Abstract
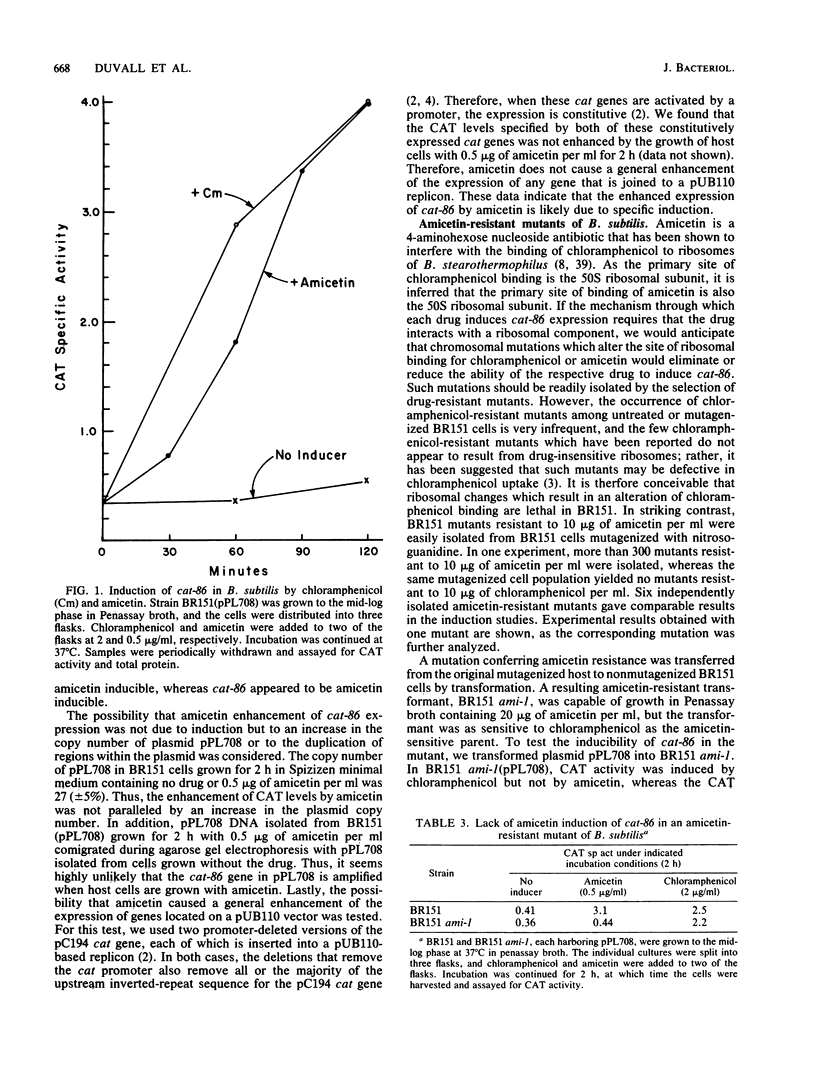
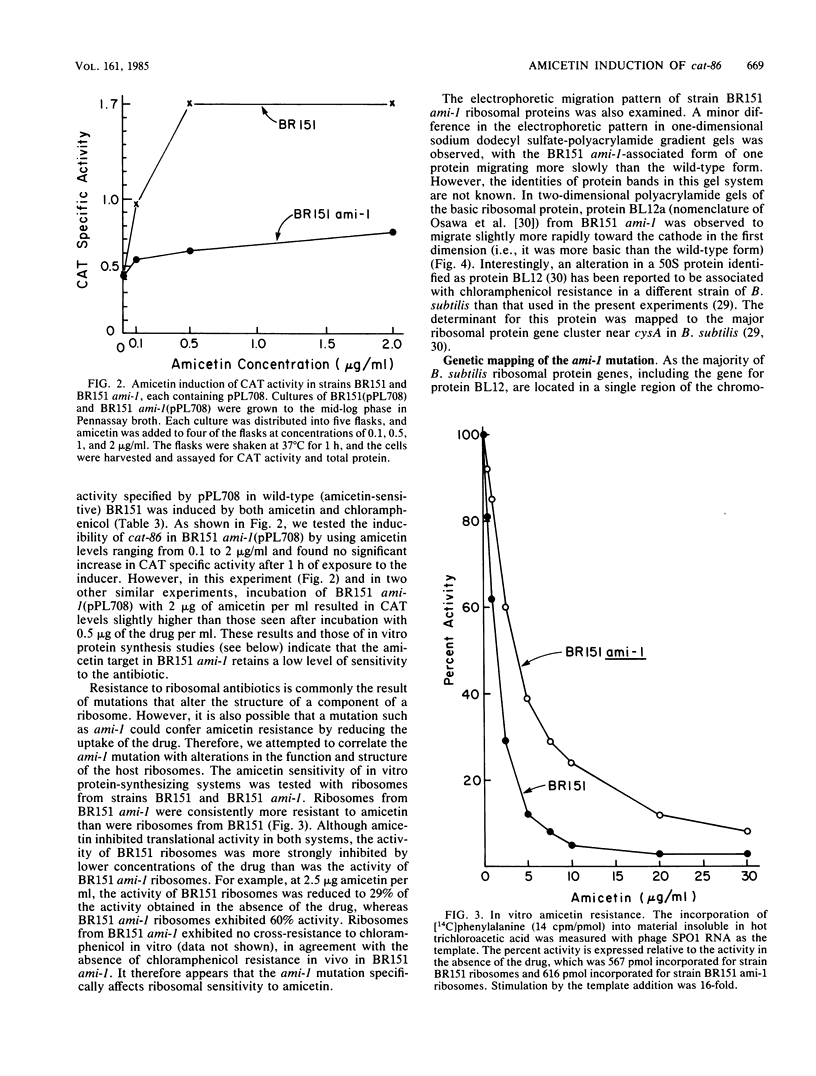
The plasmid gene cat-86 and the cat gene resident on pC194 each encode chloramphenicol-inducible chloramphenicol acetyltransferase activity in Bacillus subtilis. Chloramphenicol induction has been proposed to result from chloramphenicol binding to ribosomes, which then permits the drug-modified ribosomes to perform events essential to induction. If this proposal were correct, B. subtilis mutants containing chloramphenicol-insensitive ribosomes should not permit chloramphenicol induction of either cat-86 or pC194 cat. However, we and others have been unable to isolate chloramphenicol-resistant ribosomal mutants of B. subtilis 168. We therefore developed a simple procedure for screening other antibiotics for the potential to induce cat-86 expression. One antibiotic, amicetin, was found to be an effective inducer of cat-86 but not of the cat gene on pC194. Amicetin and chloramphenicol each interact with the 50S ribosomal subunit, and the mechanism of cat-86 induction by both drugs may be similar. Amicetin-resistant mutants of B. subtilis were readily isolated, and in none of six mutants tested was cat-86 detectably inducible by amicetin, although the chloramphenicol-inducible phenotype was retained. The ami-1 mutation which is present in one of these amicetin-resistant mutants was mapped by PBS1 transduction to the "ribosomal gene cluster" adjacent to cysA. Additionally, ribosomes from cells harboring the ami-1 mutation contained an altered BL12a protein, as detected in two-dimensional polyacrylamide gel electrophoresis. Lastly, an in vitro protein-synthesizing system that uses ribosomes from an ami-1-containing cell line was more resistant to amicetin than a system that uses ribosomes from an amicetin-sensitive but otherwise isogenic strain. These results indicate that the host mutation, ami-1, which effectively abolished the inducibility of cat-86 by amicetin, altered a ribosomal component.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adoutte-Panvier A., Davies J. E., Gritz L. R., Littlewood B. S. Studies of ribosomal proteins of yeast species and their hybrids: gel electrophoresis and immunochemical cross-reactions. Mol Gen Genet. 1980;179(2):273–282. doi: 10.1007/BF00425454. [DOI] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Chow J. H., Mongkolsuk S., Preis L. H., Vollmar W. R., 2nd, Lovett P. S. Constitutive variants of the pC194 cat gene exhibit DNA alterations in the vicinity of the ribosome binding site sequence. Gene. 1984 May;28(2):171–176. doi: 10.1016/0378-1119(84)90254-3. [DOI] [PubMed] [Google Scholar]
- Anderson L. M., Henkin T. M., Chambliss G. H., Bott K. F. New chloramphenicol resistance locus in Bacillus subtilis. J Bacteriol. 1984 Apr;158(1):386–388. doi: 10.1128/jb.158.1.386-388.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L., Yansura D. G., Henner D. J. Construction of a vector for cloning promoters in Bacillus subtilis. Gene. 1983 Dec;26(2-3):313–315. doi: 10.1016/0378-1119(83)90204-4. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Byeon W. H., Weisblum B. Post-transcriptional regulation of chloramphenicol acetyl transferase. J Bacteriol. 1984 May;158(2):543–550. doi: 10.1128/jb.158.2.543-550.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambliss G. H., Henkin T. M., Leventhal J. M. Bacterial in vitro protein-synthesizing systems. Methods Enzymol. 1983;101:598–605. doi: 10.1016/0076-6879(83)01040-x. [DOI] [PubMed] [Google Scholar]
- Chang F. N., Siddhikol C., Weisblum B. Subunit localization studies of antibiotic inhibitors of protein synthesis. Biochim Biophys Acta. 1969 Aug 20;186(2):396–398. doi: 10.1016/0005-2787(69)90020-3. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Lovett P. S., Rudolph C., Vasantha N., Guyer M. Chloramphenicol-inducible gene expression in Bacillus subtilis. Gene. 1983 Oct;24(2-3):171–177. doi: 10.1016/0378-1119(83)90077-x. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1680–1682. doi: 10.1073/pnas.74.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Israeli-Reches M., Dubnau D. Induction of macrolide-lincosamide-streptogramin B resistance requires ribosomes able to bind inducer. Mol Gen Genet. 1984;194(3):357–361. doi: 10.1007/BF00425544. [DOI] [PubMed] [Google Scholar]
- Gryczan T., Israeli-Reches M., Del Bue M., Dubnau D. DNA sequence and regulation of ermD, a macrolide-lincosamide-streptogramin B resistance element from Bacillus licheniformis. Mol Gen Genet. 1984;194(3):349–356. doi: 10.1007/BF00425543. [DOI] [PubMed] [Google Scholar]
- Harwood C. R., Williams D. M., Lovett P. S. Nucleotide sequence of a Bacillus pumilus gene specifying chloramphenicol acetyltransferase. Gene. 1983 Oct;24(2-3):163–169. doi: 10.1016/0378-1119(83)90076-8. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Chambliss G. H. Genetic mapping of a mutation causing an alteration in Bacillus subtilis ribosomal protein S4. Mol Gen Genet. 1984;193(2):364–369. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Byeon W. H., Weisblum B. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol. 1983 Jun;154(3):1252–1262. doi: 10.1128/jb.154.3.1252-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G. A., Traut R. R. Separation and radioautography of microgram quantities of ribosomal proteins by two-dimensional polyacrylamide gel electrophoresis. FEBS Lett. 1973 Jan 15;29(2):177–180. doi: 10.1016/0014-5793(73)80555-1. [DOI] [PubMed] [Google Scholar]
- Jamet C., Anagnostopoulos C. Etude d'une mutation très faiblement transformable au locus de la thréonine désaminase de Bacillus subtilis. Mol Gen Genet. 1969;105(3):225–242. doi: 10.1007/BF00337474. [DOI] [PubMed] [Google Scholar]
- Keggins K. M., Lovett P. S., Duvall E. J. Molecular cloning of genetically active fragments of Bacillus DNA in Bacillus subtilis and properties of the vector plasmid pUB110. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1423–1427. doi: 10.1073/pnas.75.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Keggins K. M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- Lovett P. S. Plasmid in Bacillus pumilus and the enhanced sporulation of plasmid-negative variants. J Bacteriol. 1973 Jul;115(1):291–298. doi: 10.1128/jb.115.1.291-298.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Young F. E. Genetic analysis in Bacillus pumilus by PBSI-mediated transduction. J Bacteriol. 1970 Feb;101(2):603–608. doi: 10.1128/jb.101.2.603-608.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Ambulos N. P., Jr, Lovett P. S. Chloramphenicol-inducible gene expression in Bacillus subtilis is independent of the chloramphenicol acetyltransferase structural gene and its promoter. J Bacteriol. 1984 Oct;160(1):1–8. doi: 10.1128/jb.160.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S., Takata R., Tanaka K., Tamaki M. Chloramphenicol resistant mutants of Bacillus subtilis. Mol Gen Genet. 1973 Dec 20;127(2):163–173. doi: 10.1007/BF00333664. [DOI] [PubMed] [Google Scholar]
- Osawa S., Tokui A., Saito H. Mapping by interspecies transformation experiments of several ribosomal protein genes near the replication origin of Bacillus subtilis chromosome. Mol Gen Genet. 1978 Aug 17;164(2):113–129. doi: 10.1007/BF00267376. [DOI] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J Biol Chem. 1967 Feb 25;242(4):687–693. [PubMed] [Google Scholar]
- Tanaka H., Oiwa R., Matsukura S., Omura S. Amphomycin inhibits phospho-N-acetylmuramyl-pentapeptide translocase in peptidoglycan synthesis of Bacillus. Biochem Biophys Res Commun. 1979 Feb 14;86(3):902–908. doi: 10.1016/0006-291x(79)91797-2. [DOI] [PubMed] [Google Scholar]
- Williams D. M., Duvall E. J., Lovett P. S. Cloning restriction fragments that promote expression of a gene in Bacillus subtilis. J Bacteriol. 1981 Jun;146(3):1162–1165. doi: 10.1128/jb.146.3.1162-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. M., Schoner R. G., Duvall E. J., Preis L. H., Lovett P. S. Expression of Escherichia coli trp genes and the mouse dihydrofolate reductase gene cloned in Bacillus subtilis. Gene. 1981 Dec;16(1-3):199–206. doi: 10.1016/0378-1119(81)90076-7. [DOI] [PubMed] [Google Scholar]
- Wilson C. R., Skinner S. E., Shaw W. V. Analysis of two chloramphenicol resistance plasmids from Staphylococcus aureus: insertional inactivation of Cm resistance, mapping of restriction sites, and construction of cloning vehicles. Plasmid. 1981 May;5(3):245–258. doi: 10.1016/0147-619x(81)90002-0. [DOI] [PubMed] [Google Scholar]




