Abstract
During the early development of Xenopus laevis, we followed in individual nuclei the formation of a nucleolus by examining simultaneously its structural organization and its transcriptional competence. Three distinct situations were encountered with different frequencies during development. During the first period of general transcriptional quiescence, the transcription factor UBF of maternal origin, was present in most nuclei at the ribosomal gene loci. In contrast, fibrillarin, a major protein of the processing machinery, was found in multiple prenucleolar bodies (PNBs) whereas nucleolin was dispersed largely in the nucleoplasm. During the second period, for most nuclei these PNBs had fused into two domains where nucleolin concentrated, generating a structure with most features expected from a transcriptionally competent nucleolus. However, RNA polymerase I–dependent transcription was not detected using run-on in situ assays whereas unprocessed ribosomal RNAs were observed. These RNAs were found to derive from a maternal pool. Later, during a third period, an increasing fraction of the nuclei presented RNA polymerase I–dependent transcription. Thus, the structural organization of the nucleolus preceded its transcriptional competence. We conclude that during the early development of X. laevis, the organization of a defined nucleolar structure, is not associated with the transcription process per se but rather with the presence of unprocessed ribosomal RNAs.
Keywords: fibrillarin, nucleolin, nucleologenesis, pre-rRNA, UBF transcription factor, Xenopus development
uring interphase, the nucleus of a eukaryotic cell is highly organized (Lamond and Earnshaw, 1998). Whereas the chromatin corresponding to individual chromosomes occupies defined territories (Cremer et al., 1993), the machineries supporting nuclear functions dedicated to transcription, processing, replication and repair have been localized in specific sites possibly determining functional domains in the nucleus (reviewed by Spector, 1993; Jackson, 1995; Strouboulis and Wolffe, 1996; Singer and Green, 1997; Lamond and Earnshaw, 1998). Understanding how the spatial and temporal assembly of these molecular machineries operate will be critical to decipher how they can be controlled and coordinated.
The nucleolus represents an attractive model of a functional domain involved in RNA metabolism. It is present in the nucleus as a morphologically distinct nuclear organelle and its organization has been largely documented in the literature (reviewed by Hadjiolov, 1985). Within this domain, the ribosomal genes (rDNAs) organized in multiple copies are transcribed and processed in morphologically distinct regions. The transcription machinery is localized in the fibrillar component: i.e., the fibrillar center (FC)1 and dense fibrillar component (DFC), whereas the processing machinery is found in the DFC and the granular component (GC; Shaw and Jordan, 1995). The rDNA transcription machinery, as defined in vitro, is composed of RNA polymerase I (RNA pol I) in association with the upstream binding factor (UBF), and the promoter selectivity factor designated SL1 in human cells (Bell et al., 1988, 1989; Jantzen et al., 1990), and Rib 1 in Xenopus laevis (McStay et al., 1991; Bodeker et al., 1996). UBF specifically binds to the rDNA promoter (Learned et al., 1986; Bell et al., 1989; Jantzen et al., 1990; McStay et al., 1991) as the first step of the assembly of a stable RNA pol I initiation transcription complex (reviewed by Moss and Stefanovsky, 1995). After transcription, processing, cleavage, methylation, and pseudo uridylation of the ribosomal RNAs (rRNAs), involve a complex machinery composed of a multitude of small nucleolar RNAs (snoRNAs) and several proteins (reviewed by Smith and Steitz, 1997). The function of the major protein, fibrillarin (Ochs et al., 1985b ), depends on its association with snoRNAs U3 (Gerbi et al., 1990; Filipowicz and Kiss, 1993), as well as with other snoRNAs, up to and including snoRNAs U63 (Smith and Steitz, 1997).
The development of specific antibodies against individual nucleolar proteins and the use of specific probes to localize the rDNAs as well as the rRNAs have made it possible to analyze the dynamic organization of this nuclear domain. A stable nucleolar entity appeared dependent on ongoing transcription since inhibition of rDNA transcription could severely affect nucleolar organization. Therefore, it was concluded that the existence of the nucleolar structure was strictly dependent on the nucleolar transcriptional function (Benavente et al., 1987; Scheer et al., 1993; Weisenberger et al., 1993).
It has also been established that the nucleolus is built at late telophase in cycling cells (reviewed by Scheer et al., 1993; Thiry and Goessens, 1996). This process is characterized by two major events, the activation of RNA pol I transcription and the formation of the prenucleolar bodies (PNBs) that move to sites of active transcription after mitosis (Jiménez-Garcia et al., 1994). Both events are predetermined by the nucleolar activity taking place during the interphase preceding mitosis. Indeed, during mitosis, components of the transcription machinery seem to remain associated with rDNAs (Roussel et al., 1996) and PNBs are formed of preexisting nucleolar complexes such as fibrillarin, nucleolin, protein B23, and snoRNAs. Therefore, the activity of the rDNAs and the formation of a functional nucleolar domain in telophase depend on events occurring in the preceding interphase. The question is now to understand the mechanism of de novo assembly of nucleoli in nuclei originating from parental cells in which no previous nucleolar activity existed.
During early embryogenesis of X. laevis, the timing of transcriptional activation provides an interesting situation (reviewed by Davidson, 1986) that we decided to exploit in this study. After fertilization, a series of rapid cleavages are observed and the zygotic genome is transcriptionally quiescent. Schematically, around the 12th division, at a time called the midblastula transition (MBT; Newport and Kirschner, 1982), the cell cycle lengthens and a sequential activation first of transcription of class II genes, followed by class III genes and only several cycles later class I genes is observed. However, the exact timing of these events as estimated by measuring accumulation of specific RNAs collected from a population of embryos during various time windows seemed to depend on the sensitivity of the method (Brown and Littna, 1966; Nakahashi and Yamana, 1976; Shiokawa et al., 1994). Moreover during each window of accumulation, it is impossible to assess precisely when the process started and if this activation operates by a progressive recruitment of an increasing number of nuclei that become transcriptionally competent (as proposed by Shiokawa et al., 1994) or if they increase their individual rate of transcription in a coordinated manner. These parameters are absolutely critical to correlate a structural organization potentially associated with a transcriptional event within an individual nucleus at a precise time during development.
Thus, we decided to develop an approach that would enable us to examine within the same nuclei, the transcriptional state and the organization of defined nucleolar domains at distinct time periods. This could be achieved by run-on in situ transcription assays paralleled with the detection of specific markers of the nucleolus. We found that the transcription factor UBF was associated to the zygotic rDNA in two loci before any detectable RNA pol I activity. The processing protein, fibrillarin, was dispersed in several PNB structures in the nuclei of the rapid cleavage stage embryo and later regrouped around UBF and rDNAs, forming a network of DFC as determined by electron microscopy analysis. Surprisingly, an important amount of pre-ribosomal RNAs (pre-rRNAs) was detected with this fibrillar network whereas rRNA transcription was not yet activated to a detectable level. Thus, the structurally defined nucleolus was inactive for transcription but contained pre-rRNAs from a maternal pool that was maintained during early development of X. laevis. Therefore, we propose that during the de novo nucleolar building, the presence of pre-rRNAs of maternal origin rather than the onset of rRNA transcription is critical to structurally organize the nucleolar domain.
Materials and Methods
Biological Materials
Eggs were obtained from female X. laevis as previously described (Wu and Gerhart, 1991). Embryos were produced by in vitro fertilization (Almouzni and Wolffe, 1995) and allowed to develop at 23°C in 0.1× modified Barth solution (Gurdon and Wickens, 1983) for different times after fertilization. In brief, at this temperature, early blastula could be collected 6 h after fertilization (stage 8), midblastula 7 h after fertilization (stage 8.5), late blastula 9 h (stage 9), early gastrula 10 h after fertilization (stage 10), gastrula 11 h after fertilization (stage 10.5), late gastrula 13 h after fertilization (stage 12), and neurula 18 h after fertilization (stage 15; as specified by Nieuwkoop and Faber, 1994). Nuclei were then isolated under conditions preserving functional properties such as specific import of proteins, DNA replication and transcription (Taddei, A., and G. Almouzni, personal communication). They were processed immediately after preparation for cytological studies. Xenopus A6 cells were grown as described (Smith and Tata, 1991).
Primary Antibodies and Probes
Two characterized human sera from patients suffering from scleroderma autoimmune disease were used: an anti-UBF (Roussel et al., 1993) and an anti-fibrillarin (Gautier et al., 1994) autoantibodies. Mouse monoclonal autoantibody 72B9 (Reimer et al., 1987) and rabbit polyclonal antibodies raised against Xenopus fibrillarin (a kind gift of M. Caizergues-Ferrer, Laboratorie de Biologie Moléculaire Eucaryote, Toulouse, France) were also used. Nucleolin labeling was performed using rabbit polyclonal serum directed against human nucleolin (a kind gift of C. Faucher, Toulouse, France).
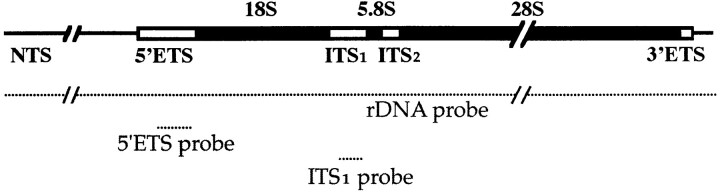
Probes for rDNA and rRNA detection correspond either to the entire Xenopus ribosomal transcription unit inserted into pBr322 (plasmid pXcr7 kindly provided by F. Amaldi, Universita di Roma Tor Vergata, Italy), or to parts of the unit. A probe designated 5′ETS (external transcribed spacer) was produced by digestion of pXcr7 with NotI and SauI at sites +176 and +632, respectively. A probe that recognizes a part of ITS1 (internal transcribed spacer) was also produced by digestion of pXcr7 with KpnI and MluI at sites +2764 and +3077, respectively. All the probes were labeled with the nick-translation kit (GIBCO BRL, Gaithersburg, MD) according to manufacturer's instructions using biotin-14-dCTP or α-[32P]dCTP.
Immunofluorescence
Isolated nuclei were fixed in paraformaldehyde in PBS and centrifuged onto a coverslip. A6 cells were cultured on coverslips and fixed. After washing, the different coverslips were postfixed in methanol, permeabilized in 0.1% Triton X-100 (IBI, New Haven, CT) in PBS and rinsed. The coverslips were then treated with 5% BSA in PBS before incubation with primary antibodies. They were then incubated with fluorescein or Texas red isothiocyanate (FITC or TRITC) conjugated secondary antibodies (anti–human, anti–mouse, or anti–rabbit IgG; Jackson ImmunoResearch Laboratories, West Grove, PA), rinsed, counterstained with DAPI (4′-6-diamidino-2-phenylindole dihydrochloride; Polysciences, Inc.,Warrington, PA) and mounted with an antifading solution (Citifluor, London, UK).
Combined Immunolocalization and In Situ Hybridization of rDNA or rRNA
After fibrillarin immunolabeling, hybridization of rDNA was performed as previously described (Junéra et al., 1995). rRNA hybridization was also performed using this protocol with the following modifications. RNase treatment was omitted and samples were not denatured before hybridization. The rRNA hybridization mixture contained 40% formamide (GIBCO BRL), 10% (wt/vol) dextran sulfate (Sigma Chemical Co., St. Louis, MO), 50 ng/μl sonicated salmon testes DNA (Sigma Chemical Co.) and the biotinylated rDNA probes diluted to a final concentration of 2 ng/ μl in 2× SSC (1× salt sodium citrate: 0.15 M NaCl, 0.015 M trisodium citrate, pH 7). As a control for the detection of RNA, hybridization was preceded by RNase digestion as described previously (Highett et al., 1993).
Assay of RNA Polymerase Activity In Situ
Cultured A6 cells or embryonic nuclei were used. Alternatively, A6 nuclei were prepared (Marzluff and Huang, 1980). All nuclei were centrifuged on coverslips. Run-on transcription was performed as previously described by incorporation of Br-UTP (Sigma Chemical Co.; Masson et al., 1996). 100 μg/ml α-amanitin, 0.2 μg/ml actinomycin D to inhibit transcription or 5 μg/ml aphidicolin to inhibit DNA synthesis were added to the run-on buffer. Before immunolabeling, cells and nuclei were fixed with paraformaldehyde in PBS and permeabilized with Triton X-100 (0.1% in PBS). A monoclonal anti-Br-deoxyuridine antibody that also recognizes Br-UTP (Boehringer Mannheim, Mannheim, Germany) was used. Human anti-fibrillarin antibody was used as a marker of a structurally defined nucleolus. Run-on and fibrillarin signals were obtained using respectively FITC-conjugated goat anti–mouse and TRITC-conjugated goat anti–human antibodies (Jackson ImmunoResearch Laboratories).
Optical Microscopy
Images were taken with a Leica epifluorescence microscope equipped with a thermoelectronically cooled charge-coupled device (CCD) camera (Leica, Lasertech, Germany). Gray scale images were collected separately with filter sets for FITC and rhodamine/TRITC using an oil immersion lens (63×, NA I.4 plan Apochromat). A Leica confocal laser scanning microscope (imaging system TCS4D) was also used. For fluorescein and Texas red excitation an argon Krypton laser operating respectively with the 488 nm and 568 lines was used. Gray scale images were pseudo colored and merged using the Adobe Photoshop 4.0 software. Quantification of the data was performed using the NIH image 1.56 software.
Electron Microscopy
Isolated nuclei of Xenopus embryos were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, at 4°C. They were washed in cacodylate buffer, post-fixed in 1% OsO4 for 1 h at 4°C, stained with 0.05% uranyl acetate en bloc, dehydrated in graded alcohol and embedded in Epon 812. Ultrathin sections were contrasted with uranyl acetate and lead citrate, and examined in a Philips EM412 electron microscope.
For immunolocalization, isolated nuclei were fixed in paraformaldehyde in PBS, washed in Sorensen buffer (sodium phosphate buffer, pH 7.4) for 30 min, dehydrated in graded alcohol and embedded in LRWhite (Polyscience, Niles, IL). Ultrathin sections were picked up on 200 mesh nickel grids and indirect immunolabeling was performed. After incubation with PBS, 0.5 M glycine, sections were blocked with PBS, 0.5% fish gelatin, and 0.1% Tween, incubated with the polyclonal serum against Xenopus fibrillarin and then with 10 nm gold conjugated anti-rabbit antibody (AuroProbe; Pharmacia Biotech, Uppsala, Sweden) or 5 nm protein A–gold. Ultrathin sections were contrasted with uranyl acetate. In control experiment, the primary antibody was omitted.
Protein Analysis by Immunoblotting
Total protein extract from 12 eggs or embryos were prepared by homogenization in 100 μl of modified RIPA buffer (20 mM Tris-HCl, pH 8, 1 mM EDTA, 150 mM NaCl, 1% deoxycholate, 0.1% SDS, 1% NP-40, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml aprotinin, and 1 mM PMSF). The samples were sonicated and centrifuged at 6,000 g for 10 min. Proteins in the yolk-free supernatants were adjusted in sample buffer (Laemmli, 1970). A6 cells were lysed directly and sonicated in this buffer. For nuclear extracts, nuclei were collected by centrifugation of nuclear suspensions before solubilizing the proteins. Western blots were performed as previously described (Roussel et al., 1993).
Northern Blot Analysis
RNAs were purified from eggs or embryos at various times after fertilization using the kit RNA now (Biogentex, Seabrook, TX), followed by a LiCl precipitation (Almouzni and Wolffe, 1995). RNAs were denatured in 50% formamide, 1.84 M formaldehyde, 10 mM sodium phosphate buffer, pH 6.5, and fractionated on 0.8% agarose gel containing formaldehyde. RNAs were transferred onto a nitrocellulose filter (BA-S85; Biorad, Hercules, CA). After baking and cross-linking, the filter was prehybridized for 5 h at 42°C in buffer containing 50% formamide, 5× SSPE (Maniatis et al., 1982), 10× Denhardt's solution (Maniatis et al., 1982), 0.1% SDS, and 50 μg/ml salmon sperm DNA. It was hybridized with the appropriate 32P-labeled probe in the same buffer for 20 h at 42°C. After hybridization, the filter was washed twice for 10 min each in 2× SSC, 0.1% SDS at 65°C, and finally once for 5 min in 0.1× SSC, and 0.1% SDS at 65°C. Autoradiography was performed with a PhosphoImager (Molecular Dynamics, Inc., Sunnyvale, CA). The size of the RNAs was determined by comparison to an RNA ladder (GIBCO BRL).
Results
The Maintenance and Subcellular Localization of the Maternal Pool of Nucleolar Proteins during Early Development of X. laevis
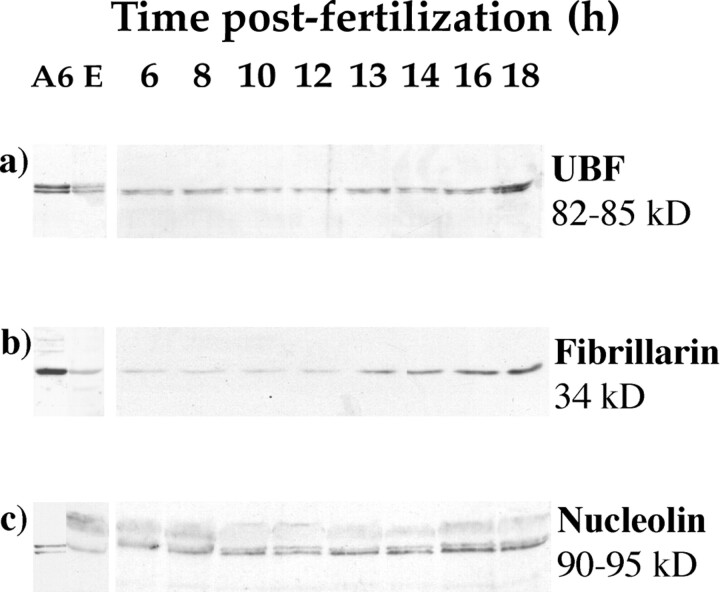
The presence of three nucleolar proteins (UBF, fibrillarin, and nucleolin) was investigated at different stages of development of X. laevis (Fig. 1). Western blot analysis using specific antibodies and performed with A6 cell lysates or whole protein extracts from eggs or embryos at different times after fertilization revealed two Xenopus UBF forms of 82 and 85 kD (Guimond and Moss, 1992; Fig. 1 a), fibrillarin of 34 kD (Lapeyre et al., 1990; Fig. 1 b) and the two forms of Xenopus nucleolin of 90 and 95 kD (Caizergues-Ferrer et al., 1989; Messmer and Dreyer, 1993; Fig. 1 c). Whereas the equivalent of 105 A6 cells was needed to detect each nucleolar protein, the material extracted from the equivalent of 1.5 eggs was sufficient to obtain a signal (Fig. 1, compare lanes A6 and E). This is consistent with the excess amount of RNA pol I activity found in an egg when compared with a somatic cell (Gurdon and Wickens, 1983). The relative amount of the three antigens was determined in embryos at later stages of development. No detectable change was noticed until 12 h after fertilization consistent with the maintenance of the maternal pool throughout early development. Increasing amounts of the three proteins were measured at later stages, possibly reflecting new synthesis. Accumulation of fibrillarin was already observed 13 h after fertilization while UBF and nucleolin increased only slightly. At 18 h after fertilization, the three nucleolar proteins were already present in large quantities.
Figure 1.
The maternal pool of nucleolar proteins is conserved until zygotic activation. Total protein extracts from A6 cells (A6), eggs (E), or embryos at different times after fertilization were prepared. Proteins extracted from 105 A6 cells or 1.5 eggs or embryos were loaded onto each lane of an SDS-8% polyacrylamide gel. The gel was blotted onto nitrocellulose and probed with anti-human UBF (a), anti-human fibrillarin (b), and anti-human nucleolin (c) sera. Detection of the complexes was achieved using an appropriated second antibody coupled to peroxidase and revealed by ECL.
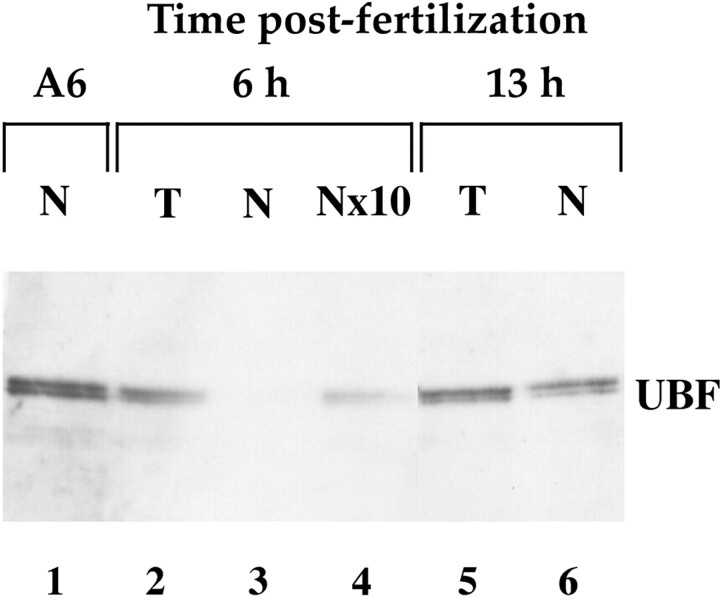
The subcellular distribution of these proteins was examined. Nuclei were isolated and both nuclear and total extracts were compared (Fig. 2) from embryos 6 h after fertilization (inactive RNA pol I transcription) and 13 h after fertilization (active RNA pol I transcription; Brown and Littna, 1964; Brown and Littna, 1966; Shiokawa et al., 1989). The results are presented for UBF; however similar results were obtained for the pool of fibrillarin and nucleolin (data not shown). Using embryos 6 h after fertilization, a strong signal was detected with the total protein extract prepared from 1.5 embryos (Fig. 2, lane 2). The extract corresponding to the equivalent number of nuclei (1,500 nuclei according to Nieuwkoop and Faber, 1994; Fig. 2, lane 3) was not sufficient to detect a UBF signal: up to an equivalent of 15,000 nuclei was required to detect a weak signal (Fig. 2, lane 4). These data are consistent with nuclei containing only a limited fraction of the UBF pool at this stage, the majority being in the cytoplasm. Using embryos 13 h after fertilization, the difference of signal intensity between total and nuclear extracts was greatly reduced as compared with 6 h after fertilization (Fig. 2, lanes 5 and 6). This is in agreement with previous results (Messmer and Dreyer, 1993) reporting nuclear translocation of nucleolin starting at early blastula. Our results indicate that the maternal pool of these nucleolar proteins is recruited late in nuclei formed during early development.
Figure 2.
Subcellular distribution of the maternal pool of UBF during development. Total (T) or nuclear (N) protein extracts were prepared and analyzed by SDS-8% polyacrylamide gel, blotted onto nitrocellulose and probed with anti-human UBF serum. For A6 cells, an equivalent to 66,000 nuclei was loaded (A6, N, lane 1). Lanes 2 and 3 contain respectively total and nuclear proteins from 1.5 embryos 6 h after fertilization. Lane 4 contains nuclear proteins from 15 embryos 6 h after fertilization (an equivalent to 15,000 nuclei since one embryo contains only 1,000 nuclei at this stage). Lanes 5 and 6 contain, respectively, total and nuclear proteins from 1.5 embryos 13 h after fertilization (one embryo contains ∼15,000 nuclei at this stage).
Subnuclear Distribution of the Nucleolar Proteins, UBF, Fibrillarin, and Nucleolin
The subnuclear localization of these proteins was followed by immunofluorescence on embryonic nuclei isolated at different stages of development. To ensure that the data collected were statistically significant, labeling of each nucleolar protein was determined on more than 1,500 nuclei at a specific stage, prepared from at least four distinct batches of fertilization.
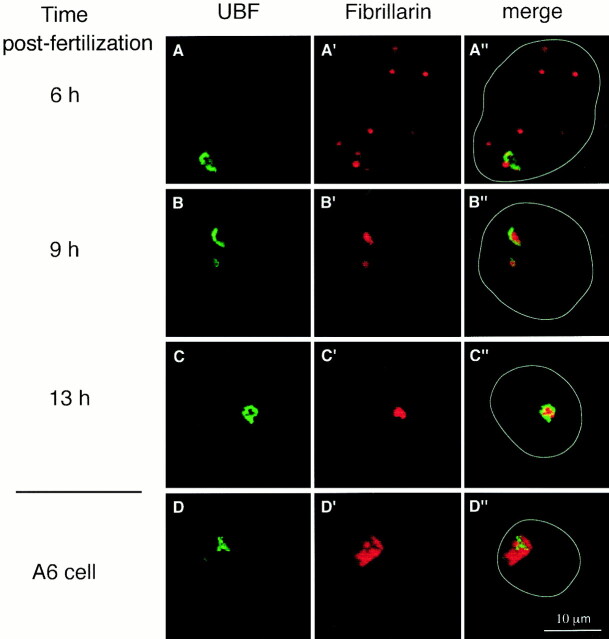
UBF labeling did not vary significantly at the different times of development examined (Fig. 3, A–C) or at later stages (not shown). It was always distributed in several beads aligned as a folded filament and confined to two sites in the nuclei, one of them being more predominant than the other. A single confocal optical section corresponding to the predominant site is shown for each stage (Fig. 3, A–C). Interestingly, the distribution of UBF at each embryonic stage was similar to that of somatic A6 cells (Fig. 3 D) known for their high nucleolar activity (Masson et al., 1996). In somatic cells, it was previously reported that the distribution of UBF as an alignment of small beads corresponded to actively transcribing genes while UBF clustered in large spots reflected rather absence of transcription (Jordan et al., 1996). Thus, it was surprising to find that during development, the subnuclear distribution of UBF was similar at all stages (Fig. 3, A–C) whether or not rRNA synthesis was active. The precise determination of active and inactive stages is presented below.
Figure 3.
Localization by confocal microscopy of UBF and fibrillarin in nuclei from embryos at 6 h (A–A″), 9 h (B–B″), and 13 h (C–C″) after fertilization and in A6 cell (D–D″). Alignments of small beads are seen in green (A–D) using anti-human UBF serum. Fibrillarin labeling with anti-mouse fibrillarin serum in red appears on dot-like structures scattered throughout the nuclei at 6 h after fertilization (A′). Regroupment of fibrillarin is observed around UBF 9 h (B′) and 13 h (C′) after fertilization and in A6 cells (D′). A″–D″ are merged images of both signals on one optical section. The white line indicates the nuclear contour (A″–D″).
Fibrillarin as revealed by immunostaining showed a dot-like distribution in embryonic nuclei isolated 6 h after fertilization (Figs. 3 A′ and 4 A′). Labeling was associated with dense particles of different sizes visible in phase contrast and dispersed throughout the nucleoplasm. At later stages, the fibrillarin redistributed in particles clustered in two sites within the nucleus (Figs. 3 B′ and 4 B′). This clustering was first observed in 3% of the nuclei in embryos 7 h after fertilization, increased rapidly to reach 75% of the population of nuclei isolated 9 h after fertilization to finally become generalized to 90% of embryonic nuclei 12 h after fertilization (see below, Fig. 12).
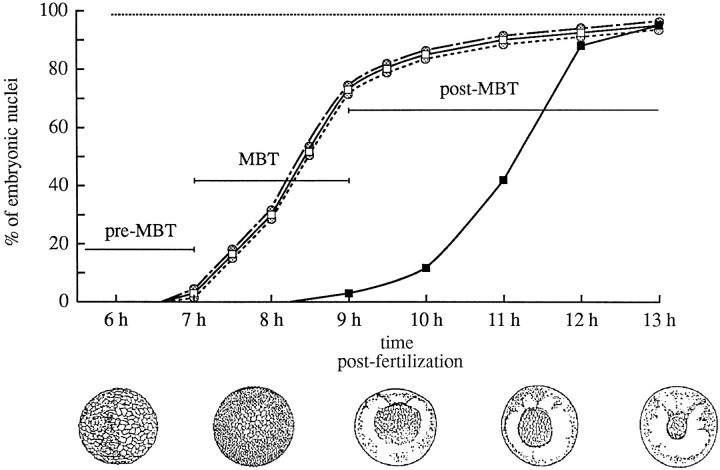
Figure 12.
Nuclear events described in this work. Percentage of embryonic nuclei are expressed as a function of time after fertilization. Schematic representations of the embryos at the different times are presented below the graph. The first event described was UBF association with rDNAs in nearly all embryonic nuclei at every time after fertilization ( ). Regroupment of fibrillarin and nucleolin to the nucleolar domain (
). Regroupment of fibrillarin and nucleolin to the nucleolar domain ( ) as well as maternal pre-rRNAs (
) as well as maternal pre-rRNAs ( ) were observed in a growing fraction of embryonic nuclei between 7 and 13 h after fertilization. The curve corresponding to RNA pol II or pol III activities detected by run-on assays in nuclei (
) were observed in a growing fraction of embryonic nuclei between 7 and 13 h after fertilization. The curve corresponding to RNA pol II or pol III activities detected by run-on assays in nuclei (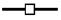 ) was identical to the two latter curves since transcription activities were detected in nuclei in that nucleolar proteins and maternal pre-rRNAs were regrouped. However, onset of RNA pol I activity was not concomitant with this regroupment. The curve expressing the percentage of nuclei with RNA pol I activity (
) was identical to the two latter curves since transcription activities were detected in nuclei in that nucleolar proteins and maternal pre-rRNAs were regrouped. However, onset of RNA pol I activity was not concomitant with this regroupment. The curve expressing the percentage of nuclei with RNA pol I activity ( ) shows a delay of ∼2.5 h. For simplification, the period that comprises the recruitment of nuclei competent for RNA pol II transcription will be referred to as MBT and the periods before or after as pre-MBT or post-MBT, respectively.
) shows a delay of ∼2.5 h. For simplification, the period that comprises the recruitment of nuclei competent for RNA pol II transcription will be referred to as MBT and the periods before or after as pre-MBT or post-MBT, respectively.
Fibrillarin clustered on the same nuclear sites as UBF, and localized in dense structures in phase contrast (Fig. 4 B″). Merging fibrillarin and UBF signals from the same optical section (Fig. 3, B″–D″) showed their tight intrication.
Figure 4.
Localization of nucleolin (Nuc) and fibrillarin (Fib) in nuclei from embryos 6 h (A–A″) and 9 h (B–B″) after fertilization. While dot-like structures are seen using anti-human fibrillarin serum at 6 h after fertilization (A′), nucleolin labeling with anti- human nucleolin serum is diffuse in the nucleoplasm (A). Both proteins are colocalized 9 h after fertilization (B and B′). (A″ and B″) are phase contrast microscopy.
The distribution of nucleolin, a major nucleolar protein that associates with rRNAs (Serin et al., 1996), was examined. Nucleolin was mainly diffusely distributed throughout the nucleoplasm in embryonic nuclei 6 h after fertilization and was also weakly present in foci corresponding to fibrillarin dots (Fig. 4 A). Later, while fibrillarin started to cluster, colocalization of both nucleolin and fibrillarin was observed. Fibrillarin and nucleolin were colocalized on two large structures that appeared dense in phase contrast when the regroupment was complete (Fig. 4, B–B″).
Fibrillarin Regrouped Near rDNA Early during X. laevis Embryogenesis
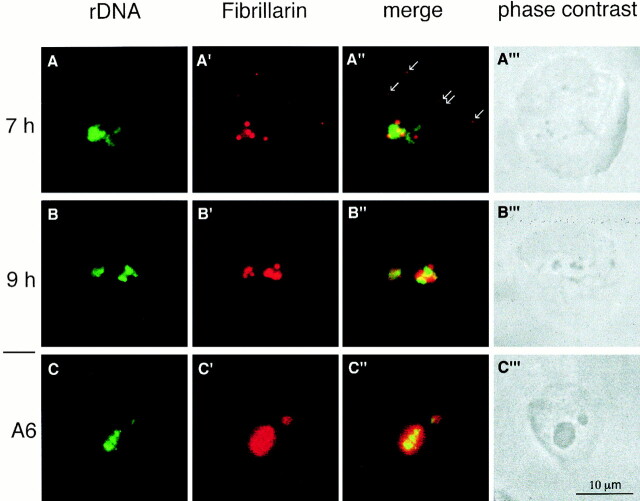
To determine whether fibrillarin regroupment occurred near ribosomal genes, rDNA was revealed by in situ hybridization and fibrillarin was detected on the same nuclei. Whereas, the dot-like pattern distribution of fibrillarin did not show any preferential localization compared with rDNA sites at 6 h after fertilization (not shown), a small fraction of dots began to distribute around the rDNA affecting 30% of the nuclei at 7 h after fertilization (Fig. 5 A″). Partial regroupment became complete in an increasing number of nuclei between 7 and 9 h after fertilization. At 9 h after fertilization, fibrillarin was entirely regrouped around the rDNA and no isolated dot could be seen; the two labelings were always close to one another (Fig. 5 B′). Several foci of fibrillarin were generally observed in the same site and corresponded to the dense structures observed by phase contrast microscopy (Fig. 5, A‴ and B‴). The space around and between the dense structures was occupied by rDNAs. In A6 cells (Fig. 5, C–C‴), one of the two rDNA sites was preferentially amplified and occupied a central position in the large nucleolus. Fibrillarin was present around each rDNA site but accumulated preferentially at the periphery of the larger one.
Figure 5.
Fibrillarin becomes clustered near the rDNAs during development. Embryonic nuclei and A6 cells were labeled with anti-fibrillarin human serum (red, A′–C′) before performing rDNA hybridization in situ with an rDNA biotinylated probe (green, A–C). Images were directly numerized from a camera on a fluorescence microscope and then both labelings were merged (A″–C″). At 7 h after fertilization, part of the fibrillarin in red was clustered close to rDNAs in green (A″) but isolated dots of fibrillarin were seen in the nucleoplasm (arrows, compare with Fig. 4 for earlier time). At 9 h after fertilization, all the fibrillarin dots were regrouped around rDNA (B″). In the A6 cell (C–C″), most rDNAs (C) were totally embedded in the largest nucleolar structure labeled by anti-fibrillarin serum (C′). Sites of fibrillarin accumulation are dense in phase contrast (A‴–C‴).
Fine Structure of the Nucleolar Domain and Nucleolar Components during X. laevis Embryogenesis
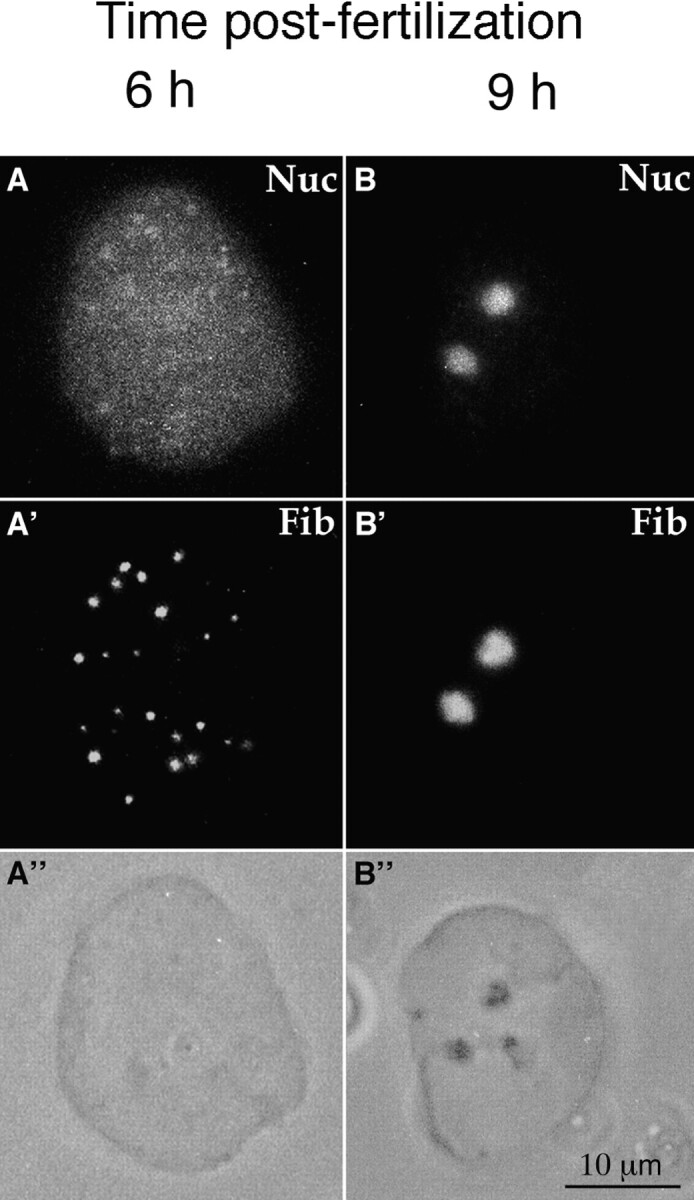
The structural organization of nuclei was also analyzed at high resolution by electron microscopy. At 6 h after fertilization, highly decondensed chromatin and round-shaped dense fibrillar structures could be observed (Fig. 6, A and B). The aspect of these dense structures was reminiscent of the PNBs previously described (Ochs et al., 1985a ; Scheer et al., 1993; Zatsepina et al., 1997) confirming previous report (Hay and Gurdon, 1967). The presence of fibrillarin in these structures (Fig. 6 C) further supports their identification as PNBs. Discrete structures in which nucleolar proteins accumulated could be observed before any typical nucleolar assembly. In nuclei from embryos 8 and 9 h after fertilization, nucleolar domains could be identified around which chromatin was not as loosely compacted as in other parts of the nuclei (Fig. 6 D). However, it was surprising to find that in these nucleolar domains, DFC formed a network (Fig. 6 D). Since DFC in a network is generally associated with transcribing nucleoli, this observation was unexpected. Immunolabeling of fibrillarin (Fig. 6 E), confirmed that this DFC was generated, at least in part, by regroupment of PNB-containing fibrillarin around the rDNAs as described above (Figs. 3 and 5). In nuclei 13 h after fertilization, the nucleoli increased in size and complexity, and were organized in DFC and in GC as seen in fully active nucleoli (Fig. 6, F and G). Since, DFC formed a network already 8 h after fertilization, it was therefore important to examine the transcriptional activity of these embryonic nuclei.
Figure 6.
Ultrastructural analysis of nuclei at different times during X. laevis development: 6 h (A–C), 8 h (D and E) and 13 h after fertilization (F and G). At 6 h after fertilization, the nuclei exhibited decondensed chromatin and numerous nuclear pore complexes (A). Round-shaped dense fibrillar structures similar to PNB were observed (A and B). When sections were incubated with anti-fibrillarin antibodies, these dense fibrillar structures were labeled by protein A gold (C). At 8 h after fertilization, the nucleolar domain was characterized by dense fibrillar component forming a network (D) and containing fibrillarin (gold particles in E). At 13 h after fertilization (F), the nucleolus was formed by dense fibrillar component (DFC) surrounded by granular component (GC).The star indicates a structure similar to fibrillar center. Fibrillarin was localized on DFC whereas no label was detected over the granular component (G). Bar: 0.2 μm.
Transcriptional Activity of the rDNA at the Time of Fibrillarin Regroupment
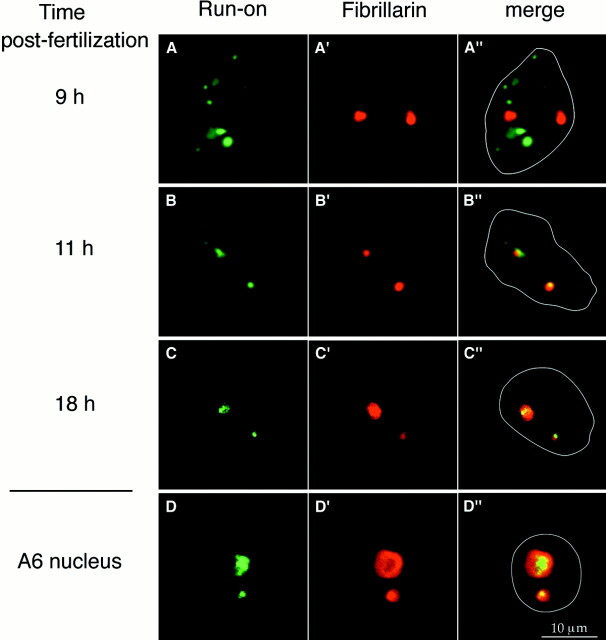
We analyzed transcription by in situ run-on assays and localized the nucleolar domain on the very same nuclei by fibrillarin labeling. For each time after fertilization, 200 embryos were collected for nuclear isolation and at least 600 nuclei were analyzed. These experiments were performed with three distinct batches of fertilization. On A6 cells, the run-on conditions used had permitted the selective detection of RNA pol I activity concomitantly with RNA pol II activity in a proportion that was representative of what was expected on the basis of biochemical estimations (Masson et al., 1996). RNA pol III activity was believed to be very low in cultured cells under these conditions (Marzluff and Huang, 1980; Wansink et al., 1993). The same conditions were used with embryonic nuclei and with nuclei isolated from A6 cells. A specific class of transcripts was further characterized using inhibitors. RNA pol I activity was abolished by 0.2 μg/ml of actinomycin D and was not affected by 100 μg/ml of α-amanitin. Interestingly, only RNA pol I activity was detected in nuclei isolated from A6 cells (Fig. 7 D). The activities detected in embryonic nuclei depended on the time after fertilization. No signal was detected in nuclei 6 h after fertilization (data not shown). Importantly, a signal was first detected only in very few nuclei (no more than 3% of the nuclei) isolated 7 h after fertilization. These nuclei displayed PNBs that had already regrouped as judged by fibrillarin labeling. The fraction of nuclei with transcriptional activity increased between 7 and 9 h after fertilization correlating with the fraction of nuclei in which the nucleolar proteins had regrouped completely. Transcription in these nuclei was scattered throughout the nucleoplasm in foci of different intensity, a distribution compatible with RNA pol II transcripts (Fig. 7 A). None of the transcripts colocalized with fibrillarin (Fig. 7 A″) and the use of inhibitors confirmed that they were not RNA pol I transcripts. RNA pol I activity colocalizing with fibrillarin was first seen only in 2% of the nuclei isolated from embryos 9 h after fertilization. In these nuclei, RNA pol I activity was the major transcription signal, whereas low levels of other transcription activities were detected outside the nucleoli. Later, RNA pol I activity was seen in 10% of the nuclei 10 h after fertilization, to reach 40% of the nuclei 11 h after fertilization (Fig. 7, B–B″), and in most nuclei 12 h after fertilization. RNA pol I activity was visible in all nuclei 18 h after fertilization: it was often higher in one of the two nucleolar structures than in the other (Fig. 7, C–C″). This difference was amplified in A6 nuclei (Fig. 7, D–D″). Thus, it appears that regroupment of nucleolar proteins occurred in a large fraction of embryonic nuclei before a significant fraction of the nuclei had detectable RNA pol I activity as illustrated by the graph summarizing the data (Fig. 12). A correlation can be established between regroupment of nucleolar proteins and the onset of RNA pol II and pol III activities. However, the regroupment of these proteins around rDNA was not associated with the onset of rDNA transcription.
Figure 7.
rDNA transcription appears after regroupment of fibrillarin. In situ run-on incorporation of Br-UTP (green) in isolated embryonic nuclei (A–C) or A6 nuclei (D) was coupled with fibrillarin labeling (red) using the anti-fibrillarin human serum (A′–D′). Images were directly numerized from a camera on a fluorescence microscope and both labelings combined (A″–D″). Superposition showed that nucleolar transcription was not yet detected 9 h after fertilization (A″) while rDNA transcription was the major transcription observed on nuclei at 11 h (B″) and 18 h (C″) after fertilization, and in A6 nuclei (D″). The white line indicates the nuclear contour (A″–D″).
The Presence and Localization of Pre-rRNAs before the Activation of RNA Pol I–dependent Transcription
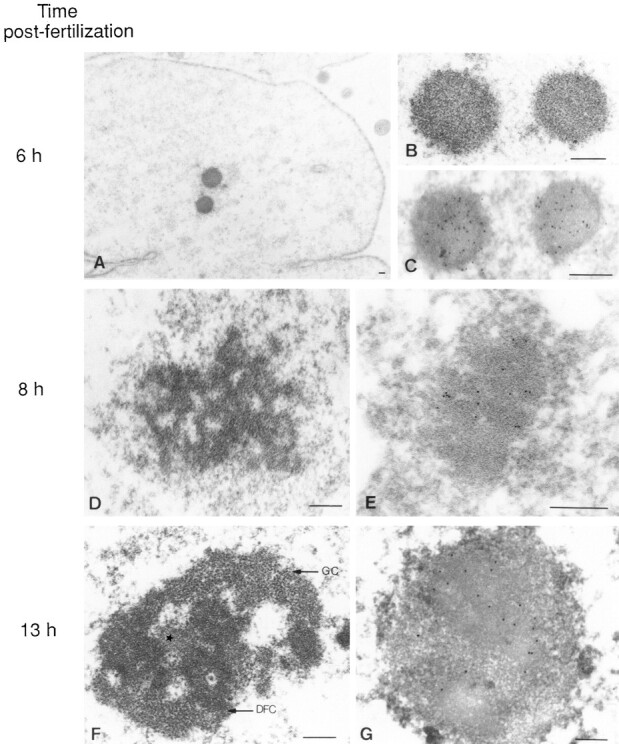
The nucleolar domain observed after nucleolar protein regroupment exhibited a structural organization compatible with nucleolar activity, whereas no RNA pol I transcription was detected in most nuclei at that time. As the role of rRNAs seemed crucial for nucleolar organization (Weisenberger et al., 1993), we looked for the presence of rRNAs in these structures. Thus, in situ hybridization was performed using the rDNA probe (Fig. 8) on nondenatured embryonic nuclei at early stages (Figs. 9 and 10) and at later stages (not illustrated). rRNAs were first detected in nuclei 7 h after fertilization that had initiated the regroupment of fibrillarin (Fig. 9 a, A–A″). The signal was associated with these sites of regroupment but not with the multiple fibrillarin foci that were still dispersed. It was abolished by RNase treatment arguing in favor of an RNA-derived hybridization signal (Fig. 9 a, B–B″). At 9 h after fertilization, relatively high rRNA signals were detected in the nuclei where the nucleolar domains were formed (Fig. 9 b, A–D, 2) while no signal was detected in nuclei where fibrillarin was still dispersed (Fig. 9 b, A–D, 1).
Figure 8.
Representation of the specific probes corresponding to the X. laevis ribosomal gene (rDNA). Map of the X. laevis 40 S pre-rRNA transcription region (boxed region; rRNA coding region are black and transcribed spacer regions are white) and surrounding rDNA regions designed non transcribed spacer (NTS, black bar). The dotted line indicates the position of the hybridization probes. The rDNA probe is complementary to the entire rDNA, and the two 5′ETS and ITS1 probes are complementary to a part of the transcribed spacer regions. The 5′ETS probe extends between positions +176 and +632 relative to the transcription start site. The ITS1 probe extends between positions +2764 and +3077.
Figure 9.
(a) Presence of rRNAs at the time of fibrillarin regroupment. Embryonic nuclei isolated 7 h after fertilization were labeled with anti-fibrillarin serum (red) before performing rRNA FISH with the rDNA probe (green). All the bright and large foci stained by the anti-fibrillarin (A′) contained rRNAs (A). The rRNA signal was abolished by RNase treatment (B). (b) Presence of rRNAs in nucleolar domain 9 h after fertilization. rRNAs (green) were present on the sites of regroupment of fibrillarin (red; A and B, 2). However, no rRNA was detected when fibrillarin was still dispersed (A and B, 1). C is merge image of rRNA and fibrillarin counterstained by DAPI for DNA. D corresponds to phase contrast microscopy.
Figure 10.
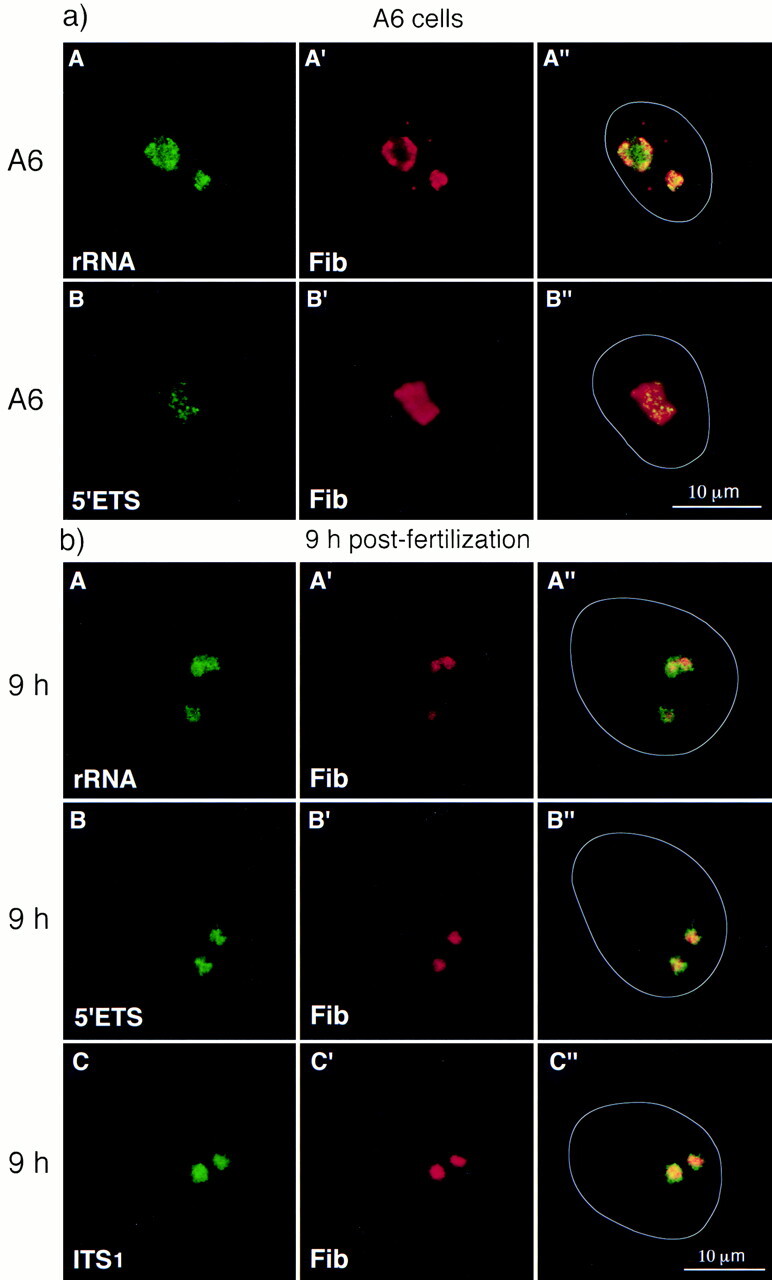
(a) Distribution of rRNAs in cycling cells. A6 cells were labeled with anti-fibrillarin serum (red) before performing rRNA FISH (green) with the rDNA probe (A) or the 5′ETS probe (B). Pre-rRNAs hybridized with the 5′ETS probe occupy a limited region of the functional nucleolus (B–B″) whereas the rRNAs hybridized with the rDNA probe distributed all over the nucleolar domain (A–A″). The white line indicates the nuclear contour (A″–B″). (b) Unprocessed rRNAs are associated with the formation of structurally defined nucleoli. At 9 h after fertilization (A–A″), rRNAs (green) were colocalized with fibrillarin (red). Some rRNAs are present in a region that surrounds the fibrillarin-labeled area (A″). The same localization was obtained using the 5′ETS (B–B″) and ITS1 probes (C–C″) indicating that the rRNAs detected 9 h after fertilization are incompletely processed rRNAs. The white line indicates the nuclear contour (A″–C″).
To determine whether these rRNAs were processed, probes that hybridize specifically with the processed sequences (5′ETS and ITS1 probes) were used (Fig. 8). Strong signals were obtained on A6 cells with the 5′ETS (Fig. 10 a, B–B″) and ITS1 probes (not shown) but the entire nucleolar domain was not stained in contrast to what was observed with the rDNA probe (Fig. 10 a, A–A″) arguing in favor of the specificity of these two probes. In embryonic nuclei 7 and 9 h after fertilization, the 5′ETS and ITS1 probes gave the same pattern of hybridization as the entire rDNA probe (compare Fig. 10 b, B–B″ and C–C″ with Fig. 10 b, A–A″), i.e., colocalizing and also surrounding the nucleolar fibrillarin signal. It is noteworthy that the distribution of the 5′ETS and ITS1 probes was homogeneous in embryonic nuclei as opposed to the punctuated distribution in A6 transcribing nucleoli (compare Fig. 10 b, B–B″ and C–C″ with Fig. 10 a, B–B″). Therefore, the homogenous distribution of rRNA over the nucleolar domain found in embryonic nuclei between 7 and 9 h after fertilization can be identified as incompletely processed pre-rRNAs.
Quantification of fluorescence signals obtained after fluorescent in situ hybridization (FISH with rDNA and 5′ETS probes) and run-on assays on A6 cells or embryonic nuclei 9 h after fertilization was performed using the NIH image 1.56 software (Table I). Unprocessed transcripts were specifically labeled by FISH with the use of 5′ETS probe or by incorporation of Br-UTP in a run-on assay. The signal obtained by FISH with rDNA probe that labeled all rRNAs was also quantified. In A6 transcribing cells, unprocessed transcripts represent a fraction of 1/15 of the total rRNA as determined by FISH (ratio 5′ETS/ rRNA). Performing the same quantification with embryonic nuclei 9 h after fertilization gave a ratio 5′ETS/rRNA of 1. Therefore, nearly all the rRNAs detected are unprocessed pre-rRNAs. By run-on assays, the elongating rRNA transcripts gave a signal that was 6.5 times the signal obtained by FISH with the 5′ETS probe. If these pre-rRNAs were newly transcribed in nuclei 9 h after fertilization, the signal obtained by run-on assays should be detected at a mean value of integrated density 6.5 times stronger (expected value 2,167 instead of 0). Since no signal was detected, we conclude that nearly all pre-rRNAs detected by FISH are imported to nucleolar sites.
Table I.
Quantification of Fluorescent Signals
| A6 cells | Embryonic nuclei | |||||
|---|---|---|---|---|---|---|
| FISH | 5′ETS | 75 ± 8 | 335 ± 60 | |||
| FISH | rRNAs | 1,180 ± 300 | 331 ± 60 | |||
| Ratio | 1/15 | 1/1 | ||||
| FISH | 5′ETS | 75 ± 8 | 335 ± 60 | |||
| run-on | elongating transcripts | 483 ± 100 | no nucleolar signal (expected 2,167) | |||
| Ratio | 1/6.5 | — |
FISH is less sensitive than run-on assays to detect unprocessed transcripts. Quantification of fluorescence signals was performed with NIH image 1.56 on acquisitions obtained after FISH with rDNA and 5′ETS probes and run-on assays on A6 cells or embryonic nuclei 9 h after fertilization. For each method, 20 images were chosen at random and the same thresholding was applied. The signal was quantified leading to an integrated density value. The mean of 20 values is given for each case and the ratios between the different methods are established (Ratio).
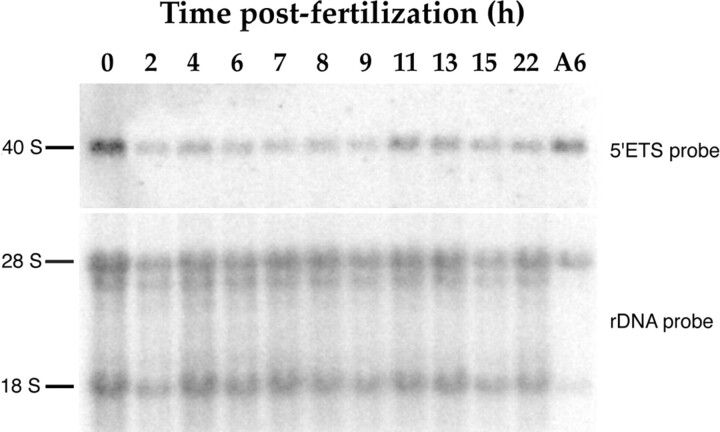
If rRNAs were not produced by activation of zygotic rDNA transcription, they might possibly originate from the maternal pool that was maintained during the early development of X. laevis. Performing Northern blot analysis using the 5′ETS probe, we showed that embryos between 2 and 9 h after fertilization contained a stable level of 40 S pre-rRNAs (Fig. 11), in agreement with previous reports (Busby and Reeder, 1982). At the time of the initial nucleolar building, we demonstrated that this pool of 40 S pre-rRNAs was present. Since transcription was not detected before 11 h after fertilization, the pre-rRNAs found in nuclei between 7 and 9 h after fertilization by in situ method could be of maternal origin.
Figure 11.
A maternal pool of 40 S pre-rRNAs is maintained during early development of X. laevis. A Northern blot was performed using sample of RNAs prepared with embryos at various time after fertilization. Each lane was loaded with RNAs from two embryo equivalents excepted the last one which was loaded with RNAs from 105 A6 cells. The blot was first probed with the 32P-labeled 5′ETS probe that hybridized to 40 S pre-rRNAs. An exposure of 24 h was performed after hybridization to 40 S pre-rRNAs. The same blot was further rehybridized with the 32P-labeled rDNA probe to estimate the RNA loading per lane. A short exposure of 1 h was sufficient to see hybridization to 18 S and 28 S rRNAs. Taking into account this difference, the signal obtained for the 18 S or 28 S rRNAs was 70 times stronger than the signal for the 40 S pre-rRNAs.
Discussion
Following fertilization in X. laevis, the zygotic genome remains silent during the first rapid divisions, and transcription is then progressively established with a delay between RNA pol II, RNA pol III, and RNA pol I transcription (reviewed by Kirschner et al., 1985; Gurdon and Wakefield, 1986; Shiokawa et al., 1994). The delay in the onset of transcription makes it possible to study the de novo assembly of the machineries involved in transcription and processing, independently of the active transcription process itself. In this study, we took advantage of this biological model to investigate the building of transcriptionally competent and structurally defined nucleoli.
Nuclei Establishing Transcriptionally Competent and Structurally Defined Nucleoli Are Progressively Recruited
Global estimations of RNA pol I activity in X. laevis embryogenesis have been examined by several authors. The first ones established that RNA pol I activity was initiated in the early gastrula stage (Brown and Littna, 1964; Brown and Littna, 1966; Nakahashi and Yamana, 1976). Similar timing for RNA pol I transcription activation was observed on injected rDNAs (Busby and Reeder, 1983). Using a more sensitive assay, others detected RNA pol I activity at blastula stage (Shiokawa et al., 1981a ; Shiokawa et al., 1981b ); measurement of the radioactivity incorporated by embryonic cells was performed for periods of 4 h or 2 h. In this type of assay, it is hard to establish precisely when transcription actually begins.
Data concerning the expression and nuclear localization of individual nucleolar proteins during embryonic development are available for X. laevis (Caizergues-Ferrer et al., 1989, 1991; Messmer and Dreyer, 1993) but there is presently no investigation on the building of the nucleolus that considers both the targeting to rDNAs of the nucleolar proteins and the transcriptional activity in the same nuclei. Such events should be analyzed in the same cells because cell divisions become asynchronous after the first 12 divisions in Xenopus embryos. Consequently, in certain cells, some nuclei can be engaged in nucleolar building earlier than others depending on the state of differentiation (Shiokawa and Yamana, 1979; Caizergues-Ferrer et al., 1991). Thus, global evaluation of the events is only indicative and cannot demonstrate precise correlation.
In this study and as summarized in Fig. 12, regroupment of nucleolar proteins towards the rDNA sites was correlated with the transcription activities in the same nuclei. Nuclei are progressively recruited for two main events, (a) regroupment of nucleolar proteins and (b) RNA pol I activation, and importantly these two events are delayed during development. The short labeling time (10 min) for the run-on in situ assay allows precise determination of the timing of events relative to the embryonic development. This assay has also proved to be a sensitive method capable of revealing a few transcripts (Jackson et al., 1993; Wansink et al., 1993). This is illustrated by the fact that in cell lines, the initiation of the RNA pol I activity can be demonstrated by this approach already in anaphase and telophase (Roussel et al., 1996; Gébrane-Younès et al., 1997). In this assay on individual nuclei, the RNA pol I activity was first detected in only a small subset of nuclei around midblastula (Fig. 12) but with a high level of activity. This can explain the low level of activity detected using a global approach (Brown and Littna, 1964; Brown and Littna, 1966; Nakahashi and Yamana, 1976; Shiokawa et al., 1981a , 1989, 1994) at the initial stage of activation. RNA pol I activity was found to take place in an increasing number of nuclei up to late gastrula. For a majority of nuclei, RNA pol I activation is thus a post-MBT event.
Presence of Pre-rRNAs before the Activation of the RNA Pol I
So far, it has been assumed that the structure of the nucleolus is the consequence of rRNA synthesis, thus the nucleolus would be an organelle formed by the act of building a ribosome (Mélèse and Xue, 1995).
In this study, we observed that although no RNA pol I–specific transcription was detected by the run-on in situ assay, unprocessed rRNAs with 5′ETS and ITS1 sequences were associated with the building process during MBT (Fig. 12). Had they arisen from zygotic transcription, a detectable signal would have been expected by the run-on in situ assay which was not the case. The distribution of these pre-RNAs is also in favor of the fact that they were not transcribed in the newly formed nucleolar structures. Indeed, they exhibited a homogeneous distribution instead of the punctuated distribution corresponding to foci of transcription described in transcribing cells (Puvion-Dutilleul et al., 1997) and also observed here in A6 cells. These pre-rRNAs most likely represent a fraction of the large maternal pool of pre-rRNAs we found to be maintained at early stages of development. These pre-rRNAs that were neither processed nor degraded during early development (Busby and Reeder, 1982) would be imported into nuclei in amounts sufficient to be detected by in situ hybridization. Since pre-RNAs are present before transcription, it is tempting to speculate that they might participate in the building of the nucleolar domain.
The organization of the nucleolar proteins assembled at MBT exhibited a DFC-like structure forming a complex network. This organization was surprising since no detectable RNA pol I transcription was found in the majority of the nuclei at this stage. In general, the complex network organization of the DFC, the nucleolar component in which pre-rRNA transcripts accumulate, is associated with active transcription (Scheer et al., 1993; Hernandez-Verdun and Junéra, 1995; Shaw and Jordan, 1995). In mutant X. laevis embryos (o-nu) devoid of rDNA zygotic transcription, compact DFC-like structures were found in pseudonucleoli (Hay and Gurdon, 1967). However, the incomplete genetic characterization of these mutants does not allow one to assess which are the critical components for the complete organization of a network in these pseudonucleoli. In the case of drug-inhibited transcription, DFC did not form a network, but a homogeneous segregated structure (reviewed by Hadjiolov, 1985; Shaw and Jordan, 1995) and (Puvion-Dutilleul et al., 1992; Puvion-Dutilleul et al., 1997).
Our findings showing the presence of pre-RNAs in all nucleolar domains assembled at MBT shed a new light on the current models in which the expression of genes directs an apparent reorganization of nuclear components (reviewed by Singer and Green, 1997). RNAs produced by active transcription could be the structuring element in the organization of nuclear component; this concept could be extended to RNAs previously transcribed. In X. laevis embryogenesis, pre-rRNAs of maternal origin could play a role in nucleolar organization of embryonic nuclei, especially in the three-dimensional organization of the DFC.
Building Functional Nucleoli in Xenopus Embryos
The transcription factor UBF was already detected at genomic rDNA sites in nontranscribing nuclei, i.e., before MBT. Similar results were recently reported (Bell and Scheer, 1997). This indicates that in embryonic nucleoli, the targeting of UBF to the rDNAs occurs very early, before transcription activation.
During the building of functional nucleoli, the de novo assembly of the rRNA processing machinery is an important step. At pre-MBT, the structures (Hay and Gurdon, 1967) containing fibrillarin were similar to the PNBs described in various animal and plant cells at the end of mitosis (Ochs et al., 1985a ; Jiménez-Garcia et al., 1994) and (reviewed by Zatsepina et al., 1997). It has also been reported that fibrillarin is assembled with newly synthesized U3 snoRNAs at pre-MBT (Caizergues-Ferrer et al., 1991). Thus, the assembly of PNB structures at pre-MBT may represent an initial step towards the assembly of the processing machinery.
At MBT, fibrillarin-containing PNBs regrouped around the rDNAs before the initiation of RNA pol I transcription. In somatic cells, it has been proposed that PNB regroupment is linked to rDNA transcription, as supported by the temporal order of nucleolar building at the end of mitosis and the effects of drugs inhibiting rDNA transcription (Benavente et al., 1987; Scheer et al., 1993; Weisenberger et al., 1993). In this work, we show that pre-rRNAs were present at the site of regroupment of the nucleolar proteins, fibrillarin and nucleolin. In X. laevis, it has been reported that nucleolar accumulation of nucleolin required the presence of the RNA binding domains (Messmer and Dreyer, 1993). It is noticeable that nucleolin from human and mouse was found to interact with nucleolin recognition element motifs in the 5′ETS but also in the ITS and in the 18S and 28S RNA sequences, i.e., with the pre-rRNAs (Serin et al., 1996). Therefore, it is tempting to speculate the existence of an interaction between the unprocessed rRNAs and nucleolin. Future work will address how these two events can be linked or regulated. We do not exclude the hypothesis that interaction of nucleolar proteins with rRNAs or with other proteins could play a role in the building of nucleolus.
Significance for the Establishment of a Functional Nucleolus
When comparing different models of nucleologenesis, it appears that in the majority of cases, preassembled building material is required. In this work, we found that in Xenopus embryos, cytoplasmic maternal pools were progressively associated with the newly formed nuclei. This was also substantiated in vitro, since fibrillarin, nucleolin and protein B23 present in Xenopus egg extracts accumulated in the reconstituted nuclei and gathered in PNBs (Bell et al., 1992; Bell and Scheer, 1997). In nuclei of early mammalian embryos up to four blastomeres, several nucleolar precursor bodies containing preassembled nucleolar components of maternal origin existed before the formation of an active nucleolus (Biggiogera et al., 1994; Baran et al., 1995, 1996). Similarly at the end of mitosis in cycling cells, nucleologenesis involved the preassembled RNA pol I transcription machinery from the mother cell (Roussel et al., 1996), and PNBs formed by the recruitment of preexisting nucleolar complexes (Scheer et al., 1993).
Thus, there may be general principles directing the building of a functional nucleolus. It can be proposed that preassembled complexes must be recruited around the nucleolar organizer regions and organized into functional domains that are dependent on rRNAs. In Xenopus embryos, these events were separated by several cell cycles taking place between 7 and 12 h after fertilization. The formation of PNBs occurred in nuclei of cleaving embryos in the absence of transcription, and their subsequent regroupment at MBT still did not depend on transcription. However pre-rRNAs were found in these latter structures. The presence of pre-rRNAs independent of transcription may be a common feature shared in the different types of nucleologenesis. Interestingly, it was reported in plant and animal cells that pre-rRNAs were imported from the mother to daughter cells during mitosis (Fan and Penman, 1971; Abramova and Neyfakh, 1973; Jiménez-Garcia et al., 1994; Medina et al., 1995; Beven et al., 1996), and pre- rRNAs were detected in PNBs. Since activation of transcription and PNB recruitment occurred simultaneously in the somatic cell cycle, it is difficult to attribute a role to these pre-rRNAs in the building of the new nucleolar domain. The delay in transcriptional activation during early embryonic development made it possible to unambiguously demonstrate the presence of these pre-rRNAs at nucleolar sites. Future studies will now address the question of how the nuclear import of these pre-RNAs and their targeting to the rDNAs are controlled.
Acknowledgments
The authors are grateful to D. Roche for preparing the Xenopus eggs and embryos, to A. Taddei for providing conditions to prepare nuclei from Xenopus embryos and sharing data before their publication, to G. Géraud for the confocal microscopy recording and to M. Barre for EM photographic work. We are particularly grateful to A.-L. Haenni and V. Doye for critical reading of the manuscript.
This work was supported in part by grants from the Centre National de la Recherche Scientifique (Programme Biologie Cellulaire no. 96098) and the Association pour la Recherche sur le Cancer (contracts no. 9143 and no. 1030), ATIPE no. 7 from the CNRS to G. Almouzni and the European Contract Biomed Human Genome Research Program (BMH4-CT95-1139) to D. Hernandez-Verdun. C. Verheggen was recipient of a Fellowship from “MRT” and S. Le Panse was recipient of a European contract Biomed-2.
Abbreviations used in this paper
- CCD
charge-coupled device
- DFC
dense fibrillar component
- FC
fibrillar center
- FISH
fluorescent in situ hybridization
- GC
granular component
- MBT
midblastula transition
- PNB
prenucleolar bodies
- RNA pol I
RNA polymerase I
- snoRNA
small nucleolar RNA
- UBF
upstream binding factor
Footnotes
Drs. Almouzni and Hernandez-Verdun contributed equally to this work.
References
- Abramova NB, Neyfakh AA. Migration of newly synthesized RNA during mitosis. III. Nuclear RNA in the cytoplasm of metaphase cells. Exp Cell Res. 1973;77:136–142. doi: 10.1016/0014-4827(73)90562-4. [DOI] [PubMed] [Google Scholar]
- Almouzni G, Wolffe AP. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopusembryogenesis. EMBO (Eur Mol Biol Organ) J. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran V, Flechon JE, Pivko J. Nucleologenesis in the cleaving bovine embryo: immunocytochemical aspects. Mol Reprod Dev. 1996;44:63–70. doi: 10.1002/(SICI)1098-2795(199605)44:1<63::AID-MRD7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Baran V, Vesela J, Rehak P, Koppel J, Flechon JE. Localization of fibrillarin and nucleolin in nucleoli of mouse preimplantation embryos. Mol Reprod Dev. 1995;40:305–310. doi: 10.1002/mrd.1080400306. [DOI] [PubMed] [Google Scholar]
- Bell P, Dabauvalle MC, Scheer U. In vitro assembly of prenucleolar bodies in Xenopusegg extract. J Cell Biol. 1992;118:1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P, Scheer U. Prenucleolar bodies contain coilin and are assembled in Xenopusegg extract depleted of specific nucleolar proteins and U3 RNA. J Cell Sci. 1997;110:43–54. doi: 10.1242/jcs.110.1.43. [DOI] [PubMed] [Google Scholar]
- Bell SP, Learned RM, Jantzen HM, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- Bell SP, Pikaard CS, Reeder RH, Tjian R. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell. 1989;59:489–497. doi: 10.1016/0092-8674(89)90032-9. [DOI] [PubMed] [Google Scholar]
- Benavente R, Rose KM, Reimer G, Hügle-Dörr B, Scheer U. Inhibition of nucleolar reformation after microinjection of antibodies to RNA polymerase I into mitotic cells. J Cell Biol. 1987;105:1483–1491. doi: 10.1083/jcb.105.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven AF, Lee R, Razaz M, Leader DJ, Brown JWS, Shaw PJ. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J Cell Sci. 1996;109:1241–1251. doi: 10.1242/jcs.109.6.1241. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Martin TE, Gordon J, Amalric F, Fakan S. Physiologically inactive nucleoli contain nucleoplasmic ribonucleoproteins: immunoelectron microscopy of mouse spermatids and early embryos. Exp Cell Res. 1994;213:55–63. doi: 10.1006/excr.1994.1172. [DOI] [PubMed] [Google Scholar]
- Bodeker MD, Cairns C, McStay B. Upstream binding factor stabilizes Rib 1, the TATA-binding-protein containing Xenopus laevisRNA polymerase I transcription factor, by multiple protein interactions in a DNA-independent manner. Mol Cell Biol. 1996;16:5572–5578. doi: 10.1128/mcb.16.10.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Littna E. RNA synthesis during development of Xenopus laevis,the South African clawed toad. J Mol Biol. 1964;8:669–687. doi: 10.1016/s0022-2836(64)80116-9. [DOI] [PubMed] [Google Scholar]
- Brown DD, Littna E. Synthesis and accumulation of DNA-like RNA during embryogenesis of Xenopus laevis. . J Mol Biol. 1966;20:81–94. doi: 10.1016/0022-2836(66)90119-7. [DOI] [PubMed] [Google Scholar]
- Busby S, Reeder RH. Fate of amplified nucleoli in Xenopus laevisembryos. Dev Biol. 1982;91:458–467. doi: 10.1016/0012-1606(82)90052-5. [DOI] [PubMed] [Google Scholar]
- Busby SJ, Reeder RH. Spacer sequences regulate transcription of ribosomal gene plasmids injected into Xenopus embryos. Cell. 1983;34:989–996. doi: 10.1016/0092-8674(83)90556-1. [DOI] [PubMed] [Google Scholar]
- Caizergues-Ferrer M, Mariottini P, Curie C, Lapeyre B, Gas N, Amalric F, Amaldi F. Nucleolin from Xenopus laevis: cDNA cloning and expression during development. Genes Dev. 1989;3:324–333. doi: 10.1101/gad.3.3.324. [DOI] [PubMed] [Google Scholar]
- Caizergues-Ferrer M, Mathieu C, Mariottini P, Amalric F, Amaldi F. Developmental expression of fibrillarin and U3 snRNA in Xenopus laevis. . Development. 1991;112:317–326. doi: 10.1242/dev.112.1.317. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schröck E, Speicher MR, Mathieu U, Jauch A, Emmerich P, et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- Davidson, E.H. 1986. Gene activity in early development. Academic Press, New York. 1–670.
- Fan H, Penman S. Regulation of synthesis and processing of nucleolar components in metaphase-arrested cells. J Mol Biol. 1971;59:27–42. doi: 10.1016/0022-2836(71)90411-6. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Kiss T. Structure and function of nucleolar snRNPs. Mol Biol Rep. 1993;18:149–156. doi: 10.1007/BF00986770. [DOI] [PubMed] [Google Scholar]
- Gautier T, Fomproix N, Masson C, Azum-Gélade MC, Gas N, Hernandez-Verdun D. Fate of specific nucleolar perichromosomal proteins during mitosis: cellular distribution and association with U3 snoRNA. Biol Cell (Paris) 1994;82:81–93. doi: 10.1016/s0248-4900(94)80010-3. [DOI] [PubMed] [Google Scholar]
- Gébrane-Younès J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- Gerbi, S.A., R. Savino, B. Stebbins-Boaz, C. Jeppesen, and R. Rivera-Leon. 1990. A role for U3 small nuclear ribonucleoprotein in the nucleolus. In The Ribosome—Structure, Function and Evolution. W.E. Hill, A. Dahlberg, R.A. Garrett, P.B. Moore, D. Schlessinger, and J.R. Warner, editors. American Society for Microbiology, Washington, DC. 452–469.
- Guimond A, Moss T. Variants of the Xenopus laevisribosomal transcription factor xUBF are developmentally regulated by differential splicing. Nucleic Acid Res. 1992;20:3361–3366. doi: 10.1093/nar/20.13.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon, J.B., and L. Wakefield. 1986. Microinjection of amphibian oocytes and eggs for the analysis of transcription. In Methods in Cell Biology. B.K. Kay and H.B. Peng, editors. Academic Press, San Diego. 269–299.
- Gurdon, J.B., and M.P. Wickens. 1983. Use of Xenopus oocytes for the expression of cloned genes. In Methods in Enzymology. R. Wu, L. Grossman, and K. Moldave, editors. Academic Press, New York. 101:370–386. [DOI] [PubMed]
- Hadjiolov, A.A. 1985. The nucleolus and ribosome biogenesis. In Cell Biology Monographs. Vol. 12. Springer-Verlag, Wien, New-York. 1–268.
- Hay ED, Gurdon JB. Fine structure of the nucleolus in normal and mutant Xenopusembryos. J Cell Sci. 1967;2:151–162. doi: 10.1242/jcs.2.2.151. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Junéra HR. The nucleolus. Princ Med Biol Cell Organels. 1995;2:73–92. [Google Scholar]
- Highett MI, Rawlins DJ, Shaw PJ. Different patterns of rDNA distribution in Pisum sativumnucleoli correlate with different levels of nucleolar activity. J Cell Sci. 1993;104:843–852. [Google Scholar]
- Jackson DA. Nuclear organization: uniting replication foci, chromatin domains and chromosome structure. Bioessays. 1995;17:587–591. doi: 10.1002/bies.950170704. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO (Eur Mol Biol Organ) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen HM, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Jiménez-Garcia LF, Segura-Valdez ML, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Mannervik M, Tora L, Carmofonseca M. In vivo evidence that TATA-binding protein SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junéra HR, Masson C, Geraud G, Hernandez-Verdun D. The three-dimensional organization of ribosomal genes and the architecture of the nucleoli vary with G1, S and G2 phases. J Cell Sci. 1995;108:3427–3441. doi: 10.1242/jcs.108.11.3427. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Newport J, Gerhart J. The timing of early developmental events in Xenopus. . Trends Genet. 1985;1:41–47. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lapeyre B, Mariottini P, Mathieu C, Ferrer P, Amaldi F, Amalric F, Caizergues-Ferrer M. Molecular cloning of Xenopusfibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol Cell Biol. 1990;10:430–434. doi: 10.1128/mcb.10.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned RM, Learned TK, Haltiner MM, Tjian RT. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1982. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 9.47–9.55.
- Marzluff, W.F., and R.C. Huang. 1980. Transcription of RNA in isolated nuclei. In Transcription and Translation: A Practical Approach. B.D. Hames, and S.J. Higgins, editors. 89–128.
- Masson C, Bouniol C, Szöllösi MS, Debey P, Hernandez-Verdun D. Conditions favoring RNA polymerase I transcription in permeabilized cells. Exp Cell Res. 1996;226:114–125. doi: 10.1006/excr.1996.0209. [DOI] [PubMed] [Google Scholar]
- McStay M, Hu CH, Pikaard CS, Reeder RH. xUBF and Rib 1 are both required for formation of a stable polymerase I promoter complex in X. laevis. . EMBO (Eur Mol Biol Organ) J. 1991;10:2297–2303. doi: 10.1002/j.1460-2075.1991.tb07766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina FJ, Cerdido A, Fernandez-Gomez ME. Components of the nucleolar processing complex (pre-rRNA, fibrillarin, and nucleolin) colocalize during mitosis and are incorporated to daughter cell nucleoli. Exp Cell Res. 1995;221:111–125. doi: 10.1006/excr.1995.1358. [DOI] [PubMed] [Google Scholar]
- Mélèse T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- Messmer B, Dreyer C. Requirements for nuclear translocation and nucleolar accumulation of nucleolin of Xenopus laevis. . Eur J Cell Biol. 1993;61:369–382. [PubMed] [Google Scholar]
- Moss T, Stefanovsky VY. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog Nucl Acid Res Mol Biol. 1995;50:25–65. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- Nakahashi T, Yamana K. Biochemical and cytological examination on the initiation of ribosomal RNA synthesis during gastrulation of Xenopus laevis. . Dev Growth Differ. 1976;18:329–338. doi: 10.1111/j.1440-169X.1976.00329.x. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopusembryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop, P.D., and J. Faber. 1994. Normal Table of Xenopus laevis (Daudin). Garland Publishing, Inc., New York. 1–252.
- Ochs RL, Lischwe MA, Shen E, Caroll RE, Busch H. Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma. 1985a;92:330–336. doi: 10.1007/BF00327463. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985b;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Mazan S, Nicoloso M, Pichard E, Bachellerie J-P, Puvion E. Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function. Eur J Cell Biol. 1992;58:149–162. [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Puvion E, Bachellerie J-P. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situhybridization with 5′ETS leader probe. Chromosoma. 1997;105:496–505. doi: 10.1007/BF02510486. [DOI] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe MA, Bush H, Tan EM. Monoclonal antibody from a (New Zealand black × New Zealand White)F1 mouse and some human scleroderma sera target an M r34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Roussel P, André C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel P, André C, Masson C, Géraud G, Hernandez-Verdun D. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J Cell Sci. 1993;104:327–337. doi: 10.1242/jcs.104.2.327. [DOI] [PubMed] [Google Scholar]
- Scheer U, Thiry M, Goessens G. Structure, function and assembly of nucleolus. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Amalric F, Bouvet P. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. doi: 10.1016/0300-9084(96)84759-6. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Shiokawa K, Kurashima R, Shinga J. Temporal control of gene expression from endogenous and exogenously-introduced DNAs in early embryogenesis of Xenopus laevis. . Int J Dev Biol. 1994;38:249–255. [PubMed] [Google Scholar]
- Shiokawa K, Misumi Y, Tashiro K, Nakakura N, Yamana K, Oh-uchida M. Changes in the patterns of RNA synthesis in early embryogenesis of Xenopus laevis. . Cell Differ Dev. 1989;28:17–26. doi: 10.1016/0922-3371(89)90019-1. [DOI] [PubMed] [Google Scholar]
- Shiokawa K, Misumi Y, Yamana K. Demonstration of rRNA synthesis in pre-gastrular embryos of Xenopus laevis. . Dev Growth Differ. 1981a;23:579–587. doi: 10.1111/j.1440-169X.1981.00579.x. [DOI] [PubMed] [Google Scholar]
- Shiokawa K, Tashiro K, Misumi Y, Yamana K. Non-coordinated synthesis of RNA's in pre-gastrular embryos of Xenopus laevis. . Dev Growth Differ. 1981b;23:589–597. doi: 10.1111/j.1440-169X.1981.00589.x. [DOI] [PubMed] [Google Scholar]
- Shiokawa K, Yamana K. Differential initiation of rRNA gene activity in progenies of different blastomeres of early Xenopusembryos; evidence for regulated synthesis of rRNA. Dev Growth Differ. 1979;21:501–507. doi: 10.1111/j.1440-169X.1979.00501.x. [DOI] [PubMed] [Google Scholar]
- Singer RH, Green MR. Compartimentalization of eukaryotic gene expression: causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Smith, J.C., and J.R. Tata. 1991. Xenopus cell lines. In Methods in Cell Biology. B.K. Kay, and H.B. Peng, editors. Academic Press, San Diego. 36:635–654. [PubMed]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Strouboulis J, Wolffe AP. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Thiry, M., and G. Goessens. 1996. The nucleolus during the cell cycle. Springer-Verlag, Heidelberg. 146 pp.
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D, Scheer U, Benavente R. The DNA topoisomerase I inhibitor camptothecin blocks postmitotic reformation of nucleoli in mammalian cells. Eur J Cell Biol. 1993;61:189–192. [PubMed] [Google Scholar]
- Wu, M., and J. Gerhart. 1991. Raising Xenopus in the laboratory. In Methods in Cell Biology. B.K. Kay, and H.B. Peng, editors. Academic Press, San Diego, CA. 36:3–18. [DOI] [PubMed]
- Zatsepina OV, Dudnic OA, Todorov IT, Thiry M, Spring H, Trendelenburg MF. Experimental induction of prenucleolar bodies (PNBs) in interphase cells: interphase PNBs show similar characteristics as those typically observed at telophase of mitosis in untreated cells. Chromosoma. 1997;105:418–430. doi: 10.1007/BF02510478. [DOI] [PubMed] [Google Scholar]