Abstract
Balbiani ring (BR) pre-mRNP particles reside in the nuclei of salivary glands of the dipteran Chironomus tentans and carry the message for giant-sized salivary proteins. In the present study, we identify and characterize a new protein component in the BR ribonucleoprotein (RNP) particles, designated hrp23. The protein with a molecular mass of 20 kD has a single RNA-binding domain and a glycine-arginine-serine–rich auxiliary domain. As shown by immunoelectron microscopy, the hrp23 protein is added to the BR transcript concomitant with transcription, is still present in the BR particles in the nucleoplasm, but is absent from the BR particles that are bound to the nuclear pore complex or are translocating through the central channel of the complex. Thus, hrp23 is released just before or at the binding of the particles to the nuclear pore complex. It is noted that hrp23 behaves differently from two other BR RNP proteins earlier studied: hrp36 and hrp45. These proteins both reach the nuclear pore complex, and hrp36 even accompanies the RNA into the cytoplasm. It is concluded that each BR RNA-binding protein seems to have a specific flow pattern, probably related to the particular role of the protein in gene expression.
Keywords: hnRNP protein, nonshuttling proteins, nucleocytoplasmic transport, Balbiani rings, nucleolus
Immediately upon synthesis, pre-messenger RNA (pre-mRNA)1 molecules associate with proteins to form heterogeneous nuclear ribonucleoprotein particles (hnRNPs) (Dreyfuss et al., 1993). The predominant hnRNP proteins remain bound to the transcripts while these reside in the nucleus. When the processed transcripts leave the nucleus through the nuclear pores, some of the hnRNP proteins are displaced from the transcripts, while others accompany the transcripts into the cytoplasm (Pinol-Roma and Dreyfuss, 1992; Daneholt, 1997). For a long time, no specific function could be assigned to the hnRNP proteins, but substantial information has now accumulated suggesting that they play crucial roles in pre-mRNA splicing as well as in the transport of mature mRNA into the cytoplasm.
The hnRNP proteins are among the most abundant in the cell nucleus (Michael et al., 1995b ). In human, the hnRNP particles contain more than 20 major protein species, designated hnRNP A1 to hnRNP U (Pinol-Roma et al., 1988). In Drosophila, more than 10 of the predominant hnRNP proteins have been found and characterized, some of them showing high homology to the human proteins (e.g., Haynes et al., 1990; Amero et al., 1991; Matunis et al., 1992). The hnRNP proteins have a modular structure containing one or more RNA-binding motifs and an auxiliary domain (Dreyfuss et al., 1993). The predominant RNA-binding domain (RBD) is composed of 90–100 amino acids and is usually called the RNP consensus RBD or the RNA recognition motif; within this sequence, there is a highly conserved octapeptide, RNP-1, and a hexapeptide, RNP-2. Several of the most abundant hnRNP proteins, e.g., hnRNP A1, have a glycine-rich auxiliary domain and are designated RBD-Gly proteins (Dreyfuss et al., 1993).
The hnRNP proteins are added to the growing RNA transcript concomitant with transcription (Economides and Pederson, 1983; Matunis et al. 1993; Wurtz et al., 1996), and an RNP fibril is immediately formed (Miller and Bakken, 1972; Skoglund et al., 1983; Fakan, 1994). If the transcript is long, the fibril is further folded into a compact RNP particle (Monneron and Bernhard, 1969; Skoglund et al., 1986). The packing of the transcript into an RNP fibril or RNP particle could be important to ensure efficient transport and a minimum of irrelevant interactions and entanglements. Furthermore, the RNP complex is organized in a nonrandom manner. The various hnRNP proteins bind to RNA in a sequence-dependent manner (for review see Dreyfuss et al., 1993) and are, therefore, likely to appear in a specific arrangement along an hnRNA molecule. In addition, when the basic RNP fibril is further folded, the RNP particles attain a characteristic shape (Malcolm and Sommerville, 1974; Skoglund et al., 1986). Thus, the RNA molecule is organized in a specific way in the RNP complexes, presumably exposing certain sequences for molecular interactions while others are likely to be hidden in the complex.
The hnRNP proteins are important to accomplish proper splicing of pre-mRNA. The splicing takes place on the growing RNP complex (Osheim et al., 1985; Beyer and Osheim, 1988; LeMaire and Thummel, 1990; Baurén and Wieslander, 1994) or on the completed RNP product released from the template (Nevins, 1983; see also Baurén and Wieslander, 1994). It was shown early on that antibodies to hnRNP proteins can block splicing in vitro, suggesting that the hnRNP proteins are involved in the splicing reaction (Choi et al., 1986; Sierakowska et al., 1986). There is growing evidence suggesting that SR proteins, an extensively studied family of splicing factors, could also be regarded as constitutive components of the RNP complex rather than as spliceosome components that are assembled and disassembled in conjunction with splicing (Alzhanova-Ericsson et al., 1996; see also Cáceres et al., 1998). Like the classical hnRNP proteins, the SR proteins contain one or more RNA-binding domains of the RNP consensus type and an auxiliary domain (Birney et al., 1993; Fu, 1995). The auxiliary domain of the SR proteins is characterized by a series of serine-arginine dipeptides (SR domain). The SR proteins are essential splicing factors, which can often also regulate alternative splicing (for reviews see Fu, 1995; Manley and Tacke, 1996; Valcárcel and Green, 1996). They not only take part in establishing the early commitment complex but also participate in later stages of the splicing process. The main role of the SR proteins seems to be to form molecular bridges between essential components of the spliceosome. It is interesting to note that hnRNP A/B proteins and SR proteins act antagonistically in the regulation of alternative splicing in vitro (Mayeda and Krainer, 1992; Mayeda et al., 1994). Presumably, the various SR proteins have different functions and/or targets during splicing (e.g., Manley and Tacke, 1996), and at least some of them are likely to play a regulatory role during development (Ring and Lis, 1994).
The hnRNP proteins are also directly involved in the transport process. It has been revealed that hnRNP A1, which shuttles between the nucleus and the cytoplasm (Pinol-Roma and Dreyfuss, 1992), contains a nuclear export signal (NES) (Michael et al., 1995a ). It has also been demonstrated for an hnRNP A1 homologue that it leaves the nucleus still associated with mRNA (Visa et al., 1996a ). Taken together, this information strongly supports the proposition that the export signal in hnRNP A1 mediates not only the export of A1 itself but also the exit of the entire mRNP complex (Pinol-Roma and Dreyfuss, 1992). Recent microinjection experiments have further strengthened the notion that the A1 NES plays a role in the export of mRNA; however, the export of other classes of RNA remains unaffected (Izaurralde et al., 1997a ). It seems likely that the NES domain of A1 interacts with a soluble export receptor because such receptors, designated exportins and involved in the export of proteins with leucine-rich NESs (including the Rev protein), have recently been identified (Fornerod et al., 1997; Fukuda et al., 1997; Stade et al., 1997). An A1-interacting protein called transportin mediates the reentry of A1 into the nucleus (Pollard et al., 1996; Fridell et al., 1997), but it is uncertain whether it is also the RNP export receptor looked for (Izaurralde et al., 1997a ,b; Siomi et al., 1997). It should be added that a putative export signal different from the A1 NES has recently been identified in hnRNP K (Michael et al., 1997). Because hnRNP A1 and hnRNP K are likely to be bound to the same transcript and both in many copies, the transport machinery for mRNP could be quite complex.
Finally, the hnRNP proteins could be involved in active retention of mRNAs in the nucleus. The nonshuttling hnRNP proteins, e.g., hnRNP C1, C2, and U (Pinol-Roma and Dreyfuss, 1991, 1992), are stripped off from RNP particles before mRNA is transported into the cytoplasm. A 78–amino acid–long nuclear retention signal has been discovered in the auxiliary domain of hnRNP C1 (Nakielny and Dreyfuss, 1996). This nuclear retention signal can override nuclear export signals in the shuttling hnRNP proteins, and therefore, the nonshuttling proteins have to be actively removed from the hnRNP complex before the nucleocytoplasmic translocation.
The hnRNP complexes bind to and pass through the nuclear pore complex (NPC) in an ordered, multistep process (Daneholt, 1997). The NPC consists of a spoke assembly sandwiched between a nuclear and a cytoplasmic ring (for recent review see Panté and Aebi, 1996). In the center of the NPC, there is a plug containing a central, transport-mediating channel. In addition, nuclear fibers extend from the nuclear ring into the nucleoplasm, forming a well- defined basket structure, and cytoplasmic fibers anchored in the cytoplasmic ring reach into the cytoplasm. The RNP complexes bind to the nuclear basket, are transferred to the entrance of the central channel, and translocate through the channel (Daneholt, 1997; Panté et al., 1997). On the cytoplasmic side, they enter the cytoplasm with no obvious contact with the cytoplasmic fibers. Before or in conjunction with the passage through the NPC, the hnRNP complexes lose the nonshuttling proteins (Pinol-Roma and Dreyfuss, 1992; Alzhanova-Ericsson et al., 1996), while the shuttling ones seem to remain attached to the RNA molecule (Visa et al., 1996a ). Our knowledge of the molecular structure of the NPC is rapidly increasing (Doye and Hurt, 1997; Ohno et al., 1998), but there is still only limited information at the molecular level on the nature of the initial binding of the mRNP to the NPC and the further translocation of the particle through the NPC.
To further elucidate the structure of the hnRNP complexes and the function of the various hnRNP proteins involved, it is a great advantage to be able to analyze a specific pre-mRNP complex. The Balbiani ring (BR) pre-mRNP particles in the salivary glands of the dipteran Chironomus tentans offer such a possibility (Daneholt, 1997). By electron microscopy, it is possible to follow how the BR pre-mRNP particle is assembled along a gene, and how the released BR pre-mRNP particle is transported in the nucleoplasm to and through a nuclear pore. It has been possible to define a series of discrete steps during the passage of the particle through the NPC and to reveal drastic conformational changes of the particle during the translocation. Furthermore, the fate of defined hnRNP proteins in the BR particles can be investigated by immunoelectron microscopy. Two hnRNP proteins, hrp36 and hrp45, have been analyzed in detail (Wurtz et al., 1996). The hrp36 protein shows high homology to human hnRNP A1 and Drosophila hrp40 (Visa et al., 1996a ). It is being added to BR particles concomitant with transcription and accompanies the BR-mRNA through the nuclear pore and ends up with mRNA in polysomes in cytoplasm. The hrp45 protein is an SR protein and is similar to the human splicing factor SF2/ASF and Drosophlia SRp55/B52 (Alzhanova-Ericsson et al., 1996). This protein is confined to the nucleus and is released when the particle enters the central channel of the nuclear pore complex.
In the present study, we have identified a third hnRNP protein in the BR particles, hrp23, and determined its fate. This protein contains one RNA-binding domain and a glycine-arginine-serine–rich auxiliary domain. It is being added to the BR transcript concomitant with transcription and is released from the BR particle just before or at the binding of the particle to the nuclear pore complex, i.e., hrp23 is shed late but still clearly before hrp45, the nonshuttling protein earlier studied. It seems likely, therefore, that there is not a single protein-removal step at nucleocytoplasmic transport but rather a series of preparatory steps before the actual translocation of the RNP particle through the pore. We conclude that each hnRNP protein seems to have a specific flow pattern during nucleocytoplasmic transport, presumably coupled to the particular function of the protein. It should finally be added that unlike hrp36 and hrp45, hrp23 is present also in the nucleoli.
Materials and Methods
Culturing Conditions and Drug Treatments
C. tentans was cultured as described by Lezzi et al. (1981), and salivary glands were isolated from fourth instar larvae. C. tentans tissue culture cells were grown in suspension at 24°C as previously described (Wyss, 1982). In actinomycin D treatments, the cells were first incubated for 30 min in fresh medium containing 50 μg/ml cycloheximide. The actinomycin D was subsequently added to the culture at a final concentration of 5 μg/ml, and the incubation continued for an additional 60 min. In all cases, control cells were incubated in parallel in fresh medium without drugs.
Preparation of RNP Extract from C. tentans Tissue Culture Cells
The RNP extract was prepared as earlier described (Wurtz et al., 1996). In short, C. tentans tissue culture cells were washed in PBS (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, and 2 mM NaH2PO4, pH 7.2), resuspended in TNM (10 mM triethanolamine-HCl, pH 7.0, 100 mM NaCl, and 1 mM MgCl2) containing 0.2% NP-40 and 0.1 mM PMSF, homogenized in a glass tissue grinder with a tight-fitting pestle, and centrifuged at 2,000 g for 5 min at 4°C. The pellet, containing the nuclei, was resuspended in TNM provided with E. coli tRNA (0.1 mg/ml), sonicated, and centrifuged at 7,000 g for 10 min at 4°C. The supernatant, designated the RNP extract, was used for immunoprecipitation of hnRNP complexes and for Western blot analysis.
Generation of Monoclonal Antibody 1D3
The 1D3 antibody is a mouse mAb obtained from spleen cells of BALB/c mice immunized with C. tentans hnRNP proteins as follows:
Immunoprecipitation of hnRNP Complexes from C. tentans RNP Extract.
hnRNP complexes were immunoprecipitated from the RNP extract essentially as described by Wurtz et al. (1996). Tissue culture cells from 500 ml of medium were used to prepare 4 ml of the RNP extract (in TNM). The RNP extract was supplemented with NP-40 to a final concentration of 0.1% and mAb 4F9 (an anti-hrp36 antibody) to 20 μg/ml. The mixture was incubated at 4°C with gentle rotation. After 90 min of incubation, 100 μl of rabbit anti–mouse immunoglobulin coupled to protein A–Sepharose was added to the mixture, and the incubation continued at 4°C for an additional 90 min. The immunoglobulin–protein A–Sepharose complex was prepared according to Wurtz et al. (1996). Once the incubation was finished, the Sepharose resin was sedimented, washed twice with TNM containing 0.1% NP-40, and washed once with TNM. The immunoprecipitated material was finally eluted from the resin with 0.5% SDS in water. The eluted proteins were precipitated with cold acetone and resuspended in PBS. This procedure gave 25–50 μg of hnRNP proteins as judged from Coomassie blue–stained SDS-PAGE gels.
Immunization of BALB/c Mice.
The immunization was carried out following standard procedures as described by Harlow and Lane (1988). Approximately 15–20 μg of hnRNP proteins, dissolved in PBS, were mixed with complete Freund adjuvant and injected intraperitoneally to BALB/c mice. Three boost injections with the same amount of antigen in incomplete adjuvant were given in 2-wk intervals. 10 d after the last boost, the sera of the immunized mice were tested by Western blot analysis to detect antibodies against C. tentans proteins. The mouse that gave the strongest response received an intravenous injection of antigen in PBS without adjuvant.
Fusion and Screening of Suitable mAbs.
3 d after the last injection, the spleen of the immunized mouse was macerated, and the spleen cells were fused with mouse myeloma Sp20 cells in the presence of poly-ethylene-glycol 4000 (GIBCO-BRL, Life Technologies, Inc., Grand Island, NY). The fused cells were plated onto 96-well plates and cultured in Optimum medium. The standard hypoxanthine-aminopterin-thymidine (HAT) selection procedure was applied to select the hybridoma clones (Harlow and Lane, 1988). The media from wells with growing cells were tested by Western blot analysis against C. tentans RNP extracts. To select antibodies binding to Balbiani rings, the media were also tested by immunostaining of polytene chromosome squashes (Visa et al., 1996b ). Cells from the positive wells were cloned at least three times by limiting dilution. The mAb 1D3, which recognized a 23-kD protein and bound to BRs, was chosen for further analysis.
SDS-PAGE and Western Blot Analysis
The proteins in the RNP extract were precipitated with cold acetone, resuspended in sample buffer (10% glycerol, 2% SDS, 10 mM DTT, 0.02% bromophenol blue, and 0.0625 M Tris-HCl, pH 6.8), and separated by electrophoresis in a 10% polyacrylamide gel containing 0.1% SDS. After electrophoresis, the proteins were blotted to transfer membranes (Immobilon-P; Millipore Corp., Bedford, MA) using a Trans-Blot semidry electrophoretic system (Bio-Rad Laboratories, Hercules, CA). Membranes were blocked in PBS containing 10% dry milk powder for 1 h and incubated with primary antibodies diluted in PBS containing 1% dry milk powder and 0.05% Tween-20. Labeling was visualized with alkaline phosphatase–conjugated anti–mouse immunoglobulin (DAKO A/S, Glostrup, Denmark) using the NBT/BCIP system (Promega Corp., Madison, WI).
Immunostaining of Polytene Chromosomes
Salivary glands were isolated from fourth instar larvae and prefixed for 90 s at room temperature in 2% paraformaldehyde in TKM (100 mM KCl, 1 mM MgCl2, and 10 mM triethanolamine-HCl, pH 7.0). The prefixed glands were washed in cold TKM for 5 min, kept for 60 s in TKM buffer containing 2% NP-40, and transferred into 0.025% NP-40 in TKM. The subsequent isolation of the polytene chromosomes was performed on a cold stage close to 0°C. Under a stereomicroscope, the glands were repeatedly sucked in and out of a micropipette with a narrow opening (bore diameter 250 μm) to release polytene chromosomes. Single chromosomes were identified and transferred with a pipette into a drop of TKM placed on a siliconized glass slide (for details see Björkroth et al., 1988). The NP-40– containing solution was exchanged with fresh TKM to get the chromosomes to stick to the surface of the glass slide. The isolated polytene chromosomes were postfixed with 4% paraformaldehyde in TKM for 30 min at room temperature and washed three times with fresh TKM. Subsequently, they were used immediately for the immunocytological analysis.
For preparation of RNase-treated chromosomes, the isolated polytene chromosomes were incubated with 100 μg/ml of RNase A in TKM buffer for 60 min at room temperature immediately after isolation. The RNase A–treated chromosomes were then rinsed in TKM, postfixed, and incubated with primary and secondary antibodies as described above.
For immunocytological analysis, the isolated chromosomes were blocked with 50 μl of 2% BSA in TKM for 30 min at room temperature in a humid chamber and incubated for 45 min with 40 μl of undiluted 1D3 hybridoma supernatant or 40 μl of the negative control mouse monoclonal anti–factor VIII antibody (DAKO A/S) diluted 1:50 in TKM. The slides were washed three times for 5 min in 0.1% Tween-20 in TKM and incubated for 90 min with 50 μl of the gold-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA; IgG, 6 nm in diameter) diluted 1:50 in TKM containing 0.5% BSA. The specimens were washed three times for 5 min in TKM and with distilled water, an immunogold silver enhancement solution (IntenSEM; Amersham Corp., Buckinghamshire, England) was added onto the specimens for 8–10 min at room temperature, and the specimens were rinsed in distilled water, mounted in 30% glycerol, and examined and photographed in a light microscope (Carl Zeiss, Inc., Thornwood, NY).
Isolation of 1D3-specific cDNA Clones
The hybridoma culture supernatant containing mAb 1D3 was diluted 1:100 to screen a randomly primed λgt11 C. tentans salivary gland cDNA library by the ProtoBlot Immunoscreening System (Promega Corp.). Two antibody-specific clones were purified and sequenced. One of them, pHRP23.1, was used as template for PCR amplification with λgt11 forward and reverse primers (Promega Corp.). The amplified DNA fragment was purified by the Wizard™ PCR Preps DNA Purification Kit and labeled by the DIG DNA Labeling and Detection Kit (Boehringer Mannheim GmbH, Mannheim, Germany). The labeled probe was used to screen an oligo dT–primed λZAP cDNA library from the salivary glands of C. tentans following the manufacturer's instructions. 10 positive clones were obtained and purified for sequence analysis.
DNA Sequencing and Sequence Analysis
The cDNA inserts of positive clones were amplified by PCR. The purified DNA fragments were used for sequencing with walking primers. The Taq DyeDeoxy™ Terminator Cycle Sequencing Kit (PE Applied Biosystems, Warrington, GB) was applied for the sequence reaction and sequencing gel was run on an automated DNA sequencer (model 373A; PE Applied Biosystems). The DNA sequences were analyzed with the University of Wisconsin Genetics Computer Group Sequence Analysis Programs (Devereux et al., 1984) and EGCG extensions to the Wisconsin Package Sequence Analysis Programs.
Expression of hrp23 and Identification of Expression Product
The hrp23 protein was expressed according to the Promega protocol for expression of lambda lysogen in preparative amounts. The E. coli Y1089 were infected with a λgt11 clone, pHRP23.1, containing hrp23 cDNA, and the presence of the hrp23 sequence insert was confirmed by PCR using inner primers. Half of the bacterial culture was IPTG-induced, while the other half served as control. Both cultures were lysed in 2× sample buffer at 95°C for 5 min, and the supernatants were analyzed with SDS-PAGE and Western blotting as described above with a few modifications. The electrophoresis was performed in 12% polyacrylamide gels. The 1D3 hybridoma supernatant was diluted 1:100. The membrane was subjected to ECL (Amersham Corp.) according to the manufacturer's instructions using a peroxidase-conjugated goat anti–mouse immunoglobulin (DAKO A/S) diluted 1:500 in PBS provided with 0.1% Tween.
Alkaline Phosphatase Treatment of RNP Extract
The RNP extract was divided into two equal portions: one was treated with 250 U/ml of calf intestine alkaline phosphatase (Boehringer Mannheim GmbH) for 30 min at 37°C and the other in parallel with no enzyme added but with 5 mM β-glycerophosphate. The protein was precipitated with acetone and resuspended for SDS-PAGE as described above.
Immunocytology on Cryosections
Immunostaining on semithin cryosections of tissue culture cells and salivary glands was performed as previously described (Visa et al., 1996a ). When C. tentans tissue culture cells were used, the cells were washed once in PBS and collected as a pellet. The cell pellet and salivary glands were fixed for 45–60 min in 4% formaldehyde in 0.1 M cacodylate buffer, pH 7.2, at room temperature. The fixed pellet and salivary glands were cryoprotected with 2.3 M sucrose and frozen by immersion in liquid nitrogen. Semithin sections (0.5 μm) were obtained in a cryo-ultramicrotome (Ultracut S/FC S; Reichert-Jung, Vienna, Austria) and mounted on glass slides. Before immunolabeling, the sections were incubated in PBS containing 0.1 M glycine and 2% BSA. As first antibody, we used either undiluted 1D3-hybridoma supernatant or 20 μg/ml anti–factor VIII antibody (negative control). In the second step of the immunostaining, we used an antibody against mouse IgG conjugated to 6-nm gold particles (Amersham Corp.). The immunogold labeling was silver-enhanced with IntenSE™ M (Amersham Corp.).
Immunoelectron Microscopy
Immunoelectron microscopy was essentially carried out according to the method described by Tokuyasu (1980) with a few modifications (Visa et al., 1996a ). The specimens were prepared as described above for light microscopy except that fixation was performed for 20–25 min in 4% formaldehyde and 0.1% glutaraldehyde. Ultrathin cryosections were picked up on drops of 2.3 M sucrose and deposited on nickel grids coated with formvar and carbon. The grids were floated on drops of PBS containing 10% new born calf serum before incubation with the antibody solutions. The same antibodies as described above under “Immunocytology on Cryosections” were used. After immunolabeling, the sections were stained with 2% aqueous uranyl acetate and embedded in 4% polyvinyl alcohol (9–10 kD; Aldrich Chemical Co., Milwaukee, WI). The specimens were examined and photographed in a microscope (model 100 CX; JEOL U.S.A., Peabody, MA) at 80 kV.
Results
Generation and Selection of a Monoclonal Antibody against a 23-kD hnRNP Protein in C. tentans
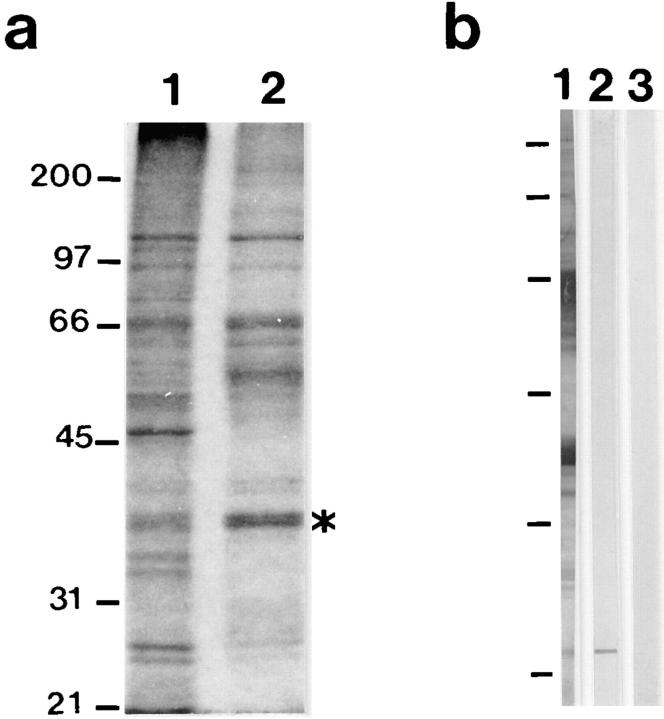
Monoclonal antibodies were raised against hnRNP proteins immunoprecipitated from a C. tentans nuclear RNP extract. C. tentans tissue culture cells were homogenized, and the intact nuclei were collected and used for preparation of the RNP extract as described by Wurtz et al. (1996). The RNP extract contained a large number of proteins as shown by SDS-PAGE (Fig. 1 a, lane 1). The hnRNP complexes were immunoprecipitated from the RNP extract by a two-step procedure, using a monoclonal antibody against hnRNP protein hrp36 (mAb 4F9) and an anti–mouse immunoglobulin coupled to protein A–Sepharose. The bound proteins were eluted, precipitated, and subjected to SDS-PAGE (Fig. 1 a, lane 2). The immunoprecipitation resulted in a less complex protein pattern suitable for production of monoclonal antibodies.
Figure 1.
Generation and characterization of monoclonal antibody 1D3. (a) SDS-PAGE of the proteins in the RNP extract from C. tentans tissue culture cells (lane 1), and of the proteins immunoprecipitated from the RNP extract by mAb 4F9 and used for subsequent immunization of BALB/c mice (lane 2). The protein recognized by mAb 4F9, hrp36, is indicated with an asterisk at lane 2. The position of molecular mass standards is shown to the left in kilodaltons. (b) Western blot analysis of the RNP extract probed with serum from an immunized mouse (lane 1), mAb 1D3 (lane 2), and negative control antibody (lane 3). The molecular mass standards are the same as in a.
The immunoprecipitated proteins were used to immunize BALB/c mice according to the standard procedure. Several major proteins in the nuclear extract reacted with mouse sera in Western blots (Fig. 1 b, lane 1). The spleen cells of the immunized mouse were fused with mouse myeloma Sp20 cells, and hybridoma clones were selected with a standard hypoxanthine-aminopterin-thymidine (HAT) procedure. Positive clones were identified by screening hybridoma supernatants against the C. tentans RNP extract in Western blot experiments (Wurtz et al., 1996). The supernatant of one hybridoma clone, 1D3, showed high affinity for a 23-kD protein in the C. tentans nuclear RNP extract (Fig. 1 b, lane 2); the same result was obtained with a C. tentans cytoplasmic extract (data not shown). No bands were detected in the control lane with the anti–factor VIII antibody (Fig. 1 b, lane 3). Furthermore, in preliminary chromosome squash experiments, the 1D3 antibody reacted with the BRs (data not shown). On the basis of these experiments, we selected the 1D3 clone for further experiments and designated the corresponding 23-kD protein Ct-hrp23 (or hrp23, for short), following earlier established nomenclature for hnRNP proteins in Diptera (Matunis et al., 1992; Wurtz et al., 1996).
hrp23 Is Present in BRs, Other Chromosomal Puffs, and in Nucleoli
To determine the chromosomal distribution of hrp23, we performed immunolabeling experiments on isolated polytene chromosomes. The chromosomes were isolated from prefixed salivary glands, immunolabeled with mAb 1D3 (or anti–factor VIII antibody as control), visualized by a gold-conjugated secondary antibody, and silver enhanced. The 1D3 antibody reacted strongly with the large BRs on chromosome IV (Fig. 2 a). However, smaller puffs were also immunostained as seen both on chromosome IV (Fig. 2 a) and chromosome III (Fig. 2 c). In addition, it was noted that nucleoli stained with 1D3 (Fig. 2 c). The staining of BRs, minor puffs, and nucleoli was sensitive to RNase (not shown). The negative control did not exhibit any significant staining (Fig. 2, b and d). In conclusion, the hrp23 protein is likely to be present in BRs, a large number of additional, smaller chromosomal puffs, and in the two nucleoli. As the immunostaining is sensitive to RNase, it is suggested that hrp23 is a protein associated with RNA.
Figure 2.
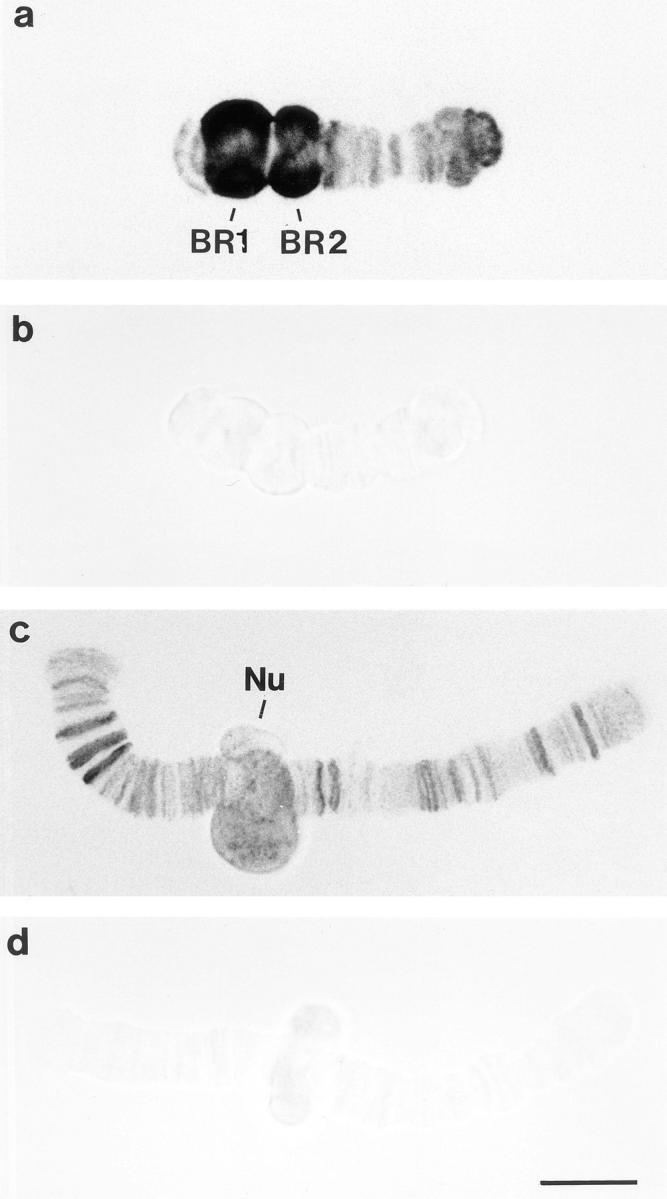
Immunolocalization of hrp23 on isolated C. tentans polytene chromosomes. The chromosomes were isolated from prefixed salivary glands and incubated with monoclonal antibody 1D3 (a and c) or with an anti–factor VIII antibody as negative control (b and d). The antibody-binding sites were visualized by a gold-conjugated secondary antibody and silver enhancement. Examples of chromosome IVs (a and b) and chromosomes IIIs (c and d) are shown. The positions of BR1, BR2, and nucleolus (Nu) have been indicated. Bar, 10 μm.
cDNA Cloning of hrp23
By screening cDNA libraries prepared from C. tentans salivary glands, the full cDNA sequence for hrp23 was found. First, the 1D3 antibody was used to screen a randomly primed λgt11 cDNA library made from salivary glands of C. tentans. Two antibody-specific clones were isolated and sequenced. One of the clones, pHRP23.1, was used to synthesize a DNA probe by PCR amplification. The probe was labeled with digoxigenin and used to screen an oligo(dT)-primed λZAP cDNA library made from salivary glands. 10 positive clones were found and sequenced (see below). All clones had the same coding sequence; no variant was found in spite of extensive screening of the salivary gland cDNA libraries.
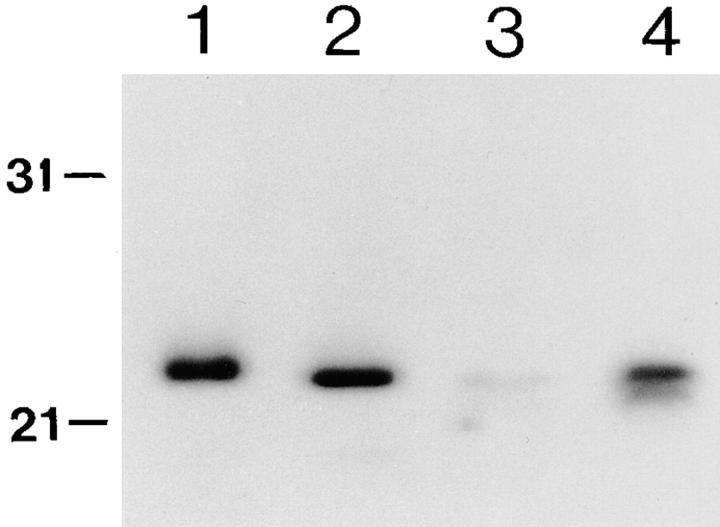
To demonstrate that the protein, encoded in the cDNA clone, is indeed recognized by 1D3 in the RNP extract, a crude lysate containing the hrp23 recombinant protein was prepared by expressing the desired λ lysogen in Escherichia coli. The expressed protein was then compared with the native hrp23 protein extract by Western blot analysis using the 1D3 antibody and the ECL detection system. In the extract from the induced bacteria, the 1D3 antibody recognized a prominent protein fraction corresponding to an apparent molecular mass of 23 kD (Fig. 3, lane 4). A very weak band at the same position was seen in the noninduced bacteria, probably because of leaky expression of the lysogen (Fig. 3, lane 3). The recombinant protein migrated somewhat more rapidly than hrp23 in the RNP extract from C. tentans tissue culture cells (Fig. 3, lane 1). When the RNP extract was treated with alkaline phosphatase, this discrepancy disappeared (Fig. 3, lane 2). We conclude that the recombinant protein reacts with the 1D3 antibody and has the expected apparent molecular mass of hrp23. In addition, it is inferred that the native hrp23 protein is likely to be phosphorylated, which has been further supported by phosphate labeling experiments in vivo (Aissouni, Y., and B. Daneholt, unpublished results).
Figure 3.
Western blot comparison of hrp23 from C. tentans tissue culture cells with hrp23 expressed in E. coli. The extracted proteins were separated by SDS-PAGE, blotted onto transfer membranes, and probed with mAb 1D3. Lane 1, RNP extract from tissue culture cells. Lane 2, alkaline phosphatase–treated RNP extract from tissue culture cells. Lane 3, extract from E. coli, containing the hrp23 cDNA clone, before IPTG induction. Lane 4, extract from E. coli, containing the hrp23 cDNA clone, after IPTG induction. The positions of two molecular mass standards are indicated to the left.
The cDNA Sequence of hrp23
The hrp23 protein contains 191 amino acids, and the predicted molecular mass is 20 kD, which is somewhat smaller than the apparent molecular mass in SDS-PAGE (23 kD). hrp23 has a modular structure like other hnRNP proteins: there is an NH2-terminal RNA-binding domain containing the RNP-1 and RNP-2 consensus sequences, and a COOH-terminal auxiliary domain rich in glycine (37%), arginine (21%), and serine (16%) (Fig. 4). It should be noted that the auxiliary domain contains clustered glycines, three Arg-Gly-Gly (RGG) tripeptides, and eight Arg-Ser or Ser-Arg dipeptides.
Figure 4.
Nucleotide sequence and deduced amino acid sequence of the hrp23 cDNA clone pHRP23.2. This HRP clone was the longest one recorded and sequenced. The RBD is boxed. Within RBD, the two most highly conserved sequence elements, RNP-1 (solid line) and RNP-2 (dashed line) are underlined. The end of the coding region is indicated by an asterisk. These sequence data are available from the EMBL DNA data base under accession number AJ003820.
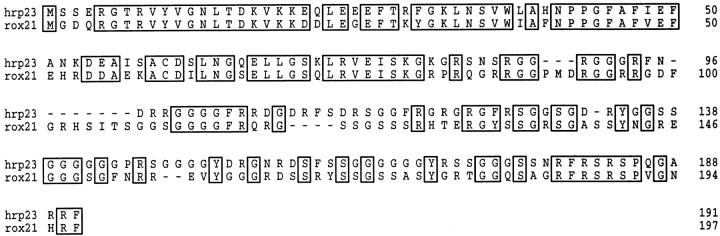
The hrp23 protein was compared with other RNA-binding proteins using the EMBL data base (Rice et al., 1993) and the Swiss-PROT data base (Bairoch and Boeckmann, 1993). A Drosophila protein, ROX21 (Brand et al., 1995), proved to be very similar to hrp23; the amino acid identity between the two proteins amounts to 74% in the RBD domain and to 48% in the auxiliary domain (Fig. 5). Furthermore, hrp23 (and ROX21) shares major features with several RNA-binding proteins in mammals, insects, and plants, which contain one or two NH2-terminal RBDs and a COOH-terminal auxiliary domain.
Figure 5.
Primary structure comparison of hrp23 with Drosophila Rox21 protein. The two complete amino acid sequences are aligned and presented by the PILEUP and PRETTYPLOT programs. Regions with sequence similarity are boxed. Gaps in the alignment are shown by dashes.
The glycine-rich auxiliary domain of hrp23 prompted comparisons between hrp23 and a subgroup of hnRNP proteins, the RBD-Gly RNA-binding proteins. These proteins contain one or two NH2-terminal RBDs and a glycine-rich COOH-terminal domain (for reviews see Birney et al. 1993; Dreyfuss et al., 1993). The RBD-Gly proteins with one RBD comprise RBM3 (Derry et al., 1995) and hnRNP G (Soulard et al., 1993) in human and a series of exceptionally Gly-rich proteins in plants (for references see Bergeron et al., 1993; van Nocker and Vierstra, 1993). The RBD-Gly proteins with two RBDs include hnRNP A1 (Buvoli et al., 1988) and hnRNP A2/B1 (Burd et al., 1989) in human, hrp 36/38, hrp40, and hrp48 in Drosophila (Haynes et al., 1990, 1991; Matunis et al., 1992), and hrp36 in Chironomus (Visa et al., 1996a ). The hrp23 and the RBD-Gly proteins all have a typical RBD with the RNP-1 and RNP-2 submotifs, and the amino acid sequence identity between the RBD of hrp23 and that of the others is in the range of 21–37%. The auxiliary domains of hrp23 and the RBD-Gly proteins display the same basic structure: clustered glycine residues being interspersed with arginine and aromatic amino acids. The sequence RGG (GRG or GGR) is also a characteristic feature. In hrp23, the aromatic amino acid is phenylalanine, while in most of the others it is tyrosine. The auxiliary domain of hrp23 shows a sequence identity of 25–39% with that of the other nonplant proteins and as high as 41–45% with the exceptionally glycine-rich (∼70%) auxiliary domains of the plant proteins. However, it should be recalled that it is difficult to compare glycine-rich domains with each other because of the very low sequence complexity (Birney et al., 1993). We conclude that hrp23 has features in common with the RBD-Gly proteins: the RBD is similar, and the auxiliary domain is glycine-rich with RGG repeats and interspersed aromatic acids.
The presence of several RS/SR dipeptides in the auxiliary domain of hrp23 necessitated comparisons between hrp23 and the SR protein family, the members of which contain one or more RBDs and an auxiliary RS domain. In SR proteins, the RS domain is characterized by a large number of RS/SR dipeptides, often in clustered arrangements. The following three SR proteins, all with one RBD and a molecular mass of ∼20 kD, showed the strongest similarities to hrp23: SRp20 (Ayane et al., 1991; Zahler et al., 1992) and 9G8 (Cavaloc et al., 1994) from human and RBP1 from Drosophila (Kim et al., 1992). The RBD region of hrp23 is 46, 49, and 41% identical to the RBD region of SRp20, 9G8, and RBP1, respectively; the RNP-1 and RNP-2 submotifs are perfectly conserved in the four proteins. Furthermore, when comparisons were made between the corresponding auxiliary domains, the following figures were obtained: 29, 25, and 41% sequence identity, respectively. The SR/RS dipeptides are, however, fewer (8) in hrp23 than in the three small SR proteins (12–31), and the dipeptides in hrp23 are interspersed rather than clustered. Furthermore, the monoclonal antibody mAb104, which is known to recognize a phosphoepitope in the RS domain of SR proteins (Roth et al., 1991), does not react with hrp23 (data not shown). We conclude that hrp23 resembles the SR proteins, as its RBD is strikingly similar to that of the SR proteins and its auxiliary domain of hrp23 contains several SR/RS dipeptides, but it is also evident that hrp23 does not fulfill all the criteria for a typical SR protein.
In summary, hrp23 is an RNA-binding protein with a single NH2-terminal RBD and a COOH-terminal auxiliary domain rich in glycine, arginine, and serine. The hrp23 has a homologous counterpart in the Drosophila protein ROX21, and it shares important features with both RBD-Gly proteins and SR proteins (see Discussion).
hrp23 Is Mainly Located in the Nucleus
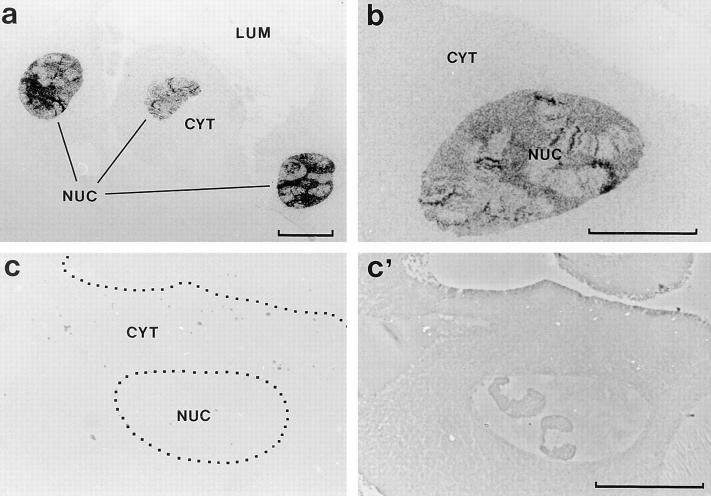
To reveal the intracellular distribution of hrp23, the salivary glands were studied with immunocytology. Semithin sections of salivary glands were incubated with the 1D3 antibody and subsequently with a gold-conjugated secondary antibody. The labeling was examined after silver enhancement. The nuclei of the salivary glands were heavily immunostained (Fig. 6, a and b). Puffs and nucleoli (cf., Fig. 2) were decorated by the antibody as well as the nucleoplasm surrounding the polytene chromosomes. The cytoplasm was weakly stained (Fig. 6, a and b). In the control (Fig. 6 c), no immunostaining could be detected either in the nucleus or in the cytoplasm; the negative control is also shown in phase contrast (Fig. 6 c′). We conclude that hrp23 is mainly located in the nucleus and that hrp23 is present both on the polytene chromosomes and in the surrounding nucleoplasm.
Figure 6.
Immunocytological localization of hrp23 in C. tentans salivary gland cells. Semithin cryosections of salivary glands were treated with mAb 1D3 (or with a negative control antibody), a gold-conjugated secondary antibody was added, and the antibody binding was visualized by silver enhancement. The 1D3-treated specimens are shown in bright field (a and b), and the negative controls are shown in bright field (c) and phase contrast (c′). The nuclei (NUC), cytoplasm (CYT), and gland lumen (LUM) are indicated. Bars, 40 μm.
hrp23 Is Present in Growing and in Nucleoplasmic BR RNP Particles but Is Absent from the BR Particles Bound to the Nuclear Pore Complex
Immunoelectron microscopy was used to investigate the behavior of hrp23 in relation to the assembly and transport of BR pre-mRNP particles. Salivary glands were fixed and frozen, and ultrathin cryosections were prepared. The sections were incubated with antibodies, stained with aqueous uranyl acetate, and embedded in polyvinyl alcohol. The specimens were examined and photographed in a transmission electron microscope.
The immunoelectron microscopy analysis was focused on the giant puffs, the BRs. The active genes appear as chromosomal loops, along which the growing BR RNP particles can be analyzed. In a single section, examples of promoter-proximal (p), middle (m), and promoter-distal (d) portions of the BR genes can be discerned (Fig. 7 a). In the proximal region, the RNP products appear as short thick fibers, while further downstream the gene, they can be described as stalked granules. This conformational change is due to the fact that the RNP fiber is being packed into a globular structure, which gradually increases in diameter along the course of transcription (for more detailed description see Kiseleva et al., 1994).
Figure 7.
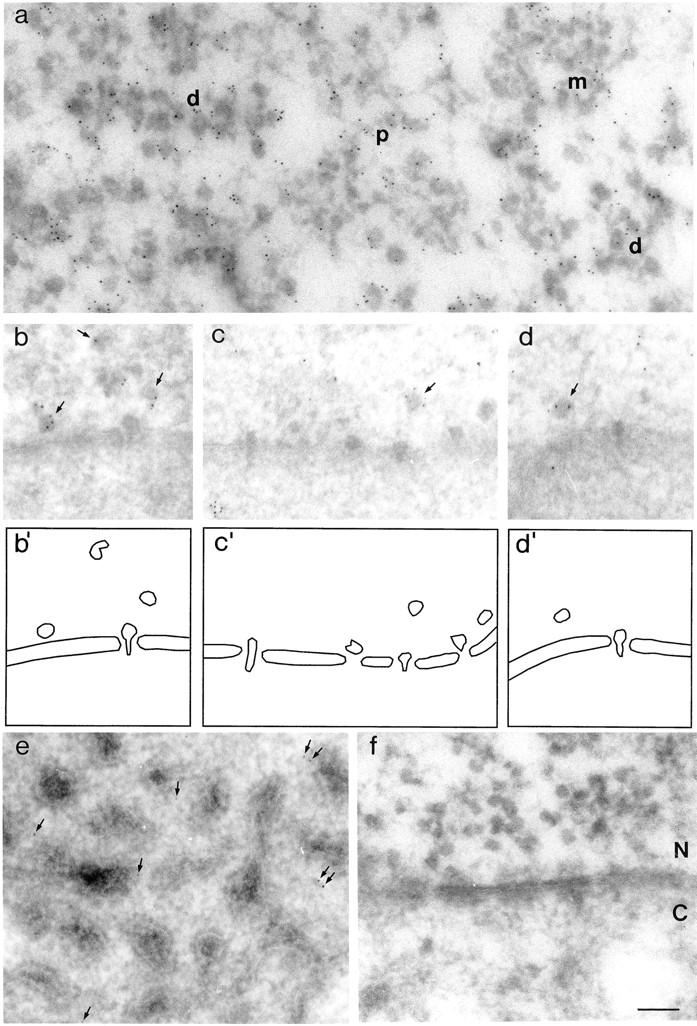
Immunoelectron microscopic localization of hrp23 in C. tentans salivary gland cells. Thin cryosections were incubated with mAb 1D3 (a–e), or with a negative control antibody (f), and a gold-conjugated secondary antibody was used to visualize the labeled sites. (a) Proximal (p), middle (m), and distal (d) segments of transcriptionally active BR genes. (b–d) Nucleoplasmic BR particles close to the nuclear envelope (arrows) and extended BR particles translocating through the nuclear pores. Schematic interpretations of the electron micrographs are shown beneath (b′–d′). The nucleus (N) is located above the nuclear envelope, and cytoplasm (C) is located beneath. The non–BR hnRNP complexes appear in the nucleoplasm as an irregular fibrous network labeled with a few gold particles (c). (e) Cytoplasm containing tubular ER with associated ribosomes. The arrows point at gold particles. (f) The negative control showing nuclear envelope with adjacent nuclear (N) and cytoplasmic (C) regions. In the nucleus, growing BR RNP particles can be discerned (cf., a). Bar, 100 nm.
As shown in Fig. 7 a, growing RNP particles in all three segments of the BR gene are labeled with gold (cf., the complete absence of gold particles in the control in Fig. 7 f). Thus, the hrp23 protein is being added to the BR transcript concomitant with transcription. The BR RNP particles can be seen released into the nucleoplasm (Fig. 7 b), and when they pass through the nuclear pore after having changed from a globular to a more extended conformation (Fig. 7, c and d). Beneath the electron micrographs of the nucleoplasmic and translocating particles, we have outlined the particles in schematic drawings, where also the nuclear envelope is indicated (Fig. 7, b′–d′). Most of the nucleoplasmic particles are decorated by gold (Fig. 7, b–d, arrows), while no gold is seen associated with the translocating particles. To further substantiate this point, we carried out a semiquantitative analysis. 400 nucleoplasmic particles were scrutinized, and 62% of them were labeled (242/400). We recorded 17 well-defined translocating particles, none of them labeled. Thus, hrp23 seems to be present in the nucleoplasmic particles, also in those close to the nuclear envelope, but once the BR particles become attached to the nuclear pore complex and translocate through the pore, the hrp23 protein is absent. In conclusion, the hrp23 protein is bound to the BR RNA concomitant with transcription, remains bound to the released BR RNA particles in the nucleoplasm, but is released from the particle before or at the binding of the particle to the nuclear pore complex for further exit into the cytoplasm.
The hnRNP particles present in chromosomal puffs other than the BRs are also decorated with gold (data not shown) (cf., the immunostaining of smaller puffs in Fig. 2, a and c). When released into the nucleoplasm, they often appear in a more or less fibrous conformation and can be seen immunolabeled (Fig. 7 c). Therefore, hrp23 seems also to associate with non–BR hnRNA during transcription, and it presumably stays with the RNA after its release into the nucleoplasm. Whether hrp23 is shed before the nucleocytoplasmic transport of the RNA cannot be decided in this case, as the non-BR hnRNP complexes cannot be recognized as well-defined entities when passing through the nuclear pores.
In the cytoplasm, the density of labeling was low, but it was significantly higher than in the negative control (Fig. 7, e vs. f). The gold particles were seen scattered in the cytoplasm between or close to the tubules of the endoplasmic reticulum (Fig. 7 e). Thus, a small amount of hrp23 can be detected in the cytoplasm. One possible interpretation would be that the low level of labeling corresponds to nascent and newly synthesized hrp23, but other explanations are conceivable (see further Discussion).
hrp23 Is Present in the Nucleolus
The analysis of immunolabeled polytene chromosomes revealed that hrp23 is present in the nucleoli of salivary gland cells. This result was confirmed and extended with immunoelectron microscopy. Fig. 8 a shows a segment of a nucleolus with its fibrillar (F) and granular (G) regions indicated. A small region (square) is presented at a higher magnification in Fig. 8 b. Gold particles appeared in both the fibrillar and granular compartments (Fig. 8 b, arrows), while no labeling could be visualized in the negative control (Fig. 8 c). As the labeling of nucleoli is RNase sensitive, we conclude that hrp23 is probably associated with RNA in the nucleolus, but the nature of this RNA remains to be determined.
Figure 8.
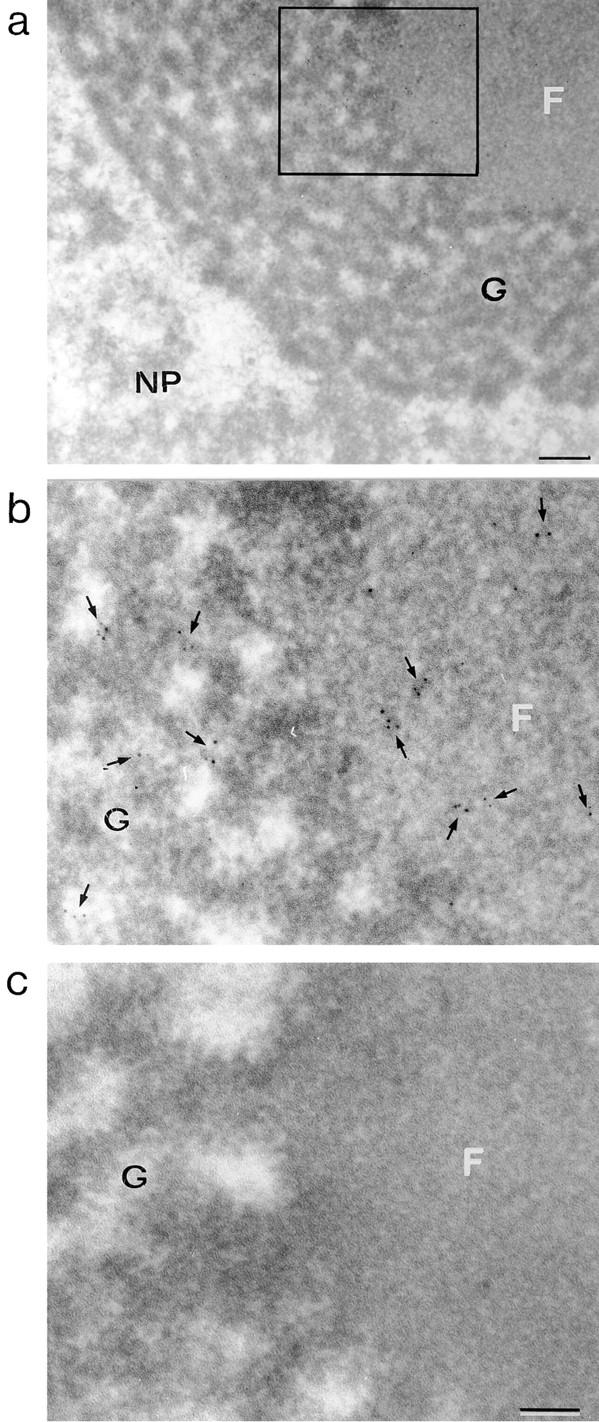
Immunoelectron microscopic localization of hrp23 in the nucleolus of C. tentans salivary glands. Semithin sections through a nucleolus in a salivary gland cell were treated with mAb 1D3, and the binding was visualized by a gold-conjugated secondary antibody. In parallel, sections were incubated with a negative control antibody. (a) A segment of a nucleolus, with its fibrillar (F) and granular (G) compartments, and the surrounding nucleoplasm (NP). (b) The area demarcated with a rectangle in a, at higher magnification. The arrows denote the position of gold particles. (c) The negative control. Bars: (a) 250 nm; (b and c) 100 nm.
hrp23 Is Likely to be a Nonshuttling Protein
To analyze whether hrp23 behaves as a shuttling protein in C. tentans, we performed an actinomycin D experiment as devised by Pinol-Roma and Dreyfuss (1991). In this type of experiment shuttling, but not nonshuttling, proteins will accumulate in the cytoplasm. C. tentans tissue culture cells were incubated for 1 h with actinomycin D in the presence of cycloheximide, fixed, sectioned, and challenged with antibodies; untreated cells were analyzed in parallel. The hrp36 protein served as control representing the behavior of a shuttling protein. The hrp23 protein was recorded mainly in the nucleus in both treated and untreated cells (Fig. 9). Thus, no accumulation of hrp23 in the cytoplasm was detected. In the control, the shuttling protein hrp36 shifted from the nucleus to the cytoplasm during the drug experiment (Fig. 9). This result shows that hrp23 is not a transcription-dependent shuttling hnRNP protein and indicates that it is a nonshuttling protein. This is in good agreement with the observation that hrp23 is shed from the BR particles before the translocation of the particles through the nuclear pores.
Figure 9.
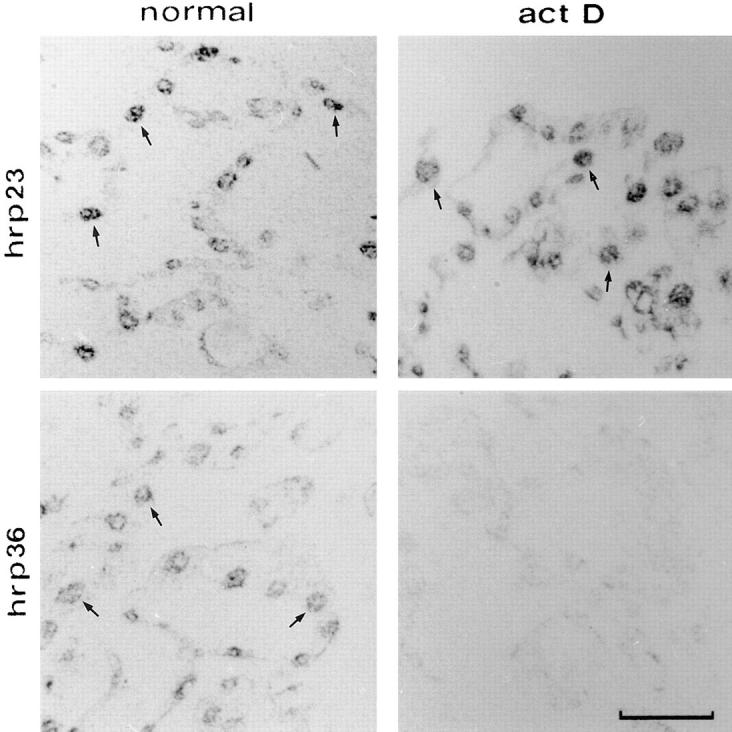
Transcription-independent nuclear localization of hrp23. C. tentans tissue culture cells were treated for 1 h with (act D) or without (normal) actinomycin D in the presence of cycloheximide. To locate hrp23, semithin cryosections were incubated with mAb 1D3 and subsequently with a gold-conjugated secondary antibody. The labeling was visualized by silver enhancement and photographed in bright field. The distribution of the shuttling protein hrp36 was analyzed in parallel with mAb 4F9. Some of the immunopositive nuclei have been indicated by arrows. Bar, 25 μm.
Discussion
Functional Implications of the Structure of the hnRNP Protein hrp23
The hrp23 protein contains a single NH2-terminal RNA-binding domain and a COOH-terminal domain rich in glycine, arginine, and serine. There is a recently described homologue in Drosophila, the ROX21 protein (Brand et al., 1995). Furthermore, sequence comparisons with RNA-binding proteins revealed that hrp23 is similar to the RBD-Gly proteins as well as to the SR proteins, all exhibiting one or two NH2-terminal RBDs and an auxiliary domain. In particular, the presence of clustered glycine residues, RGG repeats, and an interspersed aromatic amino acid (phenyalanine) in the auxiliary domain makes hrp23 appear similar to the RBD-Gly RNA-binding proteins with their glycine-rich auxiliary domain. However, the RBD in hrp23 shows a strikingly high (∼45%) identity to the RBD of low molecular weight SR proteins, and the auxiliary domain in hrp23 contains several SR/RS dipeptides. It seems, therefore, as if hrp23 and its Drosophila homologue ROX21 represent a group of proteins intermediate in structure between the RBD-Gly proteins and the SR proteins, which suggests that there is not a sharp boundary between these two groups of hnRNP proteins.
RGG-rich domains are present in a large number of proteins involved in various aspects of RNA metabolism, such as pre-rRNA processing, pre-mRNA splicing, and RNA helicase activity (for references see Steinert et al., 1991; Kiledjian and Dreyfuss, 1992). It is interesting to note that in hnRNP U protein, which lacks an RBD, a single COOH-terminal RGG-rich region, termed the RGG box, is known to be sufficient for RNA binding (Kiledjian and Dreyfuss, 1992). Likewise, a COOH-terminal fragment of nucleolin, containing an RGG-box like domain, is also able to independently accomplish binding to RNA (Ghisolfi et al., 1990). Furthermore, in hnRNP A1, the RGG-rich domain acts in a cooperative manner as to RNA binding with the two RBDs (Kumar et al., 1990). It is therefore possible that the RGG-containing auxiliary domain in hrp23 also promotes binding of the protein to the pre-mRNA molecule and perhaps also affects the specificity of the binding to the RNA.
The SR proteins are known to act as splicing factors in both in vitro and in vivo experiments (for review see Fu, 1995). For all three SR proteins most closely related to hrp23, it has been demonstrated that they can function as splicing factors. The RBP1 protein in Drosophila can both activate splicing and switch splice site selection in in vitro experiments (Kim et al., 1992). The human 9G8 protein is an essential splicing factor as also shown in experiments in vitro (Cavaloc et al., 1994). Finally, Screaton et al. (1995) could demonstrate in vivo that the human SRp20 protein can regulate alternative splicing. Thus, it seems as if this group of SR proteins is essential for splicing and can modulate alternative splicing, but it remains to be proven whether also hrp23 acts as a splicing factor.
hrp23 and Transport of BR RNP Particles
The hrp23 protein is added to BR pre-mRNA concomitant with transcription. It is present in the proximal, middle, and distal regions of the active gene. The protein is, therefore, added early during the transcription process, and most likely also along the entire BR transcript like the other two hnRNP proteins earlier studied, hrp36 (Kiseleva et al., 1994) and hrp45 (Alzhanova-Ericsson et al., 1996). It should be noted that concomitant with transcription, spliceosomes rapidly assemble and disassemble on the 5′ region of the transcript and start to assemble at the 3′ end of the transcript, leaving the middle section of the gene devoid of spliceosome components (Kiseleva et al., 1994). As hrp23 remains bound to the growing RNP particles all during transcription, hrp23 is not likely to be a true spliceosomal component. This is further supported by the observation that hrp23 is present in the released nucleoplasmic BR particles, the vast majority of which contain fully spliced BR RNA and no spliceosome components like snRNP. These results do not rule out the possibility that hrp23 is involved in splicing. However, if so, hrp23 (like the SR protein hrp45) should exert its action as a structural component of the elementary RNP fiber in the BR particles.
The nucleoplasmic BR particles are frequently labeled in the immunoelectron microscopy experiment; more than half of them are decorated with gold. This is a high figure considering that all particles are not likely to be accessible to the antibodies in the sections because of steric hindrance. It is then a striking observation that no BR particles have been seen immunolabeled when bound to the nuclear pore complex or when translocating through the central channel of the complex. Evidently, hrp23 is released from the BR particle just before the binding of the particle to the pore complex or in conjunction with the binding. It can be speculated that the shedding of hrp23 could even be a prerequisite for binding. It should be noted that this behavior of hrp23 is different from that of the SR protein hrp45. This protein is present on the particle at the pore (translocating particles are immunolabeled to almost the same extent as nucleoplasmic particles), but it is discarded when the RNP particle changes conformation and is entering the central channel, indicating that this SR protein could be linked to the translocation per se (Alzhanova-Ericsson et al., 1996).
The hrp23 protein does not seem to leave the nucleus and should therefore be classified as a nonshuttling protein. This was further substantiated in the present study by an actinomycin D experiment performed with C. tentans tissue culture cells according to Pinol-Roma and Dreyfuss (1991). It was not possible to translocate the nuclear hrp23 into cytoplasm during the drug treatment, while the shuttling protein hrp36 analyzed in parallel accumulated in the cytoplasm. This result suggested that not only is the hrp23 bound to BR RNP particles confined to the nucleus, but so is the hrp23 in all hnRNP particles. It should, however, be recalled that a shuttling experiment could be misleading because some hnRNP proteins, e.g., Npl3 in yeast (Lee et al., 1996), only shuttle in the presence of on-going transcription.
A small amount of hrp23 was detected in the cytoplasm in the immunocytology and in the immunoelectron microscopy experiments. The explanation closest at hand is that the low immunosignal corresponds to nascent and newly synthesized hrp23. However, considering the small size of hrp23, it is also possible that free hrp23 can diffuse through the nuclear pore and appear in the cytoplasm; proteins below 40–60 kD are able to do so (Peters, 1986). Finally, we cannot exclude, although we regard it as less likely, that a low amount of hrp23 is transported from the nucleus to the cytoplasm as part of an RNP complex originating from one or more of the smaller chromosomal puffs or from the nucleoli.
When the behavior of hrp23 is considered in relation to the total information available on the transport of BR particles in the salivary gland cells, a dynamic picture emerges. Until now, the following components have been studied during the assembly and transport of the BR particles: snRNP proteins (Kiseleva et al., 1994), the cap-binding protein CBP20 (Visa et al., 1996b ), hrp36 (Visa et al., 1996a ), hrp45 (Alzhanova-Ericsson et al., 1996), and now also hrp23. Each of these proteins displays its own specific behavior. The snRNP proteins show a transient association with the particle during splicing, while the CBP20 is added to the 5′ end of the transcript early during transcription and is not released until the 5′ end hits the cytoplasm. The three hnRNP proteins are all added onto and along the transcript concomitant with transcription. While hrp36, an A1-like protein, accompanies the BR RNA through the pores into cytoplasm and even into polysomes, hrp23 and hrp45 remain in the nucleus. The hrp23 protein is, however, clearly shed before or at the binding of the particle to the pore complex, while hrp45 is not displaced until the RNP is entering the central channel of the pore complex. Thus, the two nonshuttling proteins studied are not released at the same time in conjunction with the translocation of the BR particle through the nuclear pore, but do so in a consecutive fashion beginning before or at the binding of the particle to the nuclear pore complex. The finding that the BR RNA-binding proteins hrp23, hrp36, and hrp45 behave differently during nucleocytoplasmic transport suggests that each of them could play a specific role during the export of mRNA from the nucleus to the cytoplasm.
hrp23 Appears in the Nucleolus
It was surprising to find that the hrp23 protein is present in the nucleolus. It resides evidently both in the fibrillar and granular regions of the nucleolus. One hnRNP protein, hnRNP I, has earlier been detected in a discrete perinucleolar region (Ghetti et al., 1992), but hrp23 is the first hnRNP protein found to be located within the nucleolus. As the hrp23 immunolabeling was shown to be sensitive to RNase, it is likely that hrp23 is associated with RNA. One possibility would be that hrp23 is bound to ribosomal RNA, either one or more of the RNA species generated in the nucleolus or 5S RNA transported from the site of synthesis to the nucleolus. A second possibility would be that hrp23 is coupled to a nonribosomal RNA species. Typical, DRB-sensitive hnRNA has not been recorded in the nucleolus, but a rapidly labeled, DRB-insensitive, heterodisperse RNA in the 20–30 S size range has been revealed (Ringborg et al., 1970). If this specific type of putative hnRNA would be associated with hrp23, neither hrp36 nor hrp45 would be part of the RNP complex. Such a nucleolus-located hnRNP complex would then be a striking example of the concept that each type of hnRNP particle contains a specific subset of hnRNP proteins (Matunis et al., 1993; Wurtz et al., 1996).
Acknowledgments
We thank Lars Wieslander for providing us with C. tentans cDNA libraries. We are grateful to B. Björkroth and L.-M. Fjelkestam for technical assistance.
This study was supported by the Swedish Natural Science Research Council, Knut and Alice Wallenberg Foundation, Kjell and Märta Beijer Foundation, and Petrus and Augusta Hedlund Foundation. N. Visa was a recipient of a Human Capital and Mobility Individual Fellowship from the Commission of the European Communities, Y. Aaissouni received a Fellowship from the Swedish Institute, and J. Zhao received a Fellowship from the Wenner-Gren Foundation.
Abbreviations used in this paper
- BR
Balbiani ring
- hnRNP
heterogeneous ribonucleoprotein
- NES
nuclear export signal
- NPC
nuclear pore complex
- pre-mRNA
pre-messenger RNA
- RBD
RNA-binding domain
- RNP
ribonucleoprotein
Footnotes
Address all correspondence to Bertil Daneholt, Department of Cell and Molecular Biology, Medical Nobel Institute, Karolinska Institutet, S-171 77, Stockholm, Sweden. Tel.: +46-8-7287370. Fax: +46-8-313529. E-mail: bertil.daneholt@cmb.ki.se
References
- Alzhanova-Ericsson AT, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev. 1996;10:2881–2893. doi: 10.1101/gad.10.22.2881. [DOI] [PubMed] [Google Scholar]
- Amero SA, Elgin SCR, Beyer AL. A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. . Genes Dev. 1991;5:188–200. doi: 10.1101/gad.5.2.188. [DOI] [PubMed] [Google Scholar]
- Ayane M, Preuss U, Köhler G, Nielsen PJ. A differentially expressed murine RNA encoding a protein with similarities to two types of nucleic acid binding motifs. Nucleic Acids Res. 1991;19:1273–1278. doi: 10.1093/nar/19.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank, recent developments. Nucleic Acids Res. 1993;21:3093–3096. doi: 10.1093/nar/21.13.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén G, Wieslander L. Splicing of Balbiani Ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Bergeron D, Beauseigle D, Bellemar G. Sequence and expression of a gene encoding a protein with RNA-binding and glycine-rich domains in Brassica napus. . Biochim Biophys Acta. 1993;1216:123–125. doi: 10.1016/0167-4781(93)90047-h. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkroth B, Ericsson C, Lamb MM, Daneholt B. Structure of the chromatin axis during transcription. Chromosoma (Berl) 1988;96:333–340. [Google Scholar]
- Brand SF, Pichoff S, Noselli S, Bourbon H-M. Novel Drosophila melanogastergenes encoding RRM-type RNA-binding proteins identified by a degenerate PCR strategy. Gene. 1995;154:187–192. doi: 10.1016/0378-1119(94)00840-o. [DOI] [PubMed] [Google Scholar]
- Burd CG, Swanson S, Görlach M, Dreyfuss G. Primary structures of the heterogenous nuclear ribonucleoprotein A2, B1 and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc Natl Acad Sci USA. 1989;86:9788–9792. doi: 10.1073/pnas.86.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M, Biamonti G, Tsoulfas P, Bassi MT, Ghetti A, Riva S, Morandi C. cDNA cloning of human hnRNP protein A1 reveals the existence of multiple mRNA isoforms. Nucleic Acids Res. 1988;16:3751–3770. doi: 10.1093/nar/16.9.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Popielarz M, Fuchs J-P, Gattoni R, Stévenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO (Eur Mol Biol Org) J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, Grabowski PJ, Sharp PA, Dreyfuss G. Heterogenous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986;231:1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- Derry JMJ, Kerns JA, Francke U. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum Mol Genet. 1995;4:2307–2311. doi: 10.1093/hmg/4.12.2307. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Economides IV, Pedersen T. In vitro assembly of a pre-messenger ribonucleoprotein. Proc Natl Acad Sci USA. 1983;80:4296–4300. doi: 10.1073/pnas.80.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNPI, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisolfi L, Joseph G, Erard M, Escoubas JM, Mathieu C, Amalric F. Nucleolin—pre-rRNA interactions and preribosome assembly. Mol Biol Rep. 1990;14:113–114. doi: 10.1007/BF00360437. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 726 pp.
- Haynes SR, Raychaudhuri G, Beyer AL. The Drosophila Hrb98DE locus encodes four protein isoforms homologous to the A1 protein of mammalian heterogenous nuclear ribonucleoprotein complexes. Mol Cell Biol. 1990;10:316–323. doi: 10.1128/mcb.10.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SR, Johnson D, Raychaudhuri G, Beyer AL. The DrosophilaHrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 1991;19:25–31. doi: 10.1093/nar/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G. A role for the M9 transport signal of hnRNP A1 mRNA nuclear export. J Cell Biol. 1997a;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmertric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur Mol Biol Org) J. 1997b;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO (Eur Mol Biol Org) J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-J, Zuo P, Manley JL, Baker BS. The DrosophilaRNA-binding protein RBP1 is localized to transcriptionally active sites of chromosomes and shows a functional similarity to human splicing factor ASF/SF2. Genes Dev. 1992;6:2569–2579. doi: 10.1101/gad.6.12b.2569. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Wurtz T, Visa N, Daneholt B. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO (Eur Mol Biol Org) J. 1994;13:6052–6061. doi: 10.1002/j.1460-2075.1994.tb06952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Casas-Finet JR, Luneau CJ, Karpel RL, Merrill RM, Williams KR, Wilson SH. Mammalian heterogenous nuclear ribonucleoprotein A1. Nucleic acid binding properties of the COOH-terminal domain. J Biol Chem. 1990;265:17094–17100. [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- LeMaire MF, Thummel CS. Splicing precedes polyadenylation during DrosophilaE74A transcription. Mol Cell Biol. 1990;10:6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezzi M, Meyer B, Mähr R. Heat shock phenomena in Chironomus tentans. . Chromosoma (Berl) 1981;83:327–339. doi: 10.1007/BF00327356. [DOI] [PubMed] [Google Scholar]
- Malcolm DB, Sommerville J. The structure of chromosome- derived ribonucleoprotein in oocytes of Triturus cristatus carnifer(Laurenti) Chromosoma (Berl) 1974;48:137–158. doi: 10.1007/BF00283960. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Matunis EL, Matunis MJ, Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. . J Cell Biol. 1992;116:245–255. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis EL, Matunis MJ, Dreyfuss G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J Cell Biol. 1993;121:219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO (Eur Mol Biol Org) J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal mediated, temperature-dependent nuclear protein export pathway. Cell. 1995a;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Michael WM, Siomi H, Choi M, Pinol-Roma S, Nakielny S, Liu Q, Dreyfuss G. Signal sequences that target nuclear import and nuclear export of pre-mRNA binding proteins. Cold Spring Harbor Symp Quant Biol. 1995b;60:663–668. doi: 10.1101/sqb.1995.060.01.071. [DOI] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO (Eur Mol Biol Org) J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Bakken AH. Morphological studies of transcription. Karolinska Symp Res Methods Reprod Endocrinol. 1972;5:155–167. doi: 10.1530/acta.0.071s155. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Osheim, Y.N., J. Miller, O.L., and A.L. Beyer. 1985. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 43:143–151. [DOI] [PubMed]
- Panté N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- Panté N, Jarmolewski A, Izaurralde E, Sauder U, Baschong W, Mattaj IW. Visualizing nuclear export of different classes of RNA by electron microscopy. RNA. 1997;3:498–513. [PMC free article] [PubMed] [Google Scholar]
- Peters R. Fluorescence microphotolysis to measure nucleocytoplasmic transport and intracellular mobility. Biochim Biophys Acta. 1986;864:305–359. doi: 10.1016/0304-4157(86)90003-1. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Choi YD, Matunis MM, Dreyfuss G. Immunopurification of heterogenous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Rice CM, Fuchs R, Higgins DG, Stoehr PJ, Cameron GN. The EMBL data library. Nucleic Acids Res. 1993;21:2967–2971. doi: 10.1093/nar/21.13.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring HZ, Lis JT. The SR protein B52/SRp55 is essential for Drosophiladevelopment. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringborg U, Daneholt B, Edström J-E, Egyházi E, Lambert B. Electrophoretic characterization of nucleolar RNA from Chironomus tentanssalivary gland cells. J Mol Biol. 1970;51:327–340. doi: 10.1016/0022-2836(70)90146-4. [DOI] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton GR, Cáceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO (Eur Mol Biol Org) J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierakowska H, Szer W, Furdon PJ, Kole R. Antibodies to hnRNP core proteins inhibit in vitro spicing of human β-globin pre-mRNA. Nucleic Acid Res. 1986;14:5241–5254. doi: 10.1093/nar/14.13.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogenous nuclear RNP proteins. J Cell Biol. 1997;138:1–12. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Björkroth B, Lamb MM, Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983;34:847–855. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Strandberg B, Daneholt B. Three- dimensional structure of a specific pre-messenger RNP particle established by electron microscope tomography. Nature. 1986;319:560–564. doi: 10.1038/319560a0. [DOI] [PubMed] [Google Scholar]
- Soulard M, Della V, Valle, Siomi MC, Pinol-Roma S, Codogno P, Bauvy C, Bellini M, Lacroix J-C, Monod G, Dreyfuss G, Larsen C-J. hnRNP G: sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res. 1993;21:4210–4217. doi: 10.1093/nar/21.18.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export receptor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Mack JW, Korge BP, Gan SQ, Haynes SR, Steven AC. Glycine loops in proteins: their occurrence in certain intermediate filament chains, loricrins and single-stranded RNA binding proteins. Int J Biol Macromol. 1991;13:130–139. doi: 10.1016/0141-8130(91)90037-u. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. Immunocytochemistry on ultrathin frozen sections. Histochem J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Green MR. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- van Nocker S, Vierstra RD. Two cDNAs from Arabidopsis thalianaencode putative RNA binding proteins containing glycine-rich domains. Plant Mol Biol. 1993;21:695–699. doi: 10.1007/BF00014552. [DOI] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996a;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996b;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz T, Kiseleva E, Nacheva G, Alzhanova-Ericsson AT, Rosén A, Daneholt B. Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1996;16:1425–1435. doi: 10.1128/mcb.16.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. Chironomus tentansepithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp Cell Res. 1982;139:309–319. doi: 10.1016/0014-4827(82)90255-5. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]