Abstract
Proton-gated channels expressed by sensory neurons are of particular interest because low pH causes pain. Two proton-gated channels, acid-sensing ionic channel (ASIC) and dorsal root ASIC (DRASIC), that are members of the amiloride-sensitive ENaC/Degenerin family are known to be expressed by sensory neurons. Here, we describe the cloning and characterization of an ASIC splice variant, ASIC-β, which contains a unique N-terminal 172 aa, as well as unique 5′ and 3′ untranslated sequences. ASIC-β, unlike ASIC and DRASIC, is found only in a subset of small and large diameter sensory neurons and is absent from sympathetic neurons or the central nervous system. The patterns of expression of ASIC and ASIC-β transcripts in rat dorsal root ganglion neurons are distinct. When expressed in COS-7 cells, ASIC-β forms a functional channel with electrophysiological properties distinct from ASIC and DRASIC. The pH dependency and sensitivity to amiloride of ASIC-β is similar to that described for ASIC, but unlike ASIC, the channel is not permeable to calcium, nor are ASIC-β-mediated currents inhibited by extracellular calcium. The unique distribution of ASIC-β suggests that it may play a specialized role in sensory neuron function.
Tissue acidosis is a naturally occurring phenomenon that occurs in ischemic, damaged, or inflamed tissue. The reduction in pH in response to such events can be dramatic. In patients with intermittent claudication, the intramuscular pH can drop to 6.0 during exercise, whereas in an experimental model of cardiac infarction, the pH of the cardiac circulation was reduced to pH 5.7 (1). Associated with these conditions is a feeling of pain. This can be reproduced experimentally by infusion of low pH solutions into skin (2, 3) or muscle (4).
In vitro studies have shown that low extracellular pH can evoke inward currents in both central nervous system and peripheral sensory neurons. Krishtal and Pidoplichko (5) demonstrated that low pH evoked inward currents in rat trigeminal ganglion neurons, and similar observations have been made in rat dorsal root ganglion (DRG) neurons (6–8) Low pH responses from DRG are characteristically multi-phasic in nature, suggesting the existence of distinct types of channel (see, e.g., ref. 7). The molecular cloning of a number of proton-gated channels supports this conclusion.
Three mammalian proton-gated channels have been cloned recently: acid-sensing ionic channel (ASIC) (9), dorsal root ASIC (DRASIC) (10), and a mammalian degenerin homologue (MDEG-1) (11, 12). A modulatory subunit, MDEG-2, also has been found (12). All of these channels belong to the degenerin/ENaC channel superfamily, and are composed of two hydrophobic segments, intracellular N and C terminals, and a large extracellular loop (13). ASIC, DRASIC, and MDEG1 each form functional channels when expressed in COS-7 cells. Evidence for heteromultimer formation between ASIC and MDEG also has been obtained (14). The electrophysiological properties of these channels are diverse, as is their cellular localization. ASIC and MDEG-1 are widely expressed in nervous tissue (9, 12) whereas DRASIC is found not only in sensory neurons (10) but also in the brain and spinal cord. The role of proton-gated channels in the central nervous system is unknown.
We report here the cloning of a new functional splice variant of ASIC, named ASIC-β, which has electrophysiological properties distinct from other proton-gated channels and is expressed exclusively in a subset of small and large diameter DRG sensory neurons. This distribution is unique among proton-gated channels so far discovered and suggests that ASIC-β has a specialized role in sensory neuron function.
MATERIALS AND METHODS
Molecular Cloning of ASIC-Related Genes.
Sequences from the C-terminal region of rat ASIC and mouse BNaC2 (brain sodium channel) were chosen as templates to design PCR primers: 5′-ACTGTACTCCGGAGCAGTACAAGG-3′ 5′-GAGTTCCAGCACTGTGAGGATGCT-3′ that are able to amplify a 350-bp DNA fragment from DRG cDNA (9, 11). The PCR-amplified DNA fragments were labeled with 32P (GIBCO Rad-prime kit) and used as probes to screen a DRG cDNA library. Clones (200,000) from a size-fractionated (2–4 kb) oligo(dT)-primed cDNA library from neonatal rat DRG were screened by hybridization with the PCR probes (25 ng, specific activity 2 × 109 cpm/μg) in 4× SSC containing 0.5% SDS, 5× Denhardt’s solution, 100 μg/ml boiled salmon sperm DNA, 10 μg/ml poly(U), and 10 μg/ml poly(C) at 65°C for 4 h. The DRG cDNA filters were given a final wash in 0.2× SSC/0.5% SDS at 65°C. In total, 32 positive clones were picked and analyzed by sequencing.
Northern Blot Analysis.
Specific N-terminal sequences of different ASIC clones were chosen as templates to synthesized cRNA probes. ASIC (nucleotide positions 750–1,068 corresponding to amino acids 74–179) was subcloned into pGEM-3Z by using EcoRI and PstI sites; ASIC-β (320–700 corresponding to amino acids 18–163) was subcloned into pGEM-11Zf by ApaI sites. For DRASIC, a 380-bp DNA fragment was amplified by PCR by using primers 5′-GTGCGCCACTACACGCTATGCCAAGGAGC-3′ 5′-GGGGAACATGTGTTCGATGCCCATTCAAC-3′ and was subcloned into T-vector (Promega); for cyclophilin, a 300-bp DNA fragment was amplified by PCR by using primers 5′ ACCCCACCGTGTTCTTCGAC-3′ 5′ CATTTGCCATGGACAAGATG-3 and was subcloned into T-vector. Antisense labeled cRNA was synthesized from these templates by using SP6 RNA polymerase and [32P]-UTP. Such cRNAs were used to probe Northern blots with 20–50 μg of total RNA in each lane. Hybridization was carried out in 50% formaldehyde, 5× SSC containing 0.5% SDS, 5× Denhardt’s solution, 100 μg/ml boiled salmon sperm DNA, 10 μg/ml poly(U), and 10 μg/ml poly(C) at 68°C for 24 h, with a final wash in 0.1× SSC with 0.5% SDS at 75°C.
In Situ Hybridization.
The same templates used for probing Northern blots were labeled with digoxygenin-UTP (15). After in situ hybridization, sections were double-labeled with neuronal sub-population markers. mAb against peripherin or N-52 (Chemicon and Sigma) were used at a 1:500 dilution in blocking solution (1× PBS containing 10% sheep serum/0.5% Triton X-100). Fluorescein isothiocyanate-conjugated secondary antibodies (Boehringer Mannheim) were used at a 1:200 dilution in blocking solution. For IB4 staining, the IB4-fluorescein isothiocyanate (4 μg/ml, Sigma) was diluted in 1:300 PBS containing 0.1 mM CaCl2, MgCl2, MnCl2, and 0.2% Triton X-100 (16).
Expression of ASIC-β in COS Cells.
To express functional channels in COS-7 cells, ASIC-β was subcloned into the Invitrogen pTracer-CMV vector that expresses green fluorescent protein using EcoRI restriction sites. The resulting plasmids were transfected into COS-7 cells by electroporation. For electroporation, COS-7 cells cultured on 100-mm Petri dishes (80–90% confluence) were trypsinized and resuspended in 350 μl of ice-cold HEBS buffer. Plasmid (20–30 μg) then was mixed with the cells in an electroporation cuvette for a total of 5 min. After transfection, the cells were seeded at low density on 30-mm Petri dishes and cultured in 2 ml of DMEM with 10% fetal calf serum at 37°C, for 2–3 days.
Electrophysiology.
Whole-cell voltage–clamp recordings (17) were made 2–3 days after transfection. Membrane currents were recorded by using an Axopatch 200B amplifier. Currents were low pass filtered at 5 kHz (4-pole Bessel filter) and digitized by using a Digidata 1200 interface. Acquisition and analysis of currents was achieved by using pclamp6 software. Pipettes were pulled from borosilicate glass (Clark Electromedical, Reading, UK) and had DC resistances of ≈3 mΩ when filled with the pipette solution. All recordings were made at room temperature (18–22°C).
The extracellular recording solution was comprised of (in mM): NaCl 146; KCl 5; Glucose 10; MgCl2 1; CaCl2 0.01. For extracellular solutions with pH values of 7.4–6.5, 10 mM Hepes was used as the buffer, whereas for solutions of pH 6.5–4.0, 10 mM Mes was used to provide optimal buffering capacity over the wide pH range (3.5 units) required. The normal pH of the extracellular solution was 7.4. In some ion substitution experiments, extracellular sodium chloride was replaced with an equal amount of choline chloride. The effect of extracellular calcium concentration on low pH-evoked currents was investigated by substitution of choline chloride with an equal concentration of calcium chloride, in the absence of extracellular sodium ions.
Low pH solutions were applied via a U-tube (18) placed close (<1 mm) to the cell of interest. The use of the U-tube ensured that the cell was bathed completely in the test solution ensuring that no buffering by the bulk extracellular medium occurred. Low pH solutions usually were applied for 10–20 s, with at least 2 min between applications. The intracellular solution contained (in mM): KCl 120; NaCl 8; MgCl2 3; Hepes (free acid) 40; 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (free acid) 10, at pH 7.35.
RESULTS
Molecular Cloning of Splice Variants of ASIC.
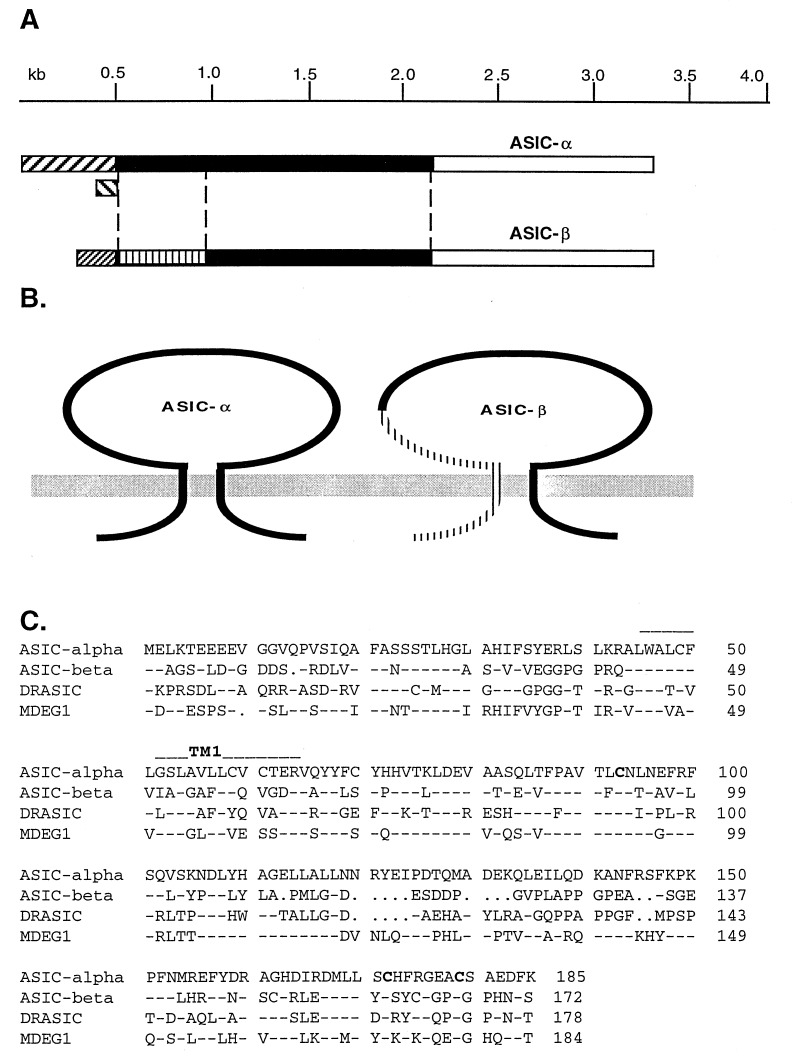
An ASIC-related clone with distinct 5′ and 3′ untranslated regions (UTRs) named ASIC-α and a splice variant with a unique 5′ region and the same C-terminal and 3′ UTR as ASIC-α named ASIC-β were identified in a rat DRG cDNA library by homology cloning (Fig. 1A). The proposed ORFs of these clones show that they have a similar molecular structure with two putative transmembrane domains, intracellular N and C terminals, and a large extracellular loop (Fig. 1B).
Figure 1.
Structure of ASIC splice variants and primary sequence alignment of ASIC-β with four other known members of the family. (A) The gene structure of ASIC splice variants. Three different transcripts are distinguished by their 5′UTRs (hatched) but share the same 3′UTR (white). The coding regions are black apart from the unique N terminus of ASIC-β (striped). (B) The proposed molecular structures of ASIC-α and ASIC-β shows that both proteins have the same two transmembrane domain structure, with intracellular N and C terminals. The striped region of AIC-β shows its unique N-terminal region including the first transmembrane domain. (C) The N-terminal sequence alignment of four ASIC-related proteins. Block letters present the cysteine resides that are conserved amongst the four proteins, implying a similar secondary structure.
The ASIC-α transcript encodes the 526-aa protein named ASIC (9) but has distinct 5′ and 3′ UTRs from the previously reported sequence. There are two types of 5′ UTR in the DRG ASIC-α clones (Fig. 1A). The major population of ASIC-α (90%) has a 5′ UTR of up to 530 bp that is GC-rich (73%). A small percentage (10%) of ASIC-α clones have a short 5′ UTR (≈100 bp) that corresponds to the sequence reported for ASIC 5′ UTRs found in brain (Fig. 1A). All of the ASIC-related clones in DRG have an identical 3′ UTR that is different from the ASIC UTR reported in brain. An L1-like repetitive sequence reported in ASIC (9) is not found in DRG ASIC-α or β transcripts.
The longest ORF of ASIC-β is 513 aa in length, sharing the same 341 aa with the C terminus of ASIC-α. The N-terminal 172 amino acids of ASIC-β are unique, with highest homology to DRASIC (43.8%), 39.7% identity to both ASIC-α and MDEG1, and 22.6% identity to the FMRFamide-gated sodium channel (FaNaC) (19) (Fig. 1C). These five proteins share three conserved cysteines within the N-terminal region of the extracellular loop. There are two additional cysteine residues in ASIC-β when compared with DRASIC, ASIC-α MDEG1, and FaNaC, suggesting that the secondary structure of ASIC-β might have some unique features. N-terminal splicing at a similar position also is found in the related MDEG2 transcript, which differs in 236 aa from MDEG1 (12), but this N-terminal sequence has no homology to the N terminus of ASIC-α. Additional homology cloning with different probes derived from ASIC-related clones showed that the major transcripts represented in our DRG library are ASIC-α, ASIC-β, and DRASIC but not MDEG1.
Tissue Distribution of Sensory Neuron-Associated, Proton-Gated Channels.
We examined the tissue distribution of ASIC-β and compared it with that of the other proton-gated channels expressed in sensory neurons. We used Northern blotting and examined the respective distributions in a variety of neuronal and nonneuronal tissues and also some cell lines.
ASIC-α was found in many neuronal tissues, including DRG, spinal cord, trigeminal ganglia, and the trigeminal mesencephalic nucleus. The cell lines PC12, ND7/23, and N-tera2 also expressed ASIC-α (Fig. 2). In contrast, ASIC-β, seen as a 3.2-kb transcript, was found only in the DRG and not in other tissues or cell lines. DRASIC has been reported to be a sensory neuron-specific, proton-gated channel (10). However, in addition to the DRG, we found low level transcripts of DRASIC in superior cervical ganglia, spinal cord, and also the brain stem. These data suggest that ASIC-β is the only proton-gated channel that is expressed exclusively in sensory neurons.
Figure 2.
Northern blots of ASIC-β distribution. The Northern blots were probed by N-terminal unique sequences of ASIC-α, ASIC-β, and DRASIC. All three proton-gated channels are expressed in sensory neurons. ASIC-α is distributed in many neural tissues and cell lines. There are three different sizes of ASIC-α transcripts in PC12 cells that are 2.5, 3.2, and 4.0 kb, but there is only one major transcript of 3.2 kb in sensory neurons. ASIC-β is only expressed in DRG as a 3.2-kb transcript. DRASIC is predominantly in DRG with two sizes of transcripts, 2.0 and 2.5 kb, but also is expressed in superior cervical ganglion, spinal cord, and brain stem. The relative amount of RNA loading is indicated by cyclophilin probe.
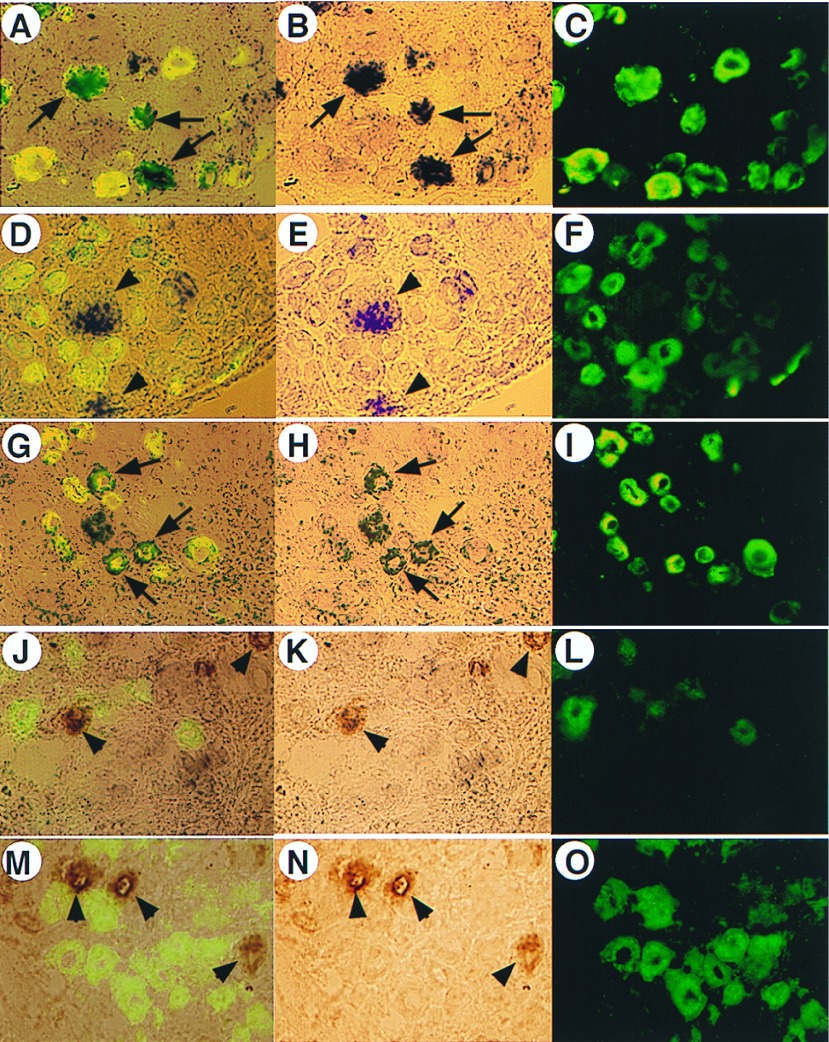
We next examined the cell-type distribution of different ASIC splice variants in DRG. We used peripherin to label small diameter sensory neurons and IB4 to label the neurotrophin-independent cells that also comprise a large proportion of nociceptors (16). We used an anti-neurofilament antibody N-52 to define the large diameter neurons that are mainly mechanoreceptors and proprioreceptors (19). By using 5′ coding region probes of ASIC-α and ASIC-β for in situ hybridization, we found that both ASIC-α and ASIC-β are expressed in 20–25% of L4 DRG neurons (Fig. 3). The ASIC-α-positive neurons are mainly small diameter cells (>90%) that coexpress peripherin but not IB4 (Fig. 3). In contrast, ASIC-β-positive neurones are comprised of both small diameter and large diameter neurones, of which 70% express neurofilaments and only 30% coexpress peripherin (Fig. 3A). These data demonstrate that ASIC-β exhibits both a tissue- and cell-specific distribution of expression that is clearly different to that shown by ASIC-α.
Figure 3.
Cell specificity of ASIC-α and ASIC-β in DRG. In situ hybridization of ASIC-α and ASIC-β, double-labeled with anti-peripherin (A–I) or isolectin B4 (J–O). The left column is overlayed images of in situ (middle column) and immunocytochemistry (right column) results. The expression of ASIC-α transcript is most (>90%) colocalized with peripherin (A–C, indicated by arrows). However, ASIC-β transcript is distributed in either peripherin-positive (30%) or peripherin-negative (70%) neurons (D–I). The colocalization of ASIC-β and peripherin is indicated by arrows (G and H). The expression of ASIC-β in peripherin-negative large diameter neurones is shown by arrowheads (D and E). Neither ASIC-α (M–O, indicated by arrowheads) nor ASIC-β (J–L, indicated by arrowheads) have colocalized staining with isolectin B4, which corresponds to GDNF-dependent small diameter neurons.
Expression of ASIC-β in COS-7 Cells.
Transfected cells were identified by the presence of green fluorescent protein. Application of low pH to ASIC-β expressing COS cells at a holding potential of −60 mV evoked rapidly activating inward currents (Fig. 4A). The threshold for activation of the current was approximately pH 6.5, and the current was maximal at approximately pH 4.0. Fig. 4B shows the mean pH response curve recorded from six ASIC-β transfected COS cells. The half-point for activation of the current in this series of experiments was pH 5.9. The inward currents evoked in response to pH 4.0 were variable in magnitude (range was 0.272 ± 8.41 nA), and the mean response was 2.39 ± 0.33 nA in 27 cells). In 83% (34 of 41) of fluorescent cells, a response was observed to the application of pH 4.0–4.5, whereas in 17 of 17 untransfected COS-7 cells, application of pH 4.0–4.5 evoked no change in membrane current.
Figure 4.
Characteristics of the pH response in COS-7 cells expressing ASIC-β. Typical response to low pH in ASIC-β transfected COS cells. The cell was voltage-clamped at −60 mV and low pH applied at the bar. Dotted line indicates zero current level. (B) pH response relationship obtained from experiments similar to that in A. Responses were normalized against the maximal response and plotted against the pH. The half-point for activation of the current was pH 5.9. (C) Time taken for the current to activate and inactivate plotted against pH. (D) Recordings made during a change in command potential by using a linear ramp protocol (duration of ramp 240 ms). Current was recorded under control conditions and during application of low pH. The current reverses at approximately +25 mV.
The pH-activated currents normally reached peak amplitude in ≈1 s in response to the lower pH solutions and rapidly inactivated (or desensitized) in the continued presence of low pH. The time taken to peak was related to the applied pH, with the quickest activation times occurring in response to the lowest pH. The effect of pH on the time taken for inactivation of the current showed a similar pH dependency but was much more marked (Fig. 4C).
Ionic Basis of ASIC-β-Mediated Inward Current.
The reversal potential for the rapid phase of the low pH activated current was established by using either a linear ramp voltage–clamp protocol or by sequentially stepping the command potential to a range of values while evoking the inward current (Fig. 4D). The command potential was ramped over a period of 240 ms between potentials of −80 and +60 mV. The speed of the ramp allowed us to make accurate recordings despite the rapidly activating and inactivating nature of the response. The reversal potential was found to be 26.1 ± 2.3 mV (n = 8). If the channel were only sodium permeable, the reversal potential would be expected to be approximately +73 mV, given the composition of the intra- and extracellular solutions used. It thus seemed likely that the channel was also permeable to other cations, most notably K+, because of its presence in such high concentration. Ion substitution studies confirmed that the channel was also permeable to K+. Replacement of extracellular sodium with choline abolished inward currents, but on some occasions, small outward currents were seen, confirming that the channel was permeable to potassium ions (data not shown). These currents were small because most recordings were made at a holding potential of −60 mV, close to the reversal potential of potassium ions with the solutions used.
Previous studies have demonstrated that increasing the extracellular calcium concentration reduced the magnitude of proton-activated inward currents passing through the ASIC channel (9). This was not the case for ASIC-β. Fig. 5A shows that, in the presence of 146 mM NaCl, increasing the extracellular calcium concentration had no effect on the magnitude of the inward currents evoked by the application of pH 4.0 buffer. In addition, ASIC-β was found not to be permeable to calcium ions. In the experiment shown in Fig. 5B, a response to pH 4.0 was obtained under control conditions (Fig. 5B, Left), and then extracellular NaCl was replaced by choline chloride and the cell was exposed to pH 4.0 buffer in the presence of a range of extracellular calcium concentrations. On removing extracellular sodium, no inward current was detected in response to low pH, and even when the calcium concentration was increased to 50 mM, no inward current was detected. Fig. 5 A and B are representative of experiments on four cells. Sodium ions were not a cofactor for calcium permeability. We recorded reversal potentials for ASIC-β-mediated currents with 140 mM Na+ in the external medium, with or without 20 mM Ca2+. The mean reversal potentials were 25.2 ± 2.3 mV (n = 4) in the absence and 24.8 ± 2.8 mV (n = 4) in the presence of Ca2+, demonstrating that the channel is impermeant to calcium, even if Na+ ions are present.
Figure 5.
Calcium dependency and pharmacology of ASIC-β-mediated currents in COS cells. (A) Responses obtained to pH 5.1 (at the closed circles) in the presence of increased extracellular calcium concentration. Recordings were made from the same cell at intervals of 3 min. (B, Left) Control response to pH 5.1 and (Right) responses to low pH in the absence of extracellular sodium and increased calcium concentration are shown. Current flowing via ASIC-β is not inhibited by extracellular calcium, nor is the channel permeable to calcium. Dotted line indicates zero current level. (C) Amiloride inhibits ASIC-β mediated current. The IC50 derived from this plot was 21 μM. (D) Capsaicin does not activate ASIC-β. Recordings made from the same cell; holding potential was −60 mV. Upper trace shows that application of capsaicin (500 nM) at the bar failed to evoke an inward current. pH 4.1, at the bar 3 min later (lower trace), evoked a robust inward current. Traces have been separated for clarity, and the dotted line indicates zero current for each recording.
Pharmacology of ASIC-β-Mediated Currents.
We investigated the effect of amiloride, a known inhibitor of other proton-gated channels, on ASIC-β-mediated currents (20). Cells were voltage-clamped at −60 mV and given a 20-s exposure to pH 4.5 solution, first in the absence and then the presence of increasing concentrations of amiloride. The threshold concentration for inhibition of low pH-evoked currents was between 1 and 10 μM. Fig. 5C shows an inhibition response curve for amiloride constructed from experiments on six cells. The data points were fitted with a single Boltzman function, giving an IC50 of 21 μM.
Capsaicin is known to have an excitatory action on small diameter sensory neurons. Moreover, it has been suggested that capsaicin and protons activate a similar ion channel (21). To investigate whether capsaicin could activate the ASIC-β channel, we exposed ASIC-β-transfected COS cells to 500 nM capsaicin. In four of four cells, capsaicin evoked no change in membrane current, whereas a subsequent application of low pH to the same cells produced characteristically large inward currents (Fig. 5D).
DISCUSSION
Protons evoke a sensation of pain, and a variety of hyperalgesic mediators potentiate the pain-inducing actions of low pH, suggesting that proton-gated channels play a central role in nociception (4). In this study, we provide detailed information on the expression of proton-gated channel transcripts in sensory neurons. The alternative splicing of ASIC results not only in two different gene products, ASIC-α and ASIC-β, but also in two different ASIC-α transcripts, each of which has a distinct 5′ UTR. The coding region of both ASIC-α transcripts corresponds to that of ASIC (9). The 5′ heterogeneity of ASIC-α and ASIC-β may be generated initially by transcription from different promoters, which may be tissue-specific (see, e.g., ref. 22). In addition, we found two other splice variants of ASIC. One has a 29-aa deletion between codons 74 and 102 of ASIC-α. The other has a 600-bp insertion between codons 236 and 237 of ASIC-α. This insert causes a premature stop in translation and results in a new ASIC-like protein that has a shorter and unique C terminus. We named this channel ASIC-γ and have shown that it forms a functional proton-gated channel when expressed in COS cells (data not shown). However, the level of expression of the ASIC-γ transcript is low, and we were not able to detect it on Northern blots.
As well as ASIC splice variants, two other proton-gated channels DRASIC and MDEG2 also are found in sensory neurons (10, 12). Such heterogeneous expression of proton-gated channels may imply a complex response of sensory neuron subpopulations to tissue acidosis. All ASIC splice variants and DRASIC alone can form functional channels when expressed in COS cells so that they may be able to function individually or combine with others to form heteromeric channels. Although the heteromultimerization of these proton-gated channels has not been demonstrated, a recently isolated modulatory subunit MDEG2 has been shown to form heteromultimers with DRASIC that result in altered channel properties (14). It is possible that this kind of modulatory subunit may interact with other proton-gated channels in sensory neurons.
At present, ASIC-β is the only proton-gated channel that has been shown to be exclusively expressed in sensory neurons. DRASIC and ASIC-α also show high levels of expression in DRG but also are found outside the spinal ganglia (refs. 9 and 10 and this study). The DRG-specific expression of ASIC-β suggests that this transcript may arise from alternative splicing of a pre-mRNA generated by a DRG-specific promoter. Similar alternative splicing also is found in MDEG1 and MDEG2, which have the same splicing site as ASIC-α and ASIC-β, but neither MDEG1 nor MDEG2 is sensory neuron-specific. It thus will be interesting to analyze the ASIC-β promoter and compare it with other DRG-specific promoters. The specific splicing of ASIC-β mainly occurs in large diameter neurons that are different from the neurons expressing ASIC-α in DRG. The unique expression pattern of ASIC-β is also different from that of other known DRG-specific genes (see, e.g., refs. 23–25).
The expression of ASIC in brain and DRG previously has been examined by using nonisotopic in situ hybridization with a probe containing an L1 repeat (9). By using the L1-containing probe to screen a DRG random-primed cDNA library, we found ≈2,500 positives from a pool of 200,000 clones, compared with nine positive clones when using an ASIC-specific probe under identical conditions. This suggests that the L1 repetitive sequence exists in both ASIC and many other transcripts. We therefore used unique N-terminal coding regions to construct specific probes to investigate the expression of different ASIC splice variants in sensory neurons. Our in situ hybridization studies reveal that 90% of ASIC-α-positive cells are small diameter peripherin-positive neurons, most of which are nociceptors. The results also show that ASIC-β is expressed in ≈20% of the total number of neurons and is found in both small and large diameter neurons.
The functional properties of the proton-gated channels so far described can be grouped broadly into two categories. First, there are those channels that show a rapid time course for activation and inactivation in response to low pH, and second, there are those that activate and inactivate much more slowly. ASIC-α (9) is typical of the former group, whereas DRASIC falls into the latter (10). We found that the kinetic properties of ASIC-β are very similar to those of ASIC-α. The currents were quick to reach a maximum and desensitized in the continued presence of low pH. The pH dependency of currents passing through the two channels was also similar (EC50 for ASIC-α was ≈pH 6 (9) vs. pH 5.9 for ASIC-β. Both channels also show a preference for Na+. In addition, reversal potential studies showed that the ASIC-β channel was also permeable to K+ ions, although much less so than to Na+.
The major difference between the electrophysiological properties of ASIC-α and ASIC-β is related to the calcium permeability of the respective channels. ASIC-α discriminates poorly between cations, and the channel will pass Ca2+ ions, although the channel is 2.5 times more permeable to Na+. However, low pH-evoked currents passing through the ASIC-α channel are inhibited at high extracellular calcium concentrations >100 μM (9). In our study, we found that the ASIC-β channel was not permeable to calcium ions, nor did raising the extracellular calcium concentration become inhibitory (Fig. 5). ASIC-β-mediated currents thus exhibit similar properties to the native fast pH-evoked current recorded from voltage-clamped DRG neurons in response to low pH (26). It seems possible that ASIC-β mediates currents that contribute to the fast proton-activated current in sensory neurons. DRASIC-mediated currents are slow and sustained (10), similar to the sustained pH-mediated currents recorded from DRG neurons (7, 26). However, we found that, in addition to DRG, transcripts for DRASIC were also present, albeit at lower levels, in superior cervical ganglia, spinal cord, and brain stem, where sustained proton-evoked currents have not been described. ASIC-β is thus the only identified proton-gated channel expressed exclusively in sensory neurons.
Amiloride inhibited ASIC-β-mediated currents in the COS cells with a similar efficiency to that seen with ASIC-α-mediated current. This may indicate that the binding site for amiloride in the respective channels is in a conserved region. This region is presumably not present in the DRASIC channel because amiloride produces potentiation of currents passing through this channel (10). It has been suggested that the selective neurotoxin capsaicin and protons may activate the same channel in sensory neurons (26). The recent molecular cloning of the capsaicin-gated channel VR1 suggests that this is not the case because VR1-mediated currents were reported not to be activated by low pH (27). We found that capsaicin was not able to activate ASIC-β when expressed in the COS cells. These findings, taken together with other studies of cloned proton-gated channels, suggest that proton-gated and capsaicin-gated channels are different molecular entities.
Almost all (80–100%) DRG neurons have been shown to respond to low pH stimulation, but only half of them appear to be nociceptors (21, 28). It thus seems likely that, in addition to sensing tissue acidosis, proton-gated channels have other roles. ASIC-β is the first cloned proton-gated channel to be found in a subset of large diameter sensory neurons, in addition to the smaller, putative nociceptors. Drawing on models derived from studies of Caenorhabditis elegans mechanosensitive mutants, it is possible that ASIC-β could be a component of mechanosensitive channels that need to be tethered to the cytoskeleton to function (13). Proton-gated channel transcripts are also present throughout the central nervous system, and it is difficult to identify a role for these channels in this region simply in terms of acid sensing. It is possible that these channels play a role as autoreceptors, given the acidic content of synaptic vesicles (29). It is also possible that the channels may be activated by other endogenous ligands or by mechanosensory stimuli. Heteromultimerization of proton-gated channel subunits may produce channels with novel properties. It is also possible that proton-gated channel subunits may combine with ligand-gated ion channels [e.g., P2X receptors that also have two-transmembrane domains (30)] yielding a completely different repertoire of channel and receptor properties.
ASIC-β can be added to the growing family of proton-gated channels (9–13). The exclusive expression of the channel in DRG neurons is consistent with a role in sensory transduction, although not necessarily in nociception. The presence of ASIC-β in large diameter sensory neurons, as well as the widespread distribution of other proton-gated channels in the central nervous system, suggests an important, as yet unknown physiological role for these channels.
Acknowledgments
We thank the Wellcome Trust for support and Samantha Ravenall for expert technical assistance.
ABBREVIATIONS
- DRG
dorsal root ganglion
- ASIC
acid-sensing ionic channel
- DRASIC
dorsal root ASIC
- MDEG-1
a mammalian degenerin homologue
- UTR
untranslated region
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper (ASIC-β) has been deposited in the GenBank database (accession no. AJ006519).
References
- 1.Jacobus W E, Taylor G J, Hollis D P, Nunnally R L. Nature (London) 1977;265:756–758. doi: 10.1038/265756a0. [DOI] [PubMed] [Google Scholar]
- 2.Steen K H, Reeh P W. Neurosci Lett. 1993;154:113–116. doi: 10.1016/0304-3940(93)90184-m. [DOI] [PubMed] [Google Scholar]
- 3.Steen K H, Reeh P W, Anton F, Handwerker H O. J Neurosci. 1992;12:86–95. doi: 10.1523/JNEUROSCI.12-01-00086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steen K H, Steen A E, Kreysel H-W, Reeh P W. Pain. 1996;66:163–170. doi: 10.1016/0304-3959(96)03034-5. [DOI] [PubMed] [Google Scholar]
- 5.Krishtal O A, Pidoplichko V I. Neuroscience. 1980;5:2325–2327. doi: 10.1016/0306-4522(80)90149-9. [DOI] [PubMed] [Google Scholar]
- 6.Kovalchuk Y N, Krishtal O A, Nowycky M C. Neurosci Lett. 1990;31:237–242. doi: 10.1016/0304-3940(90)90461-h. [DOI] [PubMed] [Google Scholar]
- 7.Bevan S, Yeats J. J Physiol (Lond) 1991;433:145–161. doi: 10.1113/jphysiol.1991.sp018419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeilhofer H U, Kress M, Swandulla D. J Physiol (Lond) 1997;503:67–78. doi: 10.1111/j.1469-7793.1997.067bi.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. Nature (London) 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 10.Waldmann R, Bassilana F, de Weille J, Champigny G, Heurteaux C, Lazdunski M. J Biol Chem. 1997;272:20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Anoveros J, Derfler B, Neville-Golden J, Hyman B T, Corey D P. Proc Natl Acad Sci USA. 1997;94:1459–1464. doi: 10.1073/pnas.94.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lingueglia E, de Weille J R, Bassilana F, Heurteaux C, Sakai H, Waldmann R, Lazdunski M. J Biol Chem. 1997;272:29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Anoveros J, Corey D P. Annu Rev Neurosci. 1997;20:567–594. doi: 10.1146/annurev.neuro.20.1.567. [DOI] [PubMed] [Google Scholar]
- 14.Bassilana F, Champigny G, Waldmann R, de Weille J R, Heurteaux C, Lazdunski M. J Biol Chem. 1997;272:28819–28822. doi: 10.1074/jbc.272.46.28819. [DOI] [PubMed] [Google Scholar]
- 15.Schaeren-Wimers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 16.Molliver D C, Wright D E, Leitner M L, Parsadanian A S, Doster K, Wen D, Yan Q, Snider W D. Neuron. 1977;19:849–861. doi: 10.1016/s0896-6273(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 17.Hammill O P, Marty A, Neher E, Sakmann B, Sigworth F J. Pflugers Arch. 1981;391:1108–1112. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 18.Bormann J. In: Practical Electrophysiological Methods. Kettenman H, Grantyn R, editors. New York: Wiley-Liss; 1992. pp. 136–140. [Google Scholar]
- 19.Guidato S, Bajaj N P, Miller C C. Neurosci Lett. 1996;217:157–160. [PubMed] [Google Scholar]
- 20.Palmer L G. Membr Biol. 1984;80:153–165. doi: 10.1007/BF01868771. [DOI] [PubMed] [Google Scholar]
- 21.Bevan S, Geppetti P. Trends Neurosci. 1994;17:509–512. doi: 10.1016/0166-2236(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 22.Elliott C E, Becker B, Oehler S, Castanon M J, Hauptmann R, Wiche G. Genomics. 1997;42:115–125. doi: 10.1006/geno.1997.4724. [DOI] [PubMed] [Google Scholar]
- 23.Akopian A N, Wood J N. J Biol Chem. 1995;270:21264–21270. doi: 10.1074/jbc.270.36.21264. [DOI] [PubMed] [Google Scholar]
- 24.Chen Chih-Cheng, Akopian A, Sivilotti L, Colquhoun D, Burnstock G, Wood J N. Nature (London) 1995;377:428–432. doi: 10.1038/377428a0. [DOI] [PubMed] [Google Scholar]
- 25.Akopian A N, Sivilotti L, Wood J N. Nature (London) 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- 26.Bevan S. Prog Brain Res. 1996;113:201–213. doi: 10.1016/s0079-6123(08)61089-4. [DOI] [PubMed] [Google Scholar]
- 27.Caterina M J, Schumacher M A, Tominaga M, Rosen T A, Levine J D, Julius D. Nature (London) 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 28.Akaike N, Ueno S. Prog Neurobiol. 1994;43:73–83. doi: 10.1016/0301-0082(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 29.Shen H, Chan J, Kass I S, Bergold P J. Neurosci Lett. 1995;158:115–118. doi: 10.1016/0304-3940(94)11238-e. [DOI] [PubMed] [Google Scholar]
- 30.Wood J N, Docherty R J. Ann Rev Physiol. 1997;59:457–482. doi: 10.1146/annurev.physiol.59.1.457. [DOI] [PubMed] [Google Scholar]