be proteolytically activated at sites that could be recognized by plasmin for multiple cycles of virus replication (2). Thus, proteolysis mediated by plasminogen-binding proteins may be relevant to the replication, and possibly the acquisition of virulence, by mammalian viruses other than influenza A viruses.
Acknowledgments
We thank Dr. Robert G. Webster for providing the antibodies used in this study and Dr. Masahiro Ueda for WSN–HK virus. We are also grateful to Clayton Naeve and the St. Jude Children’s Research Hospital Center for Biotechnology for preparing oligonucleotides and for computer support, to Mikhail Matrosovich for preparation of figures of the NA three-dimensional structure, to Ming Luo for identifying fourfold axes in the diagrams of the NA molecule, to Yuko Kawaoka for illustration, and to John Gilbert for scientific editing. This work was supported by Public Health Service Research Grants AI29599 and AI33898 from the National Institute of Allergy and Infectious Diseases, Cancer Center Support grant from the National Cancer Institute, and the American Lebanese Syrian Associated Charities.
ABBREVIATIONS
- HA
hemagglutinin
- NA
neuraminidase
- MDBK
Madin–Darby bovine kidney
- FCS
fetal calf serum
Footnotes
A commentary on this article begins on page 9713.
References
- 1.Paulson J C. In: Interactions of Animal Viruses with Cell Surface Receptors. Connor M, editor. Orlando, FL: Academic; 1985. pp. 131–219. [Google Scholar]
- 2.Klenk H-D, Garten W. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 3.Palese P, Tobita K, Ueda M, Compans R W. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 4.Air G M, Laver W G. Proteins Struct Funct Genet. 1989;6:341–356. doi: 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- 5.Lazarowitz S G, Choppin P W. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 6.Klenk H D, Rott R, Orlich M, Blodorn J. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 7.Choppin P W. Virology. 1969;38:130–134. doi: 10.1016/0042-6822(69)90354-7. [DOI] [PubMed] [Google Scholar]
- 8.Castrucci M R, Kawaoka Y. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulman J L, Palese P. J Virol. 1977;24:170–176. doi: 10.1128/jvi.24.1.170-176.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazarowitz S G, Goldberg A R, Choppin P W. Virology. 1973;56:172–180. doi: 10.1016/0042-6822(73)90296-1. [DOI] [PubMed] [Google Scholar]
- 11.Huddleston J A, Brownlee G G. Nucleic Acids Res. 1982;10:1029–1037. doi: 10.1093/nar/10.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 13.Menoud P A, Sappino N, Boudal-Khoshbeen M, Vassalli J D, Sappino A P. J Clin Invest. 1996;97:2478–2484. doi: 10.1172/JCI118694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lottenberg R, Minning-Wenz D, Boyle M D P. Trends Microbiol. 1994;2:20–24. doi: 10.1016/0966-842x(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 16.Lottenberg R, Broder C C, Boyle M D, Kain S J, Schroeder B L, Curtiss R. J Bacteriol. 1992;174:5204–5210. doi: 10.1128/jb.174.16.5204-5210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs H, Wallich R, Simon M M, Kramer M D. Proc Natl Acad Sci USA. 1994;91:12594–12598. doi: 10.1073/pnas.91.26.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miles L A, Dahlberg C M, Plescia J, Felez J, Kato K, Plow E F. Biochemistry. 1991;30:1682–1691. doi: 10.1021/bi00220a034. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Schulman J, Itamura S, Palese P. J Virol. 1993;67:6667–6673. doi: 10.1128/jvi.67.11.6667-6673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boycott R, Klenk H-D, Ohuchi M. Virology. 1994;203:313–319. doi: 10.1006/viro.1994.1489. [DOI] [PubMed] [Google Scholar]
- 21.Hajjar K A. Thromb Haemost. 1995;74:294–301. [PubMed] [Google Scholar]
- 22.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 23.Varghese J N, Laver W G, Colman P M. Nature (London) 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 24.Burmeister W P, Ruigrok R W H, Cusack S. EMBO J. 1992;11:49–56. doi: 10.1002/j.1460-2075.1992.tb05026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]