Abstract
In Saccharomyces cerevisiae, transfer of N-linked oligosaccharides is immediately followed by trimming of ER-localized glycosidases. We analyzed the influence of specific oligosaccharide structures for degradation of misfolded carboxypeptidase Y (CPY). By studying the trimming reactions in vivo, we found that removal of the terminal α1,2 glucose and the first α1,3 glucose by glucosidase I and glucosidase II respectively, occurred rapidly, whereas mannose cleavage by mannosidase I was slow. Transport and maturation of correctly folded CPY was not dependent on oligosaccharide structure. However, degradation of misfolded CPY was dependent on specific trimming steps. Degradation of misfolded CPY with N-linked oligosaccharides containing glucose residues was less efficient compared with misfolded CPY bearing the correctly trimmed Man8GlcNAc2 oligosaccharide. Reduced rate of degradation was mainly observed for mis- folded CPY bearing Man6GlcNAc2, Man7GlcNAc2 and Man9GlcNAc2 oligosaccharides, whereas Man8GlcNAc2 and, to a lesser extent, Man5GlcNAc2 oligosaccharides supported degradation. These results suggest a role for the Man8GlcNAc2 oligosaccharide in the degradation process. They may indicate the presence of a Man8GlcNAc2-binding lectin involved in targeting of misfolded glycoproteins to degradation in S. cerevisiae.
Keywords: protein degradation, endoplasmic reticulum, glycosylation, mannosidase, yeast
In Saccharomyces cerevisiae, as in other eukaryotes, the synthesis of asparagine-linked glycoproteins takes place in the ER. After transfer to protein, the N-linked oligosaccharide (NLO),1 while present in the ER, is subject to trimming reactions (see Fig. 1) involving glucosidase I, glucosidase II, and mannosidase I (Herscovics and Orlean, 1993; Moremen et al., 1994; Roth, 1995). In higher eukaryotes, a specific role of the trimming intermediate Glc1Man9GlcNAc2 oligosaccharide in the ER quality control process has been proposed (Helenius et al., 1997). An incorrectly folded glycoprotein bearing such an oligosaccharide structure is bound by specific ER resident proteins and retained in a folding competent environment. Correctly folded glycoproteins can exit the ER, enter the Golgi apparatus, and are delivered to their final destination. However, improperly folded glycoproteins are retained in the ER and are eventually degraded. In many cases, degradation occurs via the ubiquitin–proteasome pathway that requires their exit from the ER lumen to the cytosol as shown both in higher eukaryotic cells and in yeast (Jentsch and Schlenker, 1995; Bonifacino, 1996; Kopito, 1997; Sommer and Wolf, 1997; Varshavsky, 1997). For the export to the cytosol, constituents of the ER translocon play an important role (Pilon et al., 1997; Plemper et al., 1997). Additionally, ER proteins such as the chaperone Kar2p (Plemper et al., 1997), as well as the ubiquitin-conjugating proteins Ubc6p and Ubc7p, are thought to be involved in this process (Biederer et al., 1996; Hiller et al., 1996). In Saccharomyces cerevisiae, the proteolysis of nonglycosylated α-factor is ATP and cytosol-dependent (McCracken and Brodsky, 1996) and also mutated and therefore misfolded carboxypeptidase Y (prc1-1, CPY*; Wolf and Fink, 1975; Finger et al., 1993) has been shown to enter the ubiquitin–proteasome pathway (Hiller et al., 1996). The degradation of the misfolded protein appears to be glycosylation dependent, since nonglycosylated CPY* remains stable in the ER (Knop et al., 1996). Moreover, the degradation also appears to be mannosidase I–dependent (Knop et al., 1996). Despite this, the molecular signals required for the initiation of ER glycoprotein degradation are not known.
Figure 1.
Biosynthesis and trimming of oligosaccharides in the ER lumen of Saccharomyces cerevisiae. The lipid-linked oligosaccharide precursors are synthesized at the ER membrane. Some of the involved mannosyltransferases (encoded by the ALG3, ALG9, and ALG12 loci) and their products as well as the glucosyltransferases (encoded by the ALG6, ALG8 and ALG10 loci) and their respective products are depicted. The fully assembled Glc3Man9GlcNAc2 oligosaccharide precursor is transferred to asparagine residues of the N-X-S/T sequence of polypeptides. This is followed by trimming involving glucosidase I (GLS1), glucosidase II (GLS2) and mannosidase I (MNS1) to yield the protein-bound Man8GlcNAc2 oligosaccharide.
We investigated the possible role of specific oligosaccharide structures in degradation of CPY* by genetic tailoring of the protein-bound oligosaccharide structure. We found that the Man8GlcNAc2 structure as the final product of the trimming reaction in the ER in yeast (Byrd et al., 1982) was mandatory for efficient degradation. Our results suggest that the ER α1,2-mannosidase represents a key enzyme for timing the onset of degradation. The period required for complete oligosaccharide trimming appears to be the time frame for glycoproteins to fold correctly.
Materials and Methods
Materials
Strains used are detailed in Table I. Wild-type denotes a strain with both normal biosynthesis of lipid-linked oligosaccharides and trimming of protein-bound oligosaccharides but harboring the prc1-1 mutation. Oligonucleotides (Microsynth, Balgach, Switzerland) used for gene deletion and screenings are listed in Table II. The integrative plasmid pRS306-prc1-1 containing the mutated CPY gene was provided by Dr. D.H. Wolf (University of Stuttgart, Germany). The antiserum against yeast hexokinase was provided by Dr. S. Schröder (Biozentrum, University of Basel, Switzerland).
Table I.
Yeast Strains Used in This Study
| Strain | Genotype | Reference | ||
|---|---|---|---|---|
| SS328 | MATα ade2-101 ura3-52 his3Δ200 lys2-801 | Vijayraghavan et al. (1989) | ||
| YG268 | MATα ade2-101 ura3-52 his3Δ200 lys2-801 Δalg3::HIS3 | Aebi et al. (1996) | ||
| YG414 | MATα ade2-101 ura3-52 his3Δ200 lys2-801 Δalg9::KanMX | Burda et al. (1996) | ||
| YG427 | MATα ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX | Jakob et al. (1998) | ||
| YG590 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX Δalg6::HIS3 | Jakob et al. (1998) | ||
| YG424 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX Δalg8::HIS3 | Jakob et al. (1998) | ||
| YG491 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX Δalg10::KanMX | Jakob et al. (1998) | ||
| YG746 | MATa ade2-101 ura3-52 his3Δ200 tyr1 Δmns1::KanMX | This study | ||
| YG618 | MATα ade2-101 ura3-52 his3Δ200 lys2-801 prc1-1 | This study | ||
| YG619 | MATα ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX prc1-1 | This study | ||
| YG620 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δalg6::HIS3 prc1-1 | This study | ||
| YG623 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX Δalg6::HIS3 prc1-1 | This study | ||
| YG624 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX Δalg8::HIS3 prc1-1 | This study | ||
| YG696 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 Δgls2::KanMX Δalg10::KanMX prc1-1 | This study | ||
| YG796 | MATa ade2-101 ura3-52 lys2-801 his3Δ200 Δalg9::KanMX prc1-1 | This study | ||
| YG797 | MATa ade2-101 ura3-52 lys2-801 his3Δ200 Δalg3::HIS3 prc1-1 | This study | ||
| YG807 | MATα ade2-101 ura3-52 lys2-801 his3Δ200 Δalg12::KanMX prc1-1 | This study | ||
| YG777 | MATα ade2-101 ura3-52 lys2-801 his3Δ200 Δmns1::KanMX prc1-1 | This study | ||
| YG556 | MATα ura3-52 his3Δ200 lys2-801 leu2 sec18-50 | This study | ||
| YG557 | MATa ade2-101 ura3-52 his3Δ200 lys2-801 leu2 Δgls2::KanMX sec18-50 | This study | ||
| YG821 | MATα ade2-101 ura3-52 his3Δ200 lys2-801 prc1-1 YEp352 | This study | ||
| YG822 | MATa ade2-101 ura3-52 lys2-801 his3Δ200 Δalg9::KanMX prc1-1 pALG9 | This study | ||
| YG823 | MATa ade2-101 ura3-52 lys2-801 his3Δ200 Δalg3::HIS3 prc1-1 pALG3 | This study | ||
| YG824 | MATα ade2-101 ura3-52 lys2-801 his3Δ200 Δalg12::KanMX prc1-1 pALG12 | This study |
Table II.
PCR Primers
| Primer | Sequence (5′→ 3′)* | |
|---|---|---|
| Knockout primers | ||
| MNS1forKan | aaacattgaaaaaggattctatgaagaactctgtcggtattcgatgaattcgagctc | |
| MNS1revKan | ccactatatagcacactaacctacaacgaccaacctgtggcgtacgctgcaggtcgac | |
| MNS1-68u | tgccaagaaacgaaagac | |
| MNS1+431L | cggataataaaccaccta | |
| ALG12forKan | aaaagagttgaataaagccattaaacaacgattcagttgacatcgatgaattcgagctc | |
| ALG12revKan | gctcgctatatattttattggaattgacgttagctattatcacgtacgctgcaggtcgac | |
| ALG12for | caaccttttaccagccgg | |
| KanMXu | gtattgatgttggacgag | |
| Primer for prc1-1 screen | ||
| CPY462u | ggatccggtcatcctttg | |
| CPY885L | ctgagtcaatgggtcagt | |
Bold face letters represent locus-specific sequence.
Yeast Manipulations
Standard protocols were followed for growth of yeast, mating, sporulation, and ascus dissection (Guthrie and Fink, 1991). If not otherwise stated, the cells were grown at 30°C in either YPD medium (2% Bacto-Peptone, 1% Yeast extract [both from Difco Laboratories, Detroit, MI], 2% glucose) or for metabolic labeling experiments in MV medium (0.67% Yeast nitrogen base [Difco Laboratories], 2% glucose and the appropriate supplements).
Construction of Strains
Disruption of the MNS1 Locus.
The MNS1 locus ORF YJR131w (these data are available from GenBank/EMBL/DDBJ under accession number Z49631; Grondin and Herscovics, 1992) was inactivated by replacing a major part of the locus with the KanMX cassette (Wach et al., 1994). The sequence of the kanamycin resistance gene was amplified by PCR by using the template pFA6a-KanMX4 plasmid (Wach et al., 1994) and the primers MNS1forKan and MNS1revKan (Table II). The resulting DNA was transformed into strain SS328 and the cells were selected on G418 plates (200 μg/ml). Transformants were analyzed for correct integration by whole cell PCR (Sathe et al., 1991) using KanMXu and the MNS1-specific MNS1-68u and MNS1 + 431L primers.
Disruption of the ALG12 Locus.
The ALG12 locus ORF YNR030w (these data are available from GenBank/EMBL/DDBJ under accession number Z71645; Lussier et al., 1997) was inactivated by the same procedure using the primers ALG12forKan and ALG12revKan for amplifying the KanMX cassette and KanMXu and ALG12for primers for verifying the correct gene deletion (Table II).
Replacement of the PRC1 Locus with prc1-1.
The BglII-linearized plasmid pRS306-prc1-1 (Knop et al., 1996) containing the mutated form (G255R) of CPY (Wolf and Fink, 1975) was integrated into the PRC1 locus of various yeast strains, resulting in a duplication of the PRC1 locus. Strains in which an excision of the duplication by homologous recombination had occurred were selected on 5-FOA plates and the resulting colonies screened by PCR for the prc1-1 locus. A fragment of the PRC1 locus was amplified by PCR using the primers CPY462u and CPY855L (Table II) giving raise to a product of 423 bp. Due to the prc1-1 mutation, a BstXI restriction site is destroyed. Therefore, strains containing solely the prc1-1 locus were identified by the resistance of the PCR fragment towards BstXI digestion. Western blot analysis confirmed that they only expressed mutant CPY*.
Metabolic Labeling and Immunoprecipitations
Stationary grown cells from a YPD overnight culture were inoculated in minimal medium and cultivated to an OD546nm of 1.0. The cells were harvested by centrifugation, washed in minimal medium containing 0.1% glucose and then incubated in the same medium at 30°C for at least 3 h. For pulse–chase experiments, 2 × 107 cells per time point were labeled by the addition of 50 μCi [35S]methionine (Tran35S-label, 10 mCi/ml; ICN Pharmaceuticals) for 10 min and then chased with a 100-fold excess of nonradioactive methionine. The chase was terminated by the addition of NaN3 (50 mM final concentration) and immediate freezing in liquid nitrogen. Protein extractions, immunoprecipitation, and SDS-PAGE were performed as described (Franzusoff et al., 1991; te Heesen et al., 1992). The dried gels were exposed and analyzed using a PhosphoImager. The kinetics of CPY* degradation were calculated by setting the counts of time point zero as 100%. For the studies of the transport kinetics of CPY in the strains with and without Mns1p, the cells were labeled for only 5 min at 26°C. The chase, protein extraction, and immunoprecipitation were performed as described above.
Assay for Degradation of CPY* by Western Analysis
Yeast strains were grown at 30°C in YPD or minimal medium containing the appropriate supplements into stationary phase. 3 × 108 cells were harvested and broken with glass beads in 50 mM Tris-HCl, pH 7.5, 1% SDS, 2 mM PMSF (Franzusoff et al., 1991; te Heesen et al., 1992). Protein extract equivalent to 7 × 106 cells was subjected to reducing SDS-PAGE, transferred to nitrocellulose membranes, and probed with specific antibodies. Binding was visualized by chemiluminescence (SuperSignal ULTRA; Pierce Chemical Co., Rockford, IL). The x-ray films were scanned and the intensity of the protein bands was determined. The antibody conjugates on the nitrocellulose membranes were stripped by treatment in 62.5 mM Tris-HCl, pH 6.7, 2% SDS, 100 mM β-mercaptoethanol at 65°C for 45 min, and the membranes were reprobed with another antibody. As an additional control for equal protein concentrations, protein contents were determined using the method of Sailer and Weissmann (1991).
Analysis of Lipid-linked and Protein-linked Oligosaccharides
The analysis of lipid- and protein-linked oligosaccharides has been described (Cacan et al., 1993; Zufferey et al., 1995; Jakob et al., 1998). For pulse–chase labeling of the oligosaccharides, typically 3 × 109 cells of a logarithmically growing culture were pelleted, washed with YP0.1D (2% Yeast extract, 1% Bactopeptone, 0.1% glucose), and resuspended in 450 μl YP0.1D containing 400 μCi 2-[3H]mannose (30 Ci/mmol; ICN Pharmaceuticals). The oligosaccharides were labeled for 1 min at 26°C and the radioactivity was chased by adding nonradioactive (±)D-mannose (111 mM final concentration). At the given time points, 5 × 108 cells were removed, placed in 1 ml of CM 3:2 (chloroform/methanol 3:2 vol/vol) and mixed by vortexing. Extraction, work-up and analysis of lipid-linked oligosaccharides (LLO) and NLO was as described above. For detailed verification of oligosaccharide structure, endo H–released NLO were further digested with α1,2-specific mannosidase from Aspergillus saitoi (15 μU; Oxford Glycosystems, Abingdon, UK) in the supplied buffer. After the digest the NLO were separated by HPLC (see above).
Results
Trimming of Protein-bound Oligosaccharides In Vivo
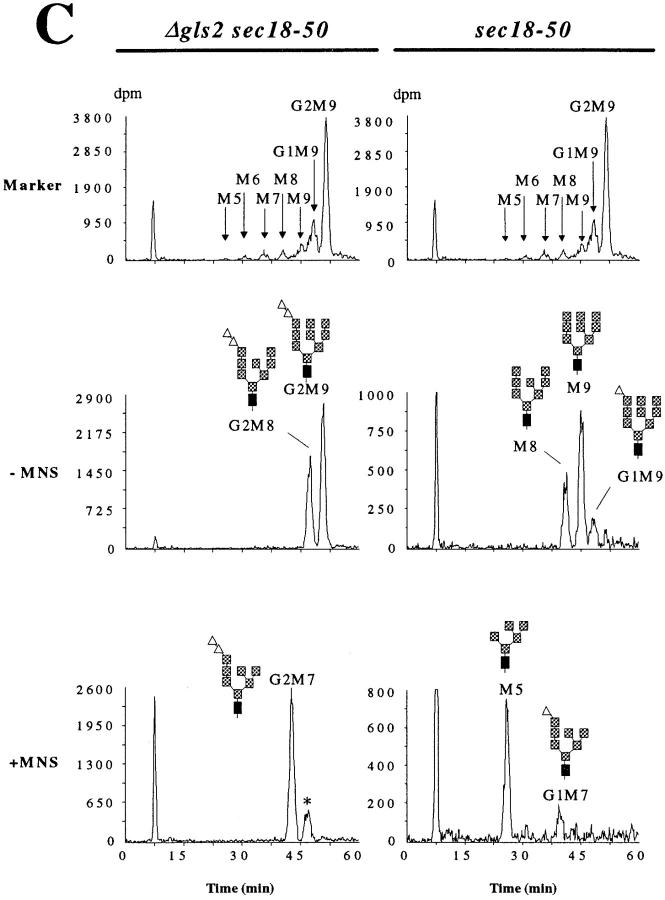
To understand in more detail the role of NLO in glycoprotein degradation, we determined the kinetics of protein-bound oligosaccharide trimming in the ER in vivo. For this, two yeast strains that carry the sec18-50 mutation were used. The sec18-50 mutation results in a temperature-sensitive phenotype and prevents the fusion of ER-derived vesicles with the Golgi apparatus at nonpermissive temperature. No processing of protein-bound oligosaccharides by Golgi glycosyltransferases was observed in sec18 mutant strains at nonpermissive conditions (Novick et al., 1980; Eakle et al., 1988). We performed the experiments at the permissive temperature for the sec18-50 mutation, nevertheless, the export rate of secretory proteins to the Golgi apparatus was slower in sec18-50 cells as compared with wild-type cells and we were able to analyze the trimming of the NLO in the ER. One of the strains carried in addition a deletion in the GLS2 locus inactivating glucosidase II. No growth phenotype was associated with the Δgls2 mutation. Cells were labeled with 3H-mannose for 1 min at 26°C and the radioactivity was chased by adding an excess of nonradioactive (±)D-mannose. At the given time points, the chase was terminated, NLO were released from protein by endoglycosidase H (endo H), and then analyzed by HPLC (see Materials and Methods section).
In the Δgls2 strain, only two trimming events occur in the ER: the removal of the terminal α1,2-linked glucose residue by glucosidase I and the cleavage of an α1,2-linked mannose residue by mannosidase I. As expected, a protein-bound oligosaccharide with the putative structure Glc2Man9GlcNAc2 (G2M9)2 was found, which was slowly converted to Glc2Man8GlcNAc2 (G2M8) with a half-life of ∼10 min (Fig. 2, A and B, left). We were unable to detect protein-bound Glc3Man9GlcNAc2.
Figure 2.

In vivo kinetics of N-linked oligosaccharide trimming. Two different yeast strains were used. Strain YG557 (Δgls2 sec18-50; left) carries a deletion of the glucosidase II–encoding locus and a mutation in the SEC18 locus resulting in a temperature-sensitive protein transport from the ER to the Golgi compartment. Strain YG556 (sec18-50; right) is fully competent in oligosaccharide trimming in the ER. Cells were labeled with a 1-min pulse of 3H-mannose, followed by a chase with an excess of unlabeled mannose. (A) Protein-bound oligosaccharides were isolated at the time indicated after initiation of the chase and analyzed by HPLC. The elution of radioactivity was monitored. The proposed oligosaccharide structure is given for the individual peaks. For explanation of the symbols, see Fig. 1. (B) Kinetics of maturation of N-linked oligosaccharides. The HPLC profiles shown in A were quantified using the FLO-ONE software (version 3.6; Packard Instrument Co., Meriden, CT). The amount of radioactivity present in each individual peak was determined, expressed as a percentage of total radioactivity present and plotted against time. The value for the G2M8 and the M8 oligosaccharide, respectively, was corrected for the absence of one mannose residue. (C) Verification of NLO species by exo-α1,2-mannosidase digestion. The NLO obtained from 5-min (sec18-50) and 10-min (Δgls2 sec18-50) chase periods (A) were treated with exo-α1,2-mannosidase from A. saitoi and separated by HPLC. Endo H–treated LLO, obtained from a Δalg10 Δgls2 (G2) strain, was used as marker for the oligosaccharides. The proposed oligosaccharide structure is given for the individual peaks. For explanation of the symbols, see Fig. 1. −MNS, NLO without exo-α1,2-mannosidase treatment; +MNS, NLO after A. saitoi exo-α1,2-mannosidase cleavage.
We analyzed the structure of the different oligosaccharides in more detail. The endo H–released Glc2Man9GlcNAc2 NLO comigrated with the Glc2Man9GlcNAc2 oligosaccharide obtained from endo H–treated LLO of a Δalg10 strain (Fig. 2 C). This strain accumulates lipid-linked Glc2Man9 GlcNAc2 due to the inactivation of the α1,2 glucosyltransferase (Burda and Aebi, 1998). Digestion of the two major protein-derived oligosaccharides from a Δgls2 sec18-50 strain by exo-α1,2-mannosidase (from Aspergillus saitoi) converted both of them to a single species (Fig. 2 C). Therefore, the two oligosaccharides found on protein in a Δgls2 sec18 strain differed by one α1,2-linked mannose. This observation, the mobility of the exo-α1,2-mannosidase digestion product and the comigration of one oligosaccharide with the Glc2Man9GlcNAc2 marker showed that protein bound Glc2Man9GlcNAc2 and the corresponding mannosidase product Glc2Man8GlcNAc2 were present in the Δgls2 sec18-50 strain. The minor peak observed after exo-α1,2-mannosidase digestion (Fig. 2 C, left) was possibly due to incomplete digestion, since the presence of glucose residues on oligosaccharides reduced the efficiency of the enzyme (Burda, P., unpublished data). Taken together, the results confirmed that release of the α1,2-linked glucose from protein-bound oligosaccharide by glucosidase I was a rapid process in the Δgls2 strain. However, trimming of the oligosaccharide by endogenous α1,2-mannosidase (Mns1p) occurred much slower.
When we analyzed the NLO processing in a strain fully competent for trimming (sec18-50; Fig. 2, right), we were again unable to detect the complete protein-bound Glc3 Man9GlcNAc2 oligosaccharide, even in preparations obtained shortly after the pulse (Fig. 2, A and B, right). The largest oligosaccharide, detected after 2 min of chase (1-min pulse), comigrated with the Glc2Man9GlcNAc2 oligosaccharide but represented a minor fraction (<10%) of the total NLO. In contrast, significant amounts of Glc1 Man9GlcNAc2 were detected at this time point (the structural analysis of this oligosaccharide is described below). However, from this oligosaccharide one or two hexose units were rapidly trimmed. To determine whether this trimming was due to glucosidase II or mannosidase I activity, we analyzed the NLO preparation from the 5-min chase point by digestion with the exo-α1,2-mannosidase (Fig. 2 C). This preparation contained small amounts of oligosaccharides comigrating with the Glc1Man9GlcNAc2 standard and significant levels of oligosaccharides that migrated as expected for Man9GlcNAc2 and Man8GlcNAc2 oligosaccharide. Indeed, digestion by exo-α1,2-mannosidase revealed that only the Glc1Man9GlcNAc2 oligosaccharides contained a protective glucose residue and was converted to Glc1Man7GlcNAc2, whereas the majority of the oligosaccharides was trimmed to Man5GlcNAc2. These results showed that the Glc1Man9GlcNAc2 protein-bound oligosaccharide was converted primarily to Man9GlcNAc2 and that the peak representing this oligosaccharide contained no significant amounts of mannosidase I–trimmed, monoglucosylated oligosaccharide.
The analysis of the structure of protein-bound oligosaccharide species as well as their temporal appearance showed that the removal of the terminal α1,2-glucose on protein-bound oligosaccharides by glucosidase I was a rapid process in vivo. Similarly, since we observed only small amounts of diglucosylated oligosaccharides (Fig. 2, A and B, 2-min chase), the hydrolysis of the first α1,3-linked glucose by glucosidase II was a rapid process. We concluded that under our experimental conditions, the monoglucosylated oligosaccharide Glc1Man9GlcNAc2 was converted to the Man9GlcNAc2 oligosaccharide with a half-life of ∼2 min and that this occurred before processing by mannosidase I, which was a relatively slow process (half-life 10 min). Evidently, removal of glucose-linked residues was not a prerequisite for mannosidase I action because mannose hydrolysis occurred with approximately the same kinetics in both glucosidase II–proficient or –deficient strains (Fig. 2 B).
Role of N-linked Oligosaccharides in Glycoprotein Processing
Removal of a mannose residue by α1,2-mannosidase concludes the trimming of NLO in the ER of S. cerevisiae (Byrd et al., 1982). Since this cleavage occurred at a slow rate, we speculated that it represents a rate-limiting step and thus is important for efficient glycoprotein transport and maturation. Therefore, we analyzed this aspect in detail by studying the processing of vacuolar proteinase CPY. In the ER, CPY receives four N-linked oligosaccharides (p1CPY, glycosylated proCPY, 67 kD) that are modified in the Golgi apparatus (p2CPY, 69 kD). Upon reaching the vacuole, CPY maturates by proteolytic cleavage of the propeptide (mCPY, 63 kD). In a pulse–chase experiment, we compared the transport rates of CPY, from ER to Golgi and to vacuole in wild-type and Δmns1 strains lacking α1,2-mannosidase activity. We observed that CPY was transported at the same rates (Fig. 3). This demonstrated that trimming of NLO by mannosidase I was not required for export of glycosylated CPY to the Golgi apparatus and the transport to the vacuole.
Figure 3.
Maturation of CPY in wild-type and mannosidase-deficient mutant cells. Logarithmically growing cells were pulsed with 35S-methionine for 5 min at 26°C and then chased for 30 min. The cells were broken, CPY immunoprecipitated, separated by SDS-PAGE and visualized by autoradiography. The processing of CPY in wild-type cells (wt, strain SS328) is shown in the top panel, that of mannosidase-deficient cells (mns1, strain YG746) in the bottom panel. The positions of the ER-modified form of proCPY (p1CPY); of the Golgi-modified form of proCPY (p2CPY) and of the vacuolar mature CPY (mCPY) are indicated.
Role of N-linked Oligosaccharides in Degradation of Misfolded CPY
Previous studies have shown that a specifically mutated form of vacuolar proteinase CPY (CPY*) is retained in the ER and degraded by the proteasome (Hiller et al., 1996) in an oligosaccharide-dependent manner (Knop et al., 1996). Moreover, deletion of the MNS1 locus affects the degradation of CPY*. These observations suggested that trimming of the oligosaccharides is required for processing of misfolded CPY in the ER. To precisely define the NLO moieties important for proteasome-dependent degradation of CPY*, we generated yeast strains containing CPY* carrying defined NLO structures on glycoproteins (for details, see Fig. 1 and Table III).
Table III.
Structure of N-linked Oligosaccharides in Mutant Yeast Strains
| Genotype | NLO after transfer | Final NLO structure* | Underglycosylation | References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Δgls2 | Glc3Man9GlcNAc2 | Glc2Man8GlcNAc2 | (G2) | No | This study | |||||
| Δalg10 Δgls2 | Glc2Man9GlcNAc2 | Glc2Man8GlcNAc2 | (G2) | Yes | Jakob et al. (1998) | |||||
| Δalg8 Δgls2 | Glc1Man9GlcNAc2 | Glc1Man8GlcNAc2 | (G1) | Yes | Jakob et al. (1998) | |||||
| Δalg6 Δgls2 | Man9GlcNAc2 | Man8GlcNAc2 | (G0) | Yes | Jakob et al. (1998) | |||||
| Δmns1 | Glc3Man9GlcNAc2 | Man9GlcNAc2 | (M9) | No | Puccia et al. (1993) | |||||
| Wild-type‡ | Glc3Man9GlcNAc2 | Man8GlcNAc2 | (M8) | No | Jakob et al. (1998), Verostek et al. (1991, 1993), | |||||
| Ziegler and Trimble (1991) | ||||||||||
| Δalg6 | Man9GlcNAc2 | Man8GlcNAc2 | (M8) | Yes | Reiss et al. (1996) | |||||
| Δalg12 | Man7GlcNAc2 | Man7GlcNAc2 | (M7) | Yes | Burda, P., C.A. Jakob, J. Beinhauer, J.H. | |||||
| Hegemann, and M. Aebi, manuscript submitted for publication | ||||||||||
| Δalg9 | Man6GlcNAc2 | Man6GlcNAc2 | (M6) | Yes | Burda et al. (1996) | |||||
| Δalg3 | Man5GlcNAc2 | Man5GlcNAc2 | (M5) | Yes | Aebi et al. (1996) |
Oligosaccharide structure on glycoproteins (NLO) after endogenous glycosidase trimming. The term in parenthesis indicates the abbreviations used throughout the figures.
The term wild-type is used with regard to oligosaccharide biosynthesis and trimming.
In a first step, we investigated the influence of glucose residues of oligosaccharides on CPY* degradation (Fig. 4). For that purpose, we constructed mutant strains that produced the following NLO structures: Glc2Man8GlcNAc2 (G2; Δalg10 Δgls2), Glc1Man8GlcNAc2 (G1; Δalg8 Δgls2), Man8GlcNAc2 (G0; wild-type and Δalg6 Δgls2). It is important to note that α1,2-mannosidase can act on glucosylated oligosaccharides in vivo (Fig. 2). When we analyzed the processing of CPY* by pulse–chase experiments, a differential degradation of CPY* was observed depending on the number of glucose residues present on the NLO (Fig. 4). The G2 CPY* (Δalg10 Δgls2) was degraded at the slowest rate, whereas the G0 CPY* was degraded at the same rate as CPY* in a strain with normal oligosaccharide biosynthesis and trimming (Fig. 4, wild-type). By Western blot analysis, another G2 CPY*, Δgls2, behaved similarly as G2 CPY* (Δalg10 Δgls2, see below and Fig. 5 C). For the G1 CPY*, we observed an intermediate degradation rate (Fig. 4). From the initial degradation rates (time points 30 and 60 min), we calculated the half-life of CPY* in the various strains (Table IV). In the wild-type and the G0 cells it was 21 min, similar to published data (Hiller et al., 1996). In comparison, the half-life of CPY* in the G1 cells was 43 min and in the G2 cells 82 min under our experimental conditions. Thus, the larger the number of glucose residues the NLO of CPY* contained, the slower its degradation rate.
Figure 4.
Degradation of misfolded CPY with glucosylated oligosaccharides. (A) Cells were labeled with a short pulse with 35S-labeled methionine, chased with an excess of unlabeled methionine, lyzed at the time indicated after the chase and CPY* was precipitated using CPY-specific antiserum. Precipitated CPY* was resolved by SDS-PAGE, autoradiography was performed using a PhosphoImager System and the level of CPY* was determined. The CPY* level at initiation of the chase was taken as 100%. Degradation rates of misfolded CPY were calculated from two independent experiments. (B) Autoradiography of immunoprecipitated misfolded CPY resolved by SDS-PAGE. The time of the chase is indicated above the lanes. The following strains indicated on the left side of A and above the different autoradiographs (B) were used in the analysis: wt: prc1-1, YG618; G0: Δalg6 Δgls2 prc1-1, YG623; G1: Δalg8 Δgls2 prc1-1, YG624; G2: Δalg10 Δgls2 prc1-1, YG625.
Figure 5.
N-linked oligosaccharide structure affects degradation of misfolded CPY. Yeast cells were grown into stationary phase (A and C, cells grown in YPD; D, growth in minimal medium), equal cell numbers harvested and their proteins were extracted, separated by SDS-PAGE, transferred to nitrocellulose and probed with antiserum for CPY. The membranes were stripped and reprobed with antiserum directed against hexokinase (HXK) which served as internal standard. All strains analyzed in this figure contained the prc1-1 mutation. The relevant genotype of the strains analyzed is given above each lane or below each column, respectively. Strains: YG618 (wt, wild-type, lanes A1, C1, and D1); YG797 (Δalg3, lanes A2 and D3); YG796 (Δalg9, lanes A3 and D5); YG807 (Δalg12, lanes A4 and D7); YG777 (Δmns1, lanes A5 and C3); YG620 (Δalg6, lane A6); YG619 (Δgls2, lane C2); YG821 (wild-type + YEp352, lane D2); YG822 (Δalg3 + pALG3, lane D4); YG823 (Δalg9 + pALG9, lane D6); YG824 (Δalg12 + pALG12, lane D8); Abbreviations: wt, wild-type; CPY*, misfolded CPY; HXK, hexokinase. (A) Degradation of misfolded CPY is dependent on core mannose residues. The position of malfolded CPY* is indicated. The mobility of CPY* in SDS-PAGE varied due to the different oligosaccharide structures. (B) Quantification of degradation of misfolded CPY. Protein amounts (mean ± SD) of three independent Western blot experiments as shown in A were quantified by using a CCD camera and the Wincam V2.1 software (Cybertech, Berlin, Germany) and normalized to the hexokinase levels. The amount of misfolded CPY of the wild-type strain was set as 1.0. (C) Degradation of misfolded CPY is reduced in Δgls2 and Δmns1 cells but not in wild-type cells. The mobility of CPY* in SDS-PAGE varied due to the different oligosaccharide structures. (D) Degradation of CPY* is dependent on the altered oligosaccharide structures. Wild-type and mutant strains with altered oligosaccharide biosynthesis (indicated above the lanes) with (+) or without (−) the corresponding complementing plasmid were analyzed for degradation of CPY* by Western blot analysis. The position of CPY* is indicated. Hexokinase served as a control protein.
Table IV.
Calculated Half-life of Mutated CPY
| Genotype of strains | Half-life* | |||
|---|---|---|---|---|
| prc1-1 (wild-type) | 20.8 ± 3.7 | |||
| Δalg6 Δgls2 prc1-1 | (G0) | 21.9 ± 1.0 | ||
| Δalg8 Δgls2 prc1-1 | (G1) | 43.3 ± 0.3 | ||
| Δalg10 Δgls2 prc1-1 | (G2) | 82.3 ± 1.1 |
The values (mean ± SD) represent the calculated half-lives from time points 0, 30, and 60 min and were derived from the experiment shown in Fig. 2.
Next, we determined whether mannose residues of the NLO influenced degradation of CPY*. The core mannose structure of LLO is synthesized by a set of sequentially acting ER mannosyltransferases (Alg3p, Alg9p, Alg12p; see Fig. 1, Orlean, 1997). Due to the fact that incompletely assembled oligosaccharides can be transferred to protein, albeit with a reduced efficiency, we were able to obtain yeast strains with incomplete, but defined NLO structures, namely Man5GlcNAc2 (M5, Δalg3; Aebi et al., 1996), Man6GlcNAc2 (M6, Δalg9; Burda et al., 1996), Man7 GlcNAc2 (M7, Δalg12, Burda and Aebi, manuscript in preparation), Man8GlcNAc2 (M8, wild-type with respect to NLO; M8, Δalg6; Reiss et al., 1996), and Man9GlcNAc2 (M9, Δmns1; Puccia et al., 1993; see also Table III).
When the half-lives of mutant CPY* bearing various oligosaccharide structures were analyzed, we found that CPY* containing Man8GlcNAc2 NLO was rapidly degraded (Fig. 5 A, lanes 1 and 6 and B). The Man5GlcNAc2 CPY* (Fig. 5, A lane 2 and B) was degraded at a reduced rate compared with the Man8GlcNAc2 CPY*. However, a greatly reduced rate of CPY* degradation was observed when the NLO were of Man6GlcNAc2 (Fig. 5 A, lane 3 and B), Man7GlcNAc2 (Fig. 5 A, lane 4 and B), and Man9 GlcNAc2 (Fig. 5 A, lane 5 and B) structures. By Western blot analysis, CPY* with diglucosylated oligosaccharides (Δgls2) was also degraded at a slower rate than Man8 GlcNAc2 CPY* (Fig. 5 C, compare with Fig. 4). However, the stabilization was not as prominent as for Man9 GlcNAc2 CPY* (Δmns1; Fig. 5 C). Taken together, our results showed that efficient degradation of CPY* was dependent on the NLO structure. Man8GlcNAc2 CPY* was rapidly degraded, whereas the trimming intermediate Man9GlcNAc2 CPY* and the incompletely assembled Man6-7GlcNAc2 CPY* were inefficiently degraded.
To demonstrate that the differences in degradation rate of CPY* were due to the altered oligosaccharide structure, we transformed the yeast cells with plasmids complementing the deleted gene loci. The Δalg3, Δalg9, and Δalg12 cells were transformed with plasmids containing the ALG3 (pALG3), ALG9 (pALG9) and ALG12 (pALG12) locus. Further, the CPY* wild-type strain was transformed with the vector plasmid (YEp352). The NLO structure-dependent CPY* degradation phenotype could be reverted by complementing the deleted alg gene loci with the appropriate plasmids (Fig. 5 D, lanes 3–8), but the degradation in wild-type cells was not influenced by the transformation with the empty plasmid (Fig. 5 D, lanes 1 and 2).
Incomplete assembly of the LLO leads to a reduced oligosaccharide transfer to protein by the oligosaccharyltransferase (Sharma et al., 1981; Silberstein and Gilmore, 1996), which is then apparent by underglycosylation of glycoproteins (Huffaker and Robbins, 1981, 1983; Stagljar et al., 1994; te Heesen et al., 1994; Burda et al., 1996; Burda and Aebi, 1998). The equal degradation of Man8GlcNAc2 CPY* in glycosylation wild-type cells and in Δalg6 cells (Fig. 5 A, lanes 1 and 6) indicated that the degradation was not due to impaired synthesis of LLO, but rather to the altered oligosaccharide structure.
Also, in contrast to wild-type CPY, CPY* was efficiently glycosylated in both Δalg3 (Man5GlcNAc2) and Δalg9 (Man6GlcNAc2) cells under the conditions used. We observed a single CPY* glycoform in Δalg3 prc1-1 and Δalg9 prc1-1 cells (Fig. 6, lanes 1 and 3), whereas correctly folded CPY was incompletely glycosylated in Δalg3 and Δalg9 cells (Fig. 6, lanes 7 and 9). This hypoglycosylation of CPY was visualized by the distinct bands upon Western blot analysis representing different glycoforms of this protein (Stagljar et al., 1994). In addition, this experiment showed that such forms of CPY* did not reach the vacuole because endo H treatment resulted in the deglycosylated pro-form of CPY* in Δalg9 cells (Fig. 6, lanes 3 and 4, compare with lanes 9 and 10). As expected, the oligosaccharides in Δalg3 cells were resistant to endo H digestion, whereas in Δalg9 cells they were endo H sensitive (Fig. 6, lanes 8 and 10).
Figure 6.
Processing of wild-type CPY occurs independent of NLO structure. Yeast cells were grown into stationary phase, equal cell numbers harvested and their proteins extracted. Extracts were analyzed before (−) or after endo H (+) treatment. The proteins were separated by SDS-PAGE, transferred to nitrocellulose and probed with antiserum to detect CPY. After exposure, membranes were stripped and reprobed with antiserum directed against hexokinase (HXK). Δalg3 strains (lanes 1, 2, 7, and 8) and Δalg9 strains (lanes 3, 4, 9, and 10) carrying a mutant prc1-1 locus expressing CPY* (lanes 1–4) or a wild-type PRC1 locus (lanes 5–10) were analyzed. The positions of the ER form p1CPY* and deglycosylated proCPY* (dpCPY*) are shown at the left (lanes 1–4). Mature, wild-type CPY (mCPY), lacking one (−1) or two (−2) oligosaccharides and deglycosylated, mature CPY (dCPY) are indicated on the right side (lanes 5–10). The following strains were used: YG797 (Δalg3 prc1-1), YG796 (Δalg9 prc1-1), SS328 (PRC1), YG228 (Δalg3 PRC1), and YG414 (Δalg9 PRC1).
In Δalg3 or Δalg9 cells, a much higher steady state level of CPY was observed compared with CPY* in various mutant cells (Fig. 6). This indicated that degradation of CPY* was not completely blocked by the altered oligosaccharide structure. However, our results demonstrated that oligosaccharide structures specifically affected the degradation of misfolded CPY accumulating in the ER, whereas processing and secretion of wild-type CPY was not altered.
Discussion
A Defined Oligosaccharide Structure Was Required for Efficient Degradation of Misfolded Glycoprotein
We observed a significant effect of the structure of N-linked oligosaccharides on degradation of misfolded CPY*, a model protein for degradation of glycoproteins retained in the ER (Wolf and Fink, 1975; Finger et al., 1993; Hiller et al., 1996; Knop et al., 1996). When expressed in Δgls2 or Δmns1 cells, we found a reduced degradation of CPY* (Fig. 5 C). The effect of the mannosidase I inactivation was significantly stronger than that of the glucosidase II deletion. A similar stabilization of CPY* as in Δmns1 cells was observed in Δalg9 and Δalg12 cells, where incompletely mannosylated oligosaccharides (Man6GlcNAc2 and Man7GlcNAc2, respectively) were transferred to protein. In Δalg3 cells, which contain protein-bound Man5Glc NAc2, degradation of CPY* was reduced, however, not to the same extent as in Δalg9 or Δalg12 cells. Interestingly, in Δalg6 cells, where nonglucosylated oligosaccharide Man9GlcNAc2 was transferred to protein and trimmed to Man8GlcNAc2, no effect on the stabilization of CPY* was observed. We concluded, that the protein-bound Man8GlcNAc2 structure was an important recognition element in the degradation pathway of CPY*. Our results were best explained by a model (Fig. 7) where the Man8GlcNAc2 oligosaccharide on a misfolded glycoprotein acted as a positive signal for degradation. This oligosaccharide structure might be recognized by a lectin, since any changes in its structure reduced the efficiency of degradation. Such a carbohydrate-binding protein has also been postulated in the degradation of glycoproteins in mammalian cells (Yang et al., 1998) and might represent “the additional signal to direct them (soluble misfolded proteins) to the dislocation and the ubiquitation machinery” (Kopito, 1997). Our results showed that glucosylated oligosaccharides also reduced the degradation rate of CPY*, albeit to a lesser extent than the Man9GlcNAc2, Man7GlcNAc2 and Man6GlcNAc2 structures. We concluded that the α1,2-α1,2-dimannose branch of the Man8 GlcNAc2 oligosaccharide was a less important structural element for oligosaccharide recognition than both the α1,6- and α1,3-branch affected by the alg3, alg9 and alg12 mutations (see Fig. 1). The observation that the Man5 GlcNAc2-producing Δalg3 mutation had a less severe effect on degradation was explained by the hypothesis that this oligosaccharide structure represents an intermediate in the degradation of glycoprotein as shown in higher eukaryotic cells (Villers et al., 1994; Ermonval et al., 1997).
Figure 7.
A role of N-linked oligosaccharides in the degradation of glycoproteins in yeast. A secreted glycoprotein folds in the lumen of the ER with the help of chaperone(s). The N-linked oligosaccharide of the glycoprotein is trimmed by glycosidases (indicated by the three arrows) to the Man8GlcNAc2 structure. The correctly folded protein is exported to the Golgi compartment. If folding of the glycoprotein is not completed within the time required for complete oligosaccharide trimming, the misfolded glycoprotein, bearing oligosaccharides of the Man8GlcNAc2 structure and associated with chaperone(s), is targeted for export to the cytosol, where degradation by the proteasome occurs. A lectin, recognizing specifically the Man8GlcNAc2 structure, is involved in the targeting of the malfolded protein to the degradation pathway.
Whether the postulated lectin additionally recognizes unfolded protein domains, as does UDP-glucose/glycoprotein glucosyltransferase, involved in the ER quality control pathway of higher eukaryotes (Sousa and Parodi, 1995), is not known. It is possible that the binding of both, the chaperones and the postulated lectin constitute a signal which targets the glycoprotein to the degradation pathway. Indeed Kar2p, the yeast homologue of BiP, transiently binds to wild-type CPY (te Heesen and Aebi, 1994; Simons et al., 1995). Furthermore, this Hsp70 protein was shown to be involved in the degradation of misfolded protein by the proteasome pathway (Plemper et al., 1997).
It has been proposed that the trimming of the protein-bound oligosaccharide in the endoplasmic reticulum represents a biological timer for the protein maturation in the ER of higher eukaryotes (Helenius et al., 1997). This timer function might be required to prevent permanent residence of misfolded glycoproteins in the ER due to the binding to calnexin and calreticulin in higher eukaryotes. Our results are fully compatible with this timer model: as in higher eukaryotic cells (Hubbard and Robbins, 1979), the protein bound oligosaccharide underwent a step-wise trimming process. Removal of the terminal α1,2-glucose by glucosidase I and the first α1,3-glucose by glucosidase II was a very rapid process, whereas the second α1,3-glucose was removed more slowly. The same difference in glucose hydrolysis was observed for glucosidase II of higher eukaryotic cells (Hubbard and Robbins, 1979) and it has been proposed that two different substrate binding sites of glucosidase II are responsible for this difference: the high affinity site would be responsible for the hydrolysis of the first glucose, the low affinity site for the hydrolysis of the second glucose (Alonso et al., 1993). Similar to the findings in higher eukaryotic cells, the mannose trimming was a slow process as compared with hydrolysis of the glucose residues of the protein-bound oligosaccharide. The removal of one specific α1,2-mannose residue by ER-α1,2-mannosidase might represent the time-point after which a misfolded protein is routed to the degradation pathway. We speculated that the processing by mannosidase I determined the time-scale in which a protein had to be correctly folded. If this was not achieved, the glycoprotein was degraded. Importantly, maturation and transport of correctly folded CPY is oligosaccharide-independent (Schwaiger et al., 1982; Winther et al., 1991) and was also not influenced by trimming or oligosaccharide structure (Figs. 3 and 6). It was also postulated that there is a selective export of proteins out of the ER in yeast (Kuehn and Schekman, 1997). Selective export of only correctly folded proteins in combination with degradation of misfolded proteins, timed by oligosaccharide trimming, might therefore represent an effective quality control system for glycoprotein folding in the ER of S. cerevisiae, where the calnexin/calreticulin cycle (reglucosylation of misfolded proteins) has not been found (Fernandez et al., 1994, Jakob et al., 1998).
In support of our model, we found that the degradation of a mutant form of the oligosaccharyltransferase component Stt3p, a glycoprotein with multiple transmembrane domains (Zufferey et al., 1995) was also controlled by oligosaccharide trimming (Bodmer, D., U. Spirig, and M. Aebi, manuscript in preparation), suggesting that resident ER glycoproteins are subject to the same degradation system as are glycoproteins that are exported from the ER.
Is there a similar role of the Man8GlcNAc2 oligosaccharide in the quality control process of glycoproteins in the ER of higher eukaryotic cells? In the trimming process of protein-bound oligosaccharides, removal of the glucose residues precedes the hydrolysis mannose trimming (Hubbard and Robbins, 1979). There is an α1,2-mannosidase activity that leads to the same Man8GlcNAc2 oligosaccharide as the yeast MNS1 enzyme in the ER of higher eukaryotic cells (Bischoff and Kornfeld, 1983). However, there are additional ER mannosidase activities in the ER of higher eukaryotic cells and a different Man8GlcNAc2 oligosaccharide isomer, where the α1,2-mannose linked to the α1,6-mannose is removed, can be produced (Weng and Spiro, 1993, 1996; Moremen et al., 1994). The trimming process in higher eukaryotes is therefore more complex than in yeast. Nevertheless, our results obtained in yeast are compatible with reports on inhibition of α-mannosidase trimming by deoxymannojirimycin that stabilizes specific misfolded glycoproteins in the ER (Su et al., 1993; Daniel et al., 1994; Liu et al., 1997; Yang et al., 1998). On the other hand, degradation of some glycoproteins in the ER was not affected by mannosidase inhibition (Yang et al., 1998).
Alternative Pathways for ER Degradation?
Previous work has shown that CPY* remains in the ER, is ubiquitinated and then degraded in a proteasome-dependent pathway (Hiller et al., 1996). When we compared the level of wild-type CPY and mutant CPY* in both the Δalg9 and Δmns1 cells (Figs. 5 A and 6), it was apparent that more mature (vacuolar) CPY was present in wild-type cells than CPY* in the prc1-1 cells. A major portion of CPY* was apparently degraded in these cells. Our results showed that alterations of the oligosaccharide structure did not completely block degradation of CPY*. Moreover, mutations reported to affect CPY* degradation do not completely block CPY* degradation either (Hiller et al., 1996; Knop et al., 1996; Plemper et al., 1997). Taken together, these results suggest an alternative, glycosylation-independent degradation pathway for misfolded glycoproteins in the ER of S. cerevisiae.
Evidence for Posttranslocational N-Glycosylation of CPY*
N-linked oligosaccharides are added co- and posttranslocationally in yeast. For CPY, competition between glycosylation and folding has been reported (Holst et al., 1996). In cells containing alg mutations, incompletely assembled oligosaccharide is transferred to nascent protein (Stagljar et al., 1994), albeit at a reduced rate (Sharma et al., 1981). This is reflected by incomplete use of potential N-glycosylation sites. However, we noticed that in both Δalg3 and Δalg9 cells, only CPY* with all four potential N-glycosylation sites occupied accumulated, whereas in the Δalg3 and Δalg9 cells, wild-type CPY lacking one or two oligosaccharides were found (Fig. 6). Therefore, we postulated that the prolonged exposure of misfolded CPY* to the oligosaccharyltransferase compensated for the reduced affinity of the oligosaccharyltransferase towards incompletely assembled oligosaccharide and resulted in fully glycosylated CPY* in both Δalg3 and Δalg9 cells.
The assembly of the lipid-linked oligosaccharide and its transfer to selected asparagine residues of polypeptides in the ER is a highly conserved process. The selective processing of the protein-bound oligosaccharide supports the idea that conservation of the transferred oligosaccharide structure is due to the function of specific trimming intermediates in glycoprotein maturation. Genetic tailoring of NLO structures will provide a useful tool to identify additional roles of the oligosaccharide in glycoprotein processing.
Acknowledgments
We thank Dr. D.H. Wolf (University of Stuttgart, Germany) for providing the prc1-1 integrative plasmid. The hexokinase antiserum was kindly provided by Dr. S. Schröder (Biozentrum, University of Basel, Switzerland). We would also like to thank Drs. A. Helenius and S. te Heesen for helpful discussions and comments on the manuscript and anonymous reviewers for invaluable remarks.
This work was supported by the Swiss National Science Foundation grants 3100-040350 (to M. Aebi) and 31-50835.97 (to J. Roth).
Abbreviations used in this paper
- CPY
carboxypeptidase Y
- CPY*
mutated, misfolded CPY
- endo H
endoglycosidase H
- LLO
lipid-linked oligosaccharides
- NLO
N-linked oligosaccharides
Footnotes
Address all correspondence to Markus Aebi, Mikrobiologisches Institut, ETH Zürich, Schmelzbergstrasse 7, CH-8092 Zürich, Switzerland. Tel.: 41 1 632 64 13. Fax: 41 1 632 11 48. E-mail: aebi@micro.biol.ethz.ch
2. Since the protein-bound oligosaccharides were released by Endo H cleavage, they are lacking one GlcNAc residue. For clarity, however, the NLO structure as found on protein is denoted.
References
- Aebi M, Gassenhuber J, Domdey H, te Heesen S. Cloning and characterization of the ALG3 gene of Saccharomyces cerevisiae. . Glycobiology. 1996;6:439–444. doi: 10.1093/glycob/6.4.439. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., A. Santa-Cecilia, and P. Calvo. Effect of bromoconduritol on glucosidase II from rat liver. Eur. J. Biochem. 215:37–42. [DOI] [PubMed]
- Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO (Eur Mol Biol Organ) J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Bischoff J, Kornfeld R. Evidence for an α-mannosidase in endoplasmic reticulum of rat liver. J Biol Chem. 1983;258:7907–7910. [PubMed] [Google Scholar]
- Bonifacino JS. Reversal of fortune for nascent proteins. Nature. 1996;384:405–406. doi: 10.1038/384405a0. [DOI] [PubMed] [Google Scholar]
- Burda P, Aebi M. The ALG10 locus of Saccharomyces cerevisiaeencodes the α-1,2 glucosyltransferase of the endoplasmic reticulum: the terminal glucose of the lipid-linked oligosaccharide is required for efficient N-linked glycosylation. Glycobiology. 1998;8:455–462. doi: 10.1093/glycob/8.5.455. [DOI] [PubMed] [Google Scholar]
- Burda P, te Heesen S, Brachat A, Wach A, Dusterhoft A, Aebi M. Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: identification of the ALG9gene encoding a putative mannosyltransferase. Proc Natl Acad Sci USA. 1996;93:7160–7165. doi: 10.1073/pnas.93.14.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Tarentino AL, Maley F, Atkinson PH, Trimble RB. Glycoprotein synthesis in yeast. Identification of Man8GlcNAc2 as an essential intermediate in oligosaccharide processing. J Biol Chem. 1982;257:14657–14666. [PubMed] [Google Scholar]
- Cacan R, Labiau O, Mir AM, Verbert A. Effect of cell attachment and growth on the synthesis and fate of dolichol-linked oligosaccharides in Chinese hamster ovary cells. Eur J Biochem. 1993;215:873–881. doi: 10.1111/j.1432-1033.1993.tb18105.x. [DOI] [PubMed] [Google Scholar]
- Daniel PF, Winchester B, Warren CD. Mammalian alpha-mannosidases-multiple forms but a common purpose? . Glycobiology. 1994;4:551–566. doi: 10.1093/glycob/4.5.551. [DOI] [PubMed] [Google Scholar]
- Eakle KA, Bernstein M, Emr SD. Characterization of a component of the yeast secretion machinery: identification of the SEC18gene product. Mol Cell Biol. 1988;8:4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermonval M, Cacan R, Gorgas K, Haas I, Verbert A, Buttin G. Differential fate of glycoproteins carrying a monoglucosylated form of truncated N-glycan in a new CHO cell line, MadIA214, selected for a thermosensitive secretory defect. J Cell Sci. 1997;110:323–336. doi: 10.1242/jcs.110.3.323. [DOI] [PubMed] [Google Scholar]
- Fernandez FS, Trombetta SE, Hellman U, Parodi AJ. Purification to homogeneity of UDP-glucose:glycoprotein glucosyltransferase from Schizosaccharomyces pombe and apparent absence of the enzyme from Saccharomyces cerevisiae. . J Biol Chem. 1994;269:30701–30706. [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Rothblatt J, Schekman R. Analysis of polypeptide transit through yeast secretory pathway. Methods Enzymol. 1991;194:662–674. doi: 10.1016/0076-6879(91)94048-h. [DOI] [PubMed] [Google Scholar]
- Grondin B, Herscovics A. Topology of ER processing alpha-mannosidase of Saccharomyces cerevisiae. . Glycobiology. 1992;2:369–372. doi: 10.1093/glycob/2.4.369. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego. 931 pp.
- Helenius A, Trombetta ES, Hebert DN, Simons JF. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7:193–200. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- Herscovics A, Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Holst B, Bruun AW, Kielland-Brandt MC, Winther JR. Competition between folding and glycosylation in the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1996;15:3538–3546. [PMC free article] [PubMed] [Google Scholar]
- Hubbard SC, Robbins PW. Synthesis and processing of protein-linked oligosaccharides in vivo. . J Biol Chem. 1979;254:4568–4576. [PubMed] [Google Scholar]
- Huffaker TC, Robbins PW. Temperature-sensitive yeast mutants deficient in asparagine-linked glycosylation. J Biol Chem. 1981;257:3203–3210. [PubMed] [Google Scholar]
- Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci USA. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Burda P, te Heesen S, Aebi M, Roth J. Genetic tailoring of N-linked oligosaccharides: the role of glucose residues in glycoprotein processing of Saccharomyces cerevisiae in vivo. . Glycobiology. 1998;8:155–164. doi: 10.1093/glycob/8.2.155. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Schlenker S. Selective protein degradation: a journey's end within the proteasome. Cell. 1995;82:881–884. doi: 10.1016/0092-8674(95)90021-7. [DOI] [PubMed] [Google Scholar]
- Knop M, Hauser N, Wolf DH. N-glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase Yscy in yeast. Yeast. 1996;12:1229–1238. doi: 10.1002/(sici)1097-0061(19960930)12:12<1229::aid-yea15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Schekman R. COPII and secretory cargo capture into transport vesicles. Curr Opin Cell Biol. 1997;9:477–483. doi: 10.1016/s0955-0674(97)80022-1. [DOI] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Intracellular disposal of incompletely folded human alpha1-antitrypsin involves release from calnexin and post-translational trimming of asparagine-linked oligosaccharides. J Biol Chem. 1997;272:7946–7951. doi: 10.1074/jbc.272.12.7946. [DOI] [PubMed] [Google Scholar]
- Lussier M, White A, Sheraton J, di Paolo T, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AFJ, Kapteyn JC, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. . Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Trimble RB, Herscovics A. Glycosidases of the asparagine-linked oligosaccharide processing pathway. Glycobiology. 1994;4:113–125. doi: 10.1093/glycob/4.2.113. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Orlean, P. 1997. Biogenesis of yeast wall and surface proteins. In Molecular and Cellular Biology of the Yeast Saccharomyces, Cell Cycle and Cell Biology. J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pilon M, Schekman R, Römisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO (Eur Mol Biol Organ) J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Böhmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Puccia R, Grondin B, Herscovics A. Disruption of the processing alpha-mannosidase gene does not prevent outer chain synthesis in Saccharomyces cerevisiae. . Biochem J. 1993;290:21–26. doi: 10.1042/bj2900021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss G, te Heesen S, Zimmerman J, Robbins PW, Aebi M. Isolation of the ALG6 locus of Saccharomyces cerevisiaerequired for glucosylation in the N-linked glycosylation pathway. Glycobiology. 1996;6:493–498. doi: 10.1093/glycob/6.5.493. [DOI] [PubMed] [Google Scholar]
- Roth, J. 1995. Biosynthesis: compartmentation of glycoprotein biosynthesis. In Glycoproteins. J. Montreuil, H. Schachter, and J.F.G. Vliegenthart, editors. Elsevier, New York. 287–312.
- Sailer A, Weissmann C. A sensitive and rapid protein assay impervious to detergents. Technique (Phila) 1991;3:37–38. [Google Scholar]
- Sathe GM, O'Brien S, McLaughlin MM, Watson F, Livi GP. Use of polymerase chain reaction for rapid detection of gene insertions in whole yeast cells. Nucleic Acids Res. 1991;19:4775. doi: 10.1093/nar/19.17.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger H, Hasilik A, von Figura K, Wiemken A, Tanner W. Carbohydrate-free carboxypeptidase Y is transferred into the lysosome-like yeast vacuole. Biochem Biophys Res Commun. 1982;104:950–956. doi: 10.1016/0006-291x(82)91341-9. [DOI] [PubMed] [Google Scholar]
- Sharma CB, Lehle L, Tanner W. N-Glycosylation of yeast proteins. Characterization of the solubilized oligosaccharyl transferase. Eur J Biochem. 1981;116:101–108. doi: 10.1111/j.1432-1033.1981.tb05306.x. [DOI] [PubMed] [Google Scholar]
- Silberstein S, Gilmore R. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 1996;10:849–858. [PubMed] [Google Scholar]
- Simons JF, Ferro-Novick S, Rose MD, Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol. 1995;130:41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Wolf DH. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1227–1233. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Parodi AJ. The molecular basis for the recognition of the UDP-Glc:glycoprotein glucosyltransferase with the acceptor glycoprotein. EMBO (Eur Mol Biol Organ) J. 1995;14:4196–4203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro RG, Zhu Q, Bhoyroo V, Soling HD. Definition of the lectin-like properties of the molecular chaperone, calreticulin, and demonstration of its copurification with endomannosidase from rat liver Golgi. J Biol Chem. 1996;271:11588–11594. doi: 10.1074/jbc.271.19.11588. [DOI] [PubMed] [Google Scholar]
- Stagljar I, te Heesen S, Aebi M. New phenotype of mutations deficient in the glycosylation of the lipid-linked oligosaccharide: the cloning of the ALG8locus. Proc Natl Acad Sci USA. 1994;91:5977–5981. doi: 10.1073/pnas.91.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K, Stoller T, Rocco J, Zemsky J, Green R. Pre-golgi degradation of yeast prepro-α-factor expressed in a mammalian cell. J Biol Chem. 1993;268:14301–14309. [PubMed] [Google Scholar]
- te Heesen S, Aebi M. The genetic interaction of kar2 and wbp1 mutations. Distinct functions of binding protein BiP and N-linked glycosylation in the processing pathway of secreted proteins in Saccharomyces cerevisiae. . Eur J Biochem. 1994;222:631–637. doi: 10.1111/j.1432-1033.1994.tb18906.x. [DOI] [PubMed] [Google Scholar]
- te Heesen S, Janetzky B, Lehle L, Aebi M. The yeast WBP1 is essential for oligosaccharyltransferase activity in vivo and in vitro. . EMBO (Eur Mol Biol Organ) J. 1992;11:2071–2075. doi: 10.1002/j.1460-2075.1992.tb05265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Heesen S, Lehle L, Weissmann A, Aebi M. Isolation of the ALG5 locus encoding the UDP-glucose:dolichyl-phosphate glucosyltransferase from Saccharomyces cerevisiae. . Eur J Biochem. 1994;224:71–79. doi: 10.1111/j.1432-1033.1994.tb19996.x. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- Verostek MF, Atkinson PH, Trimble RB. Structure of Saccharomyces cerevisiae alg3,sec18mutant oligosaccharides. J Biol Chem. 1991;266:5547–5551. [PubMed] [Google Scholar]
- Verostek MF, Atkinson PH, Trimble RB. Glycoprotein biosynthesis in the alg3 Saccharomyces cerevisiaemutant. 1. Role of glucose in the initial glycosylation of invertase in the endoplasmic reticulum. J Biol Chem. 1993;268:12095–12103. [PubMed] [Google Scholar]
- Villers C, Cacan R, Mir A-M, Labiau O, Verbert A. Release of oligomannoside-type glycans as a marker of the degradation of newly synthesized glycoproteins. Biochem J. 1994;298:135–142. doi: 10.1042/bj2980135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Weng S, Spiro RG. Demonstration that a kifunensine-resistant α-mannosidase with a unique processing action on N-linked oligosaccharides occurs in rat liver endoplasmic reticulum and various cultured cells. J Biol Chem. 1993;268:25656–25663. [PubMed] [Google Scholar]
- Weng S, Spiro RG. Evaluation of the early processing routes of N-linked oligosaccharides of glycoproteins through the characterization of Man8GlcNAc2 isomers: evidence that endomannosidase functions in vivoin the absence of a glucosidase blockade. Glycobiology. 1996;6:861–868. doi: 10.1093/glycob/6.8.861. [DOI] [PubMed] [Google Scholar]
- Winther JR, Stevens TH, Kielland-Brandt MC. Yeast carboxypeptidase Y requires glycosylation for efficient intracellular transport, but not for vacuolar sorting, in vivostability, or activity. Eur J Biochem. 1991;197:681–689. doi: 10.1111/j.1432-1033.1991.tb15959.x. [DOI] [PubMed] [Google Scholar]
- Wolf DH, Fink GR. Proteinase C (carboxypeptidase Y) mutant of yeast. J Bacteriol. 1975;123:1150–1156. doi: 10.1128/jb.123.3.1150-1156.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Oemura S, Bonifacino JS, Weissman AM. Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum in T cells: importance of oligosaccharide processing, ubiquitination, and proteasome-dependent removal from ER membranes. J Exp Med. 1998;187:835–846. doi: 10.1084/jem.187.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler FD, Trimble RB. Glycoprotein biosynthesis in yeast: purification and characterisation of the endoplasmic reticulum Man9 processing α-mannosidase. Glycobiology. 1991;1:605–614. doi: 10.1093/glycob/1.6.605. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Knauer R, Burda P, Stagljar I, te Heesen S, Lehle L, Aebi M. STT3, a highly conserved protein required for yeast oligosaccharyltransferase activity in vitro. . EMBO (Eur Mol Biol Organ) J. 1995;14:4949–4960. doi: 10.1002/j.1460-2075.1995.tb00178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]