Abstract
G protein–coupled receptors trigger the reorganization of the actin cytoskeleton in many cell types, but the steps in this signal transduction cascade are poorly understood. During Dictyostelium development, extracellular cAMP functions as a chemoattractant and morphogenetic signal that is transduced via a family of G protein–coupled receptors, the cARs. In a strain where the cAR2 receptor gene is disrupted by homologous recombination, the developmental program arrests before tip formation. In a genetic screen for suppressors of this phenotype, a gene encoding a protein related to the Wiskott-Aldrich Syndrome protein was discovered. Loss of this protein, which we call SCAR (suppressor of cAR), restores tip formation and most later development to cAR2− strains, and causes a multiple-tip phenotype in a cAR2+ strain as well as leading to the production of extremely small cells in suspension culture. SCAR−cells have reduced levels of F-actin staining during vegetative growth, and abnormal cell morphology and actin distribution during chemotaxis. Uncharacterized homologues of SCAR have also been identified in humans, mouse, Caenorhabditis elegans, and Drosophila. These data suggest that SCAR may be a conserved negative regulator of G protein-coupled signaling, and that it plays an important role in regulating the actin cytoskeleton.
Keywords: Dictyostelium, WASP, actin cytoskeleton, G protein–coupled receptors, REMI
How signals from cell surface receptors trigger reorganization of the actin cytoskeleton is poorly understood. Both tyrosine kinase and G protein– coupled receptors are known to transduce signals that cause changes in cell motility, cell shape, and cell attachment (reviewed in Zigmond, 1996). These changes are brought about, at least in part, by a reordering of actin in the cell, and form the basis of such higher order processes as morphogenesis and cell migration. One of the most useful tools in the study of any complex biological problem is an organism amenable to genetic analysis. Dictyostelium discoideum, an amoeboid eukaryotic microorganism, has been an excellent genetic model for studying how extracellular signals are translated into dynamic changes in the actin cytoskeleton.
In Dictyostelium, extracellular cAMP functions as a chemoattractant and morphogenetic signal (reviewed in Devreotes, 1994). Four receptors for this ligand have been identified (cAR1–4), and each shows a unique pattern of spatial and temporal expression (Ginsburg et al., 1995; Parent and Devreotes, 1996). These receptors are highly related to each other, and fall into the seven-transmembrane G protein–coupled class. One of the four subtypes, the cAR2 receptor, is expressed in a subset of prestalk cells that form the presumptive tip structure (Saxe et al., 1996). Disruption of the gene encoding this receptor (the carB gene) causes a morphogenetic arrest before tip formation (Saxe et al., 1993). This phenotype implies that the cAR2 receptor triggers a signaling cascade required for normal tip formation and elongation. The nature of this signaling cascade is unknown, but the more thoroughly studied cAR1 receptor pathway provides some clues as to the possible events involved.
Responses known to be mediated by cAR1 include stimulation of actin polymerization and cross-linking (Caterina and Devreotes, 1991; Dharmawardhane et al., 1989). These are the first steps in the formation of pseudopods and subsequent chemotactic movement. Disruption of the gene encoding the cAR1 receptor leads to a block at the aggregation stage of development, consistent with its crucial role in chemotactic signaling. It is unclear precisely which second messenger pathways triggered by cAR1 are involved in this response, but a heterotrimeric G-protein is clearly required (Chen et al., 1996). Whether or not cAR2 stimulates a similar pathway in the nascent tip cells is a question that remains unanswered. The involvement of the actin cytoskeleton in tip formation is strongly implied by the existence of several mutations in known components of the actin cytoskeleton that block or alter this process (see Discussion). The goal of this study was to help identify components of the signaling cascades (cytoskeletal or otherwise) that cAR2 stimulates in the tip cells, and that cause the dramatic morphogenetic changes associated with tip formation.
To understand the downstream signaling events from the cAR2 receptor, a genetic approach was used. Such approaches to G-protein–coupled signaling in other organisms have revealed both positive and negative regulators of downstream responses. Some of the positively acting regulators that have been found in other systems include components of heterotrimeric G proteins as well as various target effectors (reviewed in Bardwell et al., 1994). More recently, examples of negative regulators were found independently in C. elegans and yeast in genetic screens for negatively acting components of egg-laying and mating pheromone-signaling pathways, respectively (Koelle and Horvitz, 1996; Dohlman et al., 1996). These components, SST2 in the case of yeast and EGL-10 in the case of Caenorhabditis elegans, helped to define a new class of GTPase-accelerating proteins known as the regulators of G-protein signaling (RGSs). Based on these results in other systems, our screen began with the assumption that negative regulators of cAR2 function could be eliminated by mutation, which might lead to a partial or complete restoration of morphogenesis and development.
In this paper we describe the isolation and initial characterization of one of these suppressors. The protein encoded by the disrupted gene shows areas of striking homology to the Wiskott-Aldrich Syndrome protein (WASP).1 The protein, which we call suppressor of cAR (SCAR), does not appear to be the Dictyostelium homologue of WASP, but instead seems to define a new family of evolutionarily conserved, WASP-related proteins (see below).
Wiskott-Aldrich syndrome is an X-linked human genetic disorder that leads to a variety of defects in the immune system, including thrombocytopenia, compromised immune function, and susceptibility to leukemias and lymphomas (Kirchhausen and Rosen, 1996). Patients with this disorder fail to produce normal antibody responses to pneumococcal polysaccharide injections, and in in vitro assays, neutrophils from WAS patients have major chemotactic defects (Ochs et al., 1980). When the morphology of these immune system cells is closely examined, defects in the organization of the actin cytoskeleton are found (Kenney et al., 1986; Molina et al., 1992; Gallego et al., 1997). In 1994, the gene responsible for Wiskott-Aldrich Syndrome was identified by positional cloning (Derry et al., 1994). The protein encoded by the Wiscott-Aldrich Syndrome gene initially failed to reveal much about its function, but was recently identified in screens for Cdc42-interacting proteins by three independent labs (Aspenström et al., 1996; Kolluri et al., 1996; Symons et al., 1996). A close relative of WASP, the N-WASP protein, has been shown to potentiate filopodia formation induced by an activated Cdc42 mutant (Miki et al., 1998a ). More confirmatory evidence for WASP's role in organizing the cytoskeleton was presented in transient overexpression experiments in which accumulation of massive F-actin aggregates occurred in the cytoplasm of the transfected cells (Symons et al., 1996). In the present paper we identify a new family of WASP-related proteins, and provide genetic evidence for its involvement in G protein–coupled signaling. This work also provides further evidence for the involvement of WASP and WASP-related proteins in regulating the actin cytoskeleton.
Materials and Methods
Dictyostelium Strains, Culture, and Development
Dictyostelium discoideum strain HPS400, an axenically grown thy− strain, is the wild-type parent of all the stains described in this study. The carB null cell line, KO8-5, was derived as described previously (Saxe et al., 1996). All cell lines were grown in HL-5 media that was supplemented with 100 μg/ml thymidine where appropriate. Cells were grown at 22°C on tissue culture plates or in 250-ml flasks shaking at 220 rpm. DB-agar or filter development was initiated by washing cells twice with ice-cold developmental buffer (DB: 5 mM Na2PO4, 5 mM KH2PO4, 2 mM MgSO4, 0.2 mM CaCl2) and depositing them onto DB-agar plates or onto cellulose acetate filters (Gelman Sciences, Inc., Ann Arbor, MI) at 3 × 106 cells/cm2. Bacterial development was initiated by transferring a small volume of liquid culture with a sterile toothpick to the surface of an SM/5 agar plate that had been spread with Klebsiella aerogenes.
Isolation of Restriction Enzyme–mediated Integration (REMI) Mutants
REMI mutants were derived by a method modified from Adachi et al. (1994) and Larochelle et al. (1996). In brief, 1.2 × 107 carB null cells were pelleted and washed once with electroporation buffer (50 mM sucrose, 10 mM NaPO4, pH 6.1). These cells were resuspended in 800 μl of electroporation buffer and mixed with 100 U DpnII and 50 μg of BamH1-linearized plasmid (pUCBsrΔBam). This mixture was transferred to a 0.4-cm cuvette and electroporated in a Gene Pulser (Bio-Rad Laboratories, Hercules, CA) at 0.9 kV (3 μF, 200Ω). The cells were allowed to recover on ice for 10 min before CaCl2 and MgCl2 were added to 1 mM each. The cells were transferred to shaking culture in HL-5 media for 16–18 h before Blasticidin S was added to 10 μg/ml (ICN). The cells were then plated at 6 × 103 cells/well (50 μl per well) on 384 well plates (Nunc Inc., Naperville, IL). Under these conditions, <33% of the wells contained a Blasticidin-resistent transformant, and each could be considered clonal with 95% certainty. Cells were allowed to grow on the 384-well plates under humidified conditions for 10–14 d, or until they had reached confluency.
Clones were transferred by sterile toothpicks to agar plates spread with Klebsiella aerogenes. When the bacterial lawn had been cleared, the cells remaining at the center of a plaque initiated the developmental program and were screened visually for rescue of the carB null morphological block. The cells in the corresponding well on the 384-well plate were then expanded for further screening and analysis.
Molecular Biology Methods
Plasmid rescue was performed as described previously (Kuspa and Loomis, 1992). Amplification of the SCAR gene was achieved by using the primers 5′-CAATTATTGGACCCATACATGG and 5′-GGTGCTTGGTATTGATTGGTACC, which were derived from the flanking sequence of the plasmid rescued from the 9A/O7 strain. Genomic DNA for PCR was prepared by resuspending 1 × 106 washed cells in 100 μl of whole-cell PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.4], 15 mM MgCl2) supplemented with 100 μg/ml Proteinase K and 0.5% NP-40. This preparation was treated at 56°C for 45 min, and at 95°C for 10 min. Each reaction was performed in a total volume of 50 μl, and contained 10 μl genomic prep (1 × 105 cell equivalent), 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 250 μM dNTPs, 1 μM of each primer, and 2 U of Thermalase rec-Tbr (Ameresco Inc., Solon, OH). Reaction conditions were as follows: initial denaturation at 90°C for 5 min, followed by 40 cycles of 95°C for 2 min, 53°C for 2 min, and 72°C for 3 min. Screening the cDNA library by PCR was as Munroe et al. (1995) described, except that 100,000 pfu were screened. Recapitulation of the 9A/O7 phenotype was achieved by linearizing the plasmid rescued from 9A/O7 with Cla I and transforming it into either carB null or HPS400 cells using the conditions described above.
RNA was isolated and analyzed by Northern blot as described in Berks and Kay (1990). The probe for the Northern in Fig. 2 b was the whole plasmid containing the SCAR cDNA. The SCAR antibody was raised in rabbits with the assistance of Zymed Laboratories Inc. (S. San Francisco, CA) against the synthetic peptide (C)DSDSSEDESDDSDWDCOOH that corresponds to the COOH-terminal–most 16 residues of the SCAR protein. Affinity purification of the antisera was performed by immobilizing the peptide on a Sulfolink column (Pierce Chemical Co., Rockford, IL). Antisera was applied to the column, and peptide-specific antibodies were eluted with 100 mM glycine-HCl (pH 2.5). For Western blots, the affinity-purified antibody was used at a dilution of 1/100. Visualization of protein was achieved using protein A coupled to horseradish peroxidase as a secondary (Bio-Rad Laboratories) and chemiluminescent detection reagents (Amersham Corp., Arlington Heights, IL).
Figure 2.
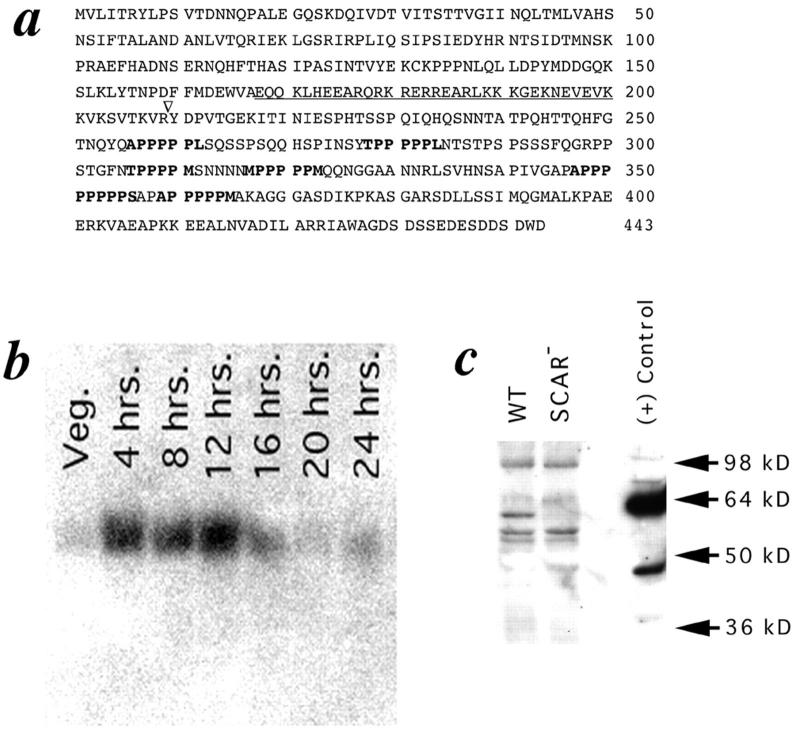
SCAR sequence, developmental Northern blot analysis, and protein expression. (a) Predicted amino acid sequence of SCAR protein derived from cDNA sequence.(∇) Site of plasmid insertion. Underlining indicates highly charged helical region, and residues in bold show polyproline repeats. (b) Developmental Northern blot analysis. At the indicated time points, poly(A)+-selected RNA was prepared from HPS400 cells developed on filter pads, subjected to electrophoresis, blotted, and probed with a 32P-labeled SCAR cDNA probe. The SCAR transcript runs as a single band at 1.8 kb. (c) SCAR protein expression. Whole-cell protein extracts were prepared from HPS400/ SCAR+ and SCAR− cells developed on filters for 17 h, subjected to SDS-PAGE, transferred, and probed with an affinity-purified SCAR polyclonal antisera. This antisera recognizes several bands, including a 60-kD band present only in SCAR+ lysates. (+) Control is a lysate of bacterial cells induced to express a recombinant SCAR fusion protein. This fusion protein includes extra sequences for purification purposes, and runs slightly higher than the endogenous SCAR protein.
Immunocytochemistry and Image Analysis
Suspension-grown vegetative cells were allowed to adhere to glass coverslips overnight. To stain cells actively involved in chemotaxis, cells were attached to coverslips as described above, and then the HL-5 media was replaced by sterile DB for 12 h. During this interval, cells began to starve, initiate the developmental program, and form robust aggregation streams visible with the unaided eye. The direction of cell movement was easily determined by observing the enlarging loose mound of cells at the center of the aggregation territory. The cells were washed once in DB and fixed in −20°C MeOH for 5 min. Cells were then washed in PBS and stained with a PBS solution containing 400 nM TRITC-phalloidin (Sigma Chemical Co.), 300 nM Oregon Green 488-DNase I (Molecular Probes, Inc., Eugene, OR), and 1% BSA for 45 min at 37°C. Cells were washed again in PBS and mounted in Vectashield mount (Vector Labs, Inc., Burlingame, CA). Specimens were viewed under a 40× or 100× objective on a 510 confocal microscope (Carl Zeiss, Inc., Thornwood, NY). Quantification of cell size was achieved by removing a small aliquot from suspension culture and immediately viewing the cells under phase-contrast optics before attachment could occur. Still video images were imported into the public domain software program, NIH Image (1.61) for analysis. Each cell in the field was outlined by hand, and areas were calculated by the software. NIH Image was also used to analyze the ratio of TRITC-phalloidin/Oregon Green 488-DNase I–bound actin. Still video images of more than 20 cells were measured for each panel, and pixel intensity per unit area was determined. Pixel intensity for all panels was adjusted simultaneously and normalized against that of SCAR-/G-actin panel, which showed the greatest signal per unit area.
Results
Isolation of REMI Suppressors of the carB Null Strain
In an effort to identify components of the cAR2-signaling pathway, we used the restriction enzyme–mediated integration (REMI) technique of insertional mutagenesis to obtain knockout suppressors of the carB null phenotype (Saxe et al., 1993; Kuspa and Loomis, 1992; Shaulsky et al., 1996). Using a novel variation of the original REMI protocol, we were able to obtain clonal transformants of a carB null strain on 384-well plates in liquid culture by limiting dilution (see Materials and Methods). This modified technique allowed us to recover partial rescue mutants that may not have been recovered with the original screening techniques. These transformants were sterilely transferred to agar plates that had been spread with Klebsiella aerogenes as a food source. As plaques grew and expanded, the Dictyostelium cells left at the center of the plaques began to starve and initiate the developmental program. Plaques with developmental structures progressing beyond the tight mound stage (i.e., the point at which carB null cells arrest development) were noted, and the original liquid cultures were expanded for further screening and analysis. In a relatively modest number of transformants (4325), eleven clones were found that proceeded beyond the carB null block to varying degrees. One of these clones, the 9A/ O7 strain, produced finger structures that fell over and became migratory slugs, some of which eventually culminated into normally proportioned fruiting bodies (Fig. 1, a and b).
Figure 1.
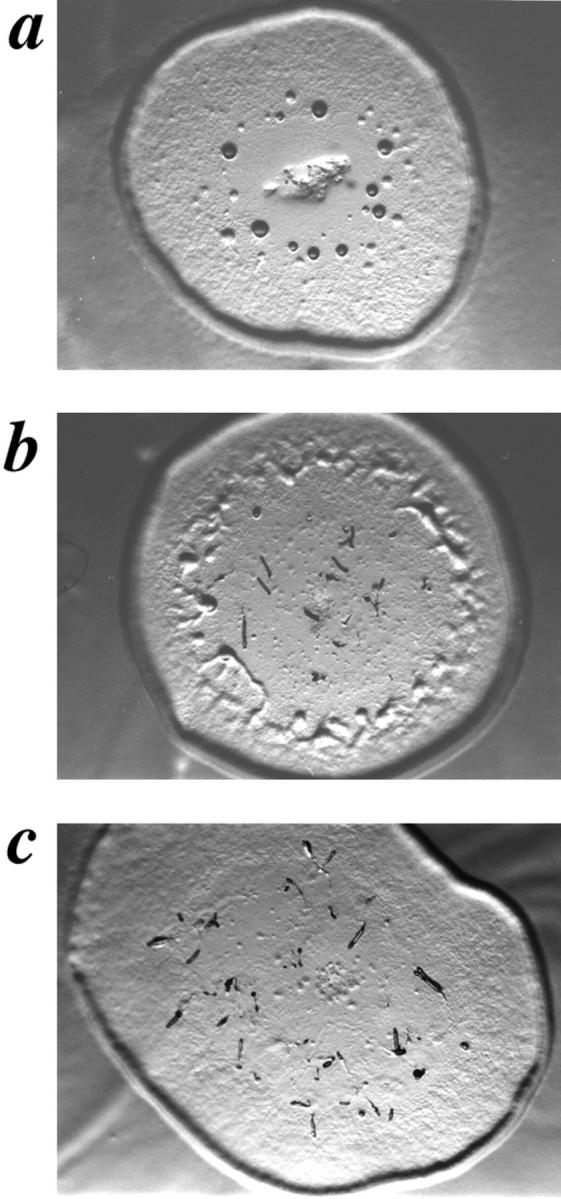
SCAR gene disruption partially rescues carB null morphological block. Developmental phenotypes of carB null strain KO8-5 (a), the original REMI mutant, 9A/O7 (b), and carB null/ SCAR−(c), grown on agar spread with Klebsiella aerogenes.
Cloning and Characterization of the SCAR Gene
To identify the gene disrupted in the 9A/O7 strain, plasmid rescue was used to clone regions of the genomic DNA flanking the insertion site. The plasmid isolated from the 9A/O7 strain contains 350 bp of flanking DNA from one side, and 3.5 kb from the other. Based on sequence obtained from these flanking regions, oligonucleotides were synthesized and used to amplify a predicted 336-bp product from genomic DNA of the carB null parental strain. These same primers did not amplify an equivalent band from the 9A/O7 strain (data not shown). This result indicates that the original plasmid insertion occurred without a deletion event. This same oligonucleotide pair was used in a PCR-based screen of a cDNA library prepared from finger-stage cells (Munroe et al., 1995). One of the clones obtained had regions of sequence identical to the original plasmid-rescue plasmid, and appeared to include a full-length–coding cDNA. The protein encoded by this gene is designated SCAR (Fig. 2 a), and the gene is designated scrA. The cDNA clone was subsequently used to probe a developmental Northern blot of poly-(A)+ mRNA prepared from the wild-type parental strain HPS400. A message of ∼1.8 kb was identified, which is expressed at low levels during vegetative growth, but which accumulates dramatically by 4 h of development. Message levels remain high through 12 h, and then appear to drop off somewhat at 16 h (Fig. 2 b). An antibody was raised against a synthetic peptide corresponding to the last 16 amino acids of the SCAR protein sequence. This antibody recognizes a protein of ∼60 kD on Western blots of wild-type (WT) lysates that appears to be entirely missing from SCAR− lysates (Fig. 2 c). Although this affinity-purified antisera recognizes other proteins in whole-cell lysates, the presence of a strong corresponding band in the lane containing a bacterially expressed SCAR fusion protein suggests that the band seen in the WT lane is indeed the SCAR protein.
Disruption of the SCAR Gene in carB Null and WT Backgrounds
To confirm that disruption of the SCAR gene is responsible for the phenotypic suppression of the carB null strain, the plasmid rescued from the 9A/O7 strain was used to disrupt the gene by homologous recombination in a fresh carB null strain. Using the same primer pair used to confirm the original disruption and isolate the cDNA clone, the genotype of transformant clones was determined by genomic PCR. In all six clones from this transformation that lacked a wild-type copy of the gene by genomic PCR, the original 9A/O7 phenotype was recreated (Fig. 1 c).
To determine the phenotype of an scrA gene disruption in the presence of a WT copy of the carB gene, a similar knockout experiment was performed in HPS400, the WT parental strain of the carB nulls. Strains that still showed a WT copy of the scrA gene were indistinguishable from the untransformed HPS400 strain (Fig. 3 a). All clones that showed a disruption of the scrA gene also displayed a multiple-tip phenotype (Fig. 3 b). The multiple tips from a single aggregate gave rise to individual fingers and slugs that migrated apart, and formed normal, albeit small fruiting bodies with viable spores. On filter pads where greater synchrony can be attained, the SCAR− cells produced tips ∼3 h earlier than the SCAR+ controls (data not shown). These accelerated structures remained ahead of the WT controls through the remainder of development, and often completed fruiting body formation within 20 h, as opposed to the 24–26 h required by the controls (data not shown).
Figure 3.
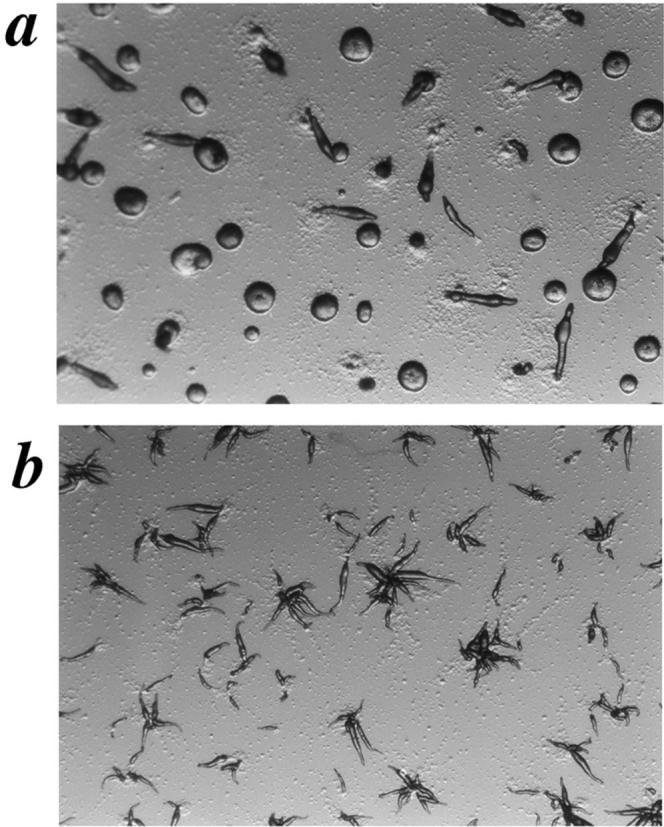
Developmental phenotypes of HPS400/SCAR+ (a) and HPS400/SCAR− (b) on DB-agar plates at 13 h of development.
The Domain Structure of the SCAR Protein and Identification of Higher Eukaryotic Homologues
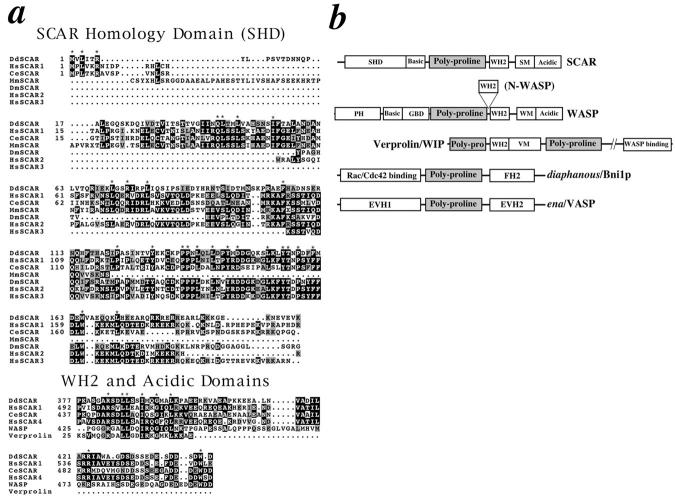
The 443–amino acid protein encoded by the SCAR gene contains a domain structure reminiscent of WASP, and shares several areas of homology with WASP in its central and COOH-terminal regions (Fig. 4 b). The domain structure of SCAR can be described as follows: the NH2-terminal 170 residues of the SCAR protein seem to define a novel structural motif found only in SCAR and the other SCAR-related sequences discussed below (Fig. 4 a). Since this domain is described here for the first time, we chose the name SCAR homology domain (SHD) for this region. The next domain found in SCAR is a region of highly basic residues that follows immediately after the SHD. WASP and N-WASP also contain areas of basic residues in their respective NH2-terminal regions. By analogy with N-WASP (Miki et al., 1998a ), this region of SCAR may serve as the intramolecular binding site for a region of acidic residues found at the COOH terminus. The third domain found in the SCAR sequence, also shared with WASP, is a region of polyproline repeats of the actin-based motility 2 (ABM-2) type (Purich and Southwich, 1997). ABM-2 repeats fit the consensus XPPPPP (X = A, L, G, S), and those repeats found in SCAR are indicated in boldface type in Fig. 2 a. ABM-2–type repeats are found in a number of other protein families known to regulate the actin cytoskeleton such as verprolin, VASP/ena, and diaphanous (Fig. 4 b). The region containing these repeats may serve as a flexible hinge region in the protein structure, or each repeat may individually serve as binding sites for specific protein ligands such as profilin or Src homology 3 (SH3) domain–containing proteins (Williamson, 1994; Purich and Southwick, 1997).
Figure 4.
Sequence alignments. (a) Sequence alignments of SHD, WH2, and acidic domains for HsSCAR1 (KIAA 0269, NCBI accession no. 1665805), CeSCAR (R06C1.b, NCBI accession no. 1628078), MmSCAR (NCBI accession no. AA035899), DmSCAR (NCBI accession no. AA567420), HsSCAR2 (NCBI accession no. AA535513), HsSCAR3 (NCBI accession no. AA445914), WASP (hWASP, NCBI accession no. 1722836), and Verprolin (NCBI accession no. 755821). Alignments were produced with the LaserGene program (DNASTAR Inc., Madison, WI). Identical residues are outlined in black, and highly conserved residues are outlined in gray. Asterisks show residues that are invariant in all sequences at that position. (b) Domain structure of proteins containing ABM-2 polyproline repeats (Poly-proline). WH2 indicates WASP homology 2 domain; PH indicates Plekstrin homology domain; GBD (also known as a CRIB motif) indicates GTPase binding domain; SM indicates SCAR-motif; WM indicates WASP-motif; VM indicates verprolin-motif; EVH1 indicates an ena/VASP homology 1 domain; EVH2 indicates an ena/VASP homology 2 domain, and FH2 indicates a Formin homology 2 domain.
The last three domains of SCAR are distal to the polyproline region in the COOH-terminal region, and tie SCAR most closely with WASP (Fig. 4 a, WH2 and Acidic Domains). The first of these is a so-called WASP homology 2 (WH2) domain that SCAR shares in common with WASP and verprolin, and has been shown to bind directly to actin (Symons et al., 1996; Vaduva et al., 1997; Miki et al., 1998b). The second domain found in the COOH terminus is a short motif that appears to be specific for SCAR and the SCAR-related sequences that are anchored by two highly conserved basic residues. The final domain of SCAR is a region of acidic residues shared with WASP that is anchored by an invariant tryptophan residue at the extreme COOH terminus. The possible function of these domains will be addressed in some detail in the Discussion.
In searching both the nonredundant protein and expressed sequence tag databases, a number of sequences were found that have striking homology to SCAR. Two of these sequences, HsSCAR1 and CeSCAR, appear to encode the human and C. elegans homologues of SCAR, respectively. These two sequences align quite well with Dictyostelium SCAR along their entire lengths, with the exception of the central polyproline repeat region (see Fig. 4 a). Although both contain multiple ABM-2 polyproline repeats, the number and spacing of the repeats is not conserved. The rest of the sequences reported in Fig. 4 a are partial sequences from the expressed sequence tag (EST) databases. These include two human, one mouse, and one Drosophila sequence with high homology to the SHD. A fourth human sequence, distinct from HsSCAR1, with high homology to SCAR in the COOH-terminal region is shown. No close SCAR homologues were identified in the completely sequenced Saccharomyces cerevisiae genome.
Effects of SCAR Gene Disruption on Cell Morphology During Growth
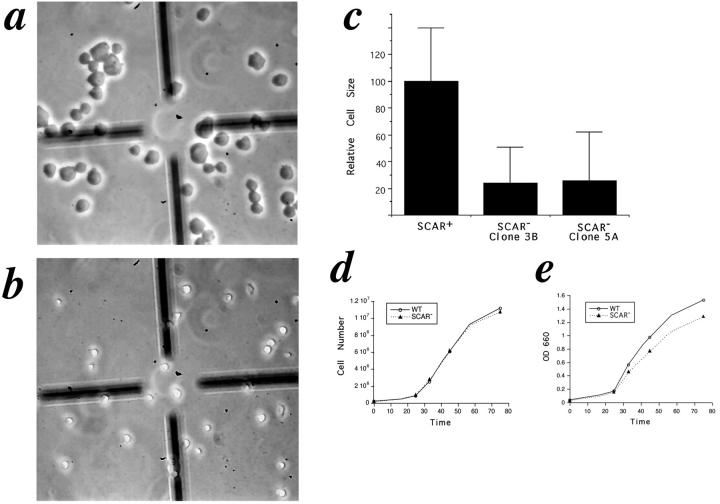
Independent of genetic background, SCAR− cells grow as very small cells in suspension culture. When these cells are viewed under phase contrast optics, their difference in size and area is readily apparent (Fig. 5, a and b). This is true for all SCAR− cell lines in both the carB null and HPS400 background (data not shown). By calculating the relative cell areas from captured video images of SCAR− and control suspension cells, an approximation of the difference in cell size was obtained. In two independent HPS400/ SCAR− strains, the cells grew to ∼25% of the relative size of a nonhomologous control transformant (Fig. 5 c). However, the growth rate of the SCAR− cells was indistinguishable from that of wild-type controls in terms of cell number (Fig. 5 d). Turbidity measurements revealed a slight reduction in absorbance for the SCAR− cells relative to that of the wild type (Fig. 5 e). Thus, in the absence of SCAR, cells growing in liquid suspension can grow and divide efficiently, but individually, cell size is dramatically reduced. The differences in cell size between SCAR− and control cells was much less pronounced when cells were grown on solid surfaces (data not shown).
Figure 5.
SCAR gene disruption causes reduction in cell size, but does not affect growth rate. HPS400/SCAR+ (a) and HPS400/SCAR− (b) cells were removed from suspension culture and immediately viewed by videomicroscopy. By quantitating the cell area from images such as a and b using the public domain software, NIH Image, an approximation of relative cell size is derived (c). SCAR− clones 3B and 5A are independently derived HPS400/SCAR− transformants. Error bars represent SD. (d and e) Growth curves of WT and SCAR−clones. Growth rates in terms of cell numbers are indistinguishable, but SCAR− cells show a consistent reduction of turbidity at 660 nm. Both curves are representative experiments.
Effects of SCAR Gene Disruption on Actin During Vegetative Growth and Aggregation
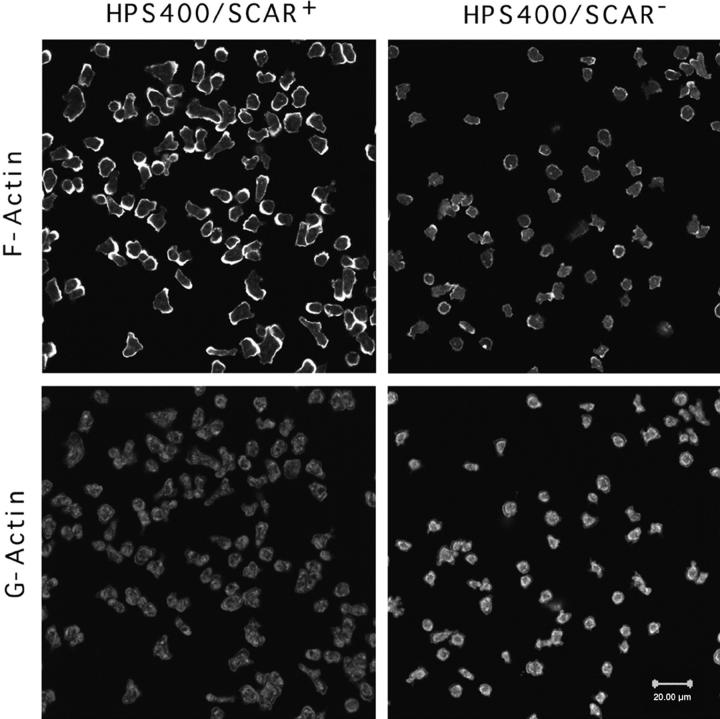
Because of the sequence similarity of SCAR to the actin regulatory protein WASP and the small cell size seen in the SCAR− cells, we proceeded to determine if SCAR gene disruption affected the actin cytoskeleton. F-actin and G-actin were simultaneously visualized in vegetative cells with TRITC-phalloidin and Oregon Green 488-DNase I using confocal microscopy. Since these probes are specific for F-actin and G-actin, respectively, the amount and distribution of each species could be monitored (Haugland et al., 1994). SCAR− cells showed a dramatic reduction of phalloidin staining, and a concomitant increase in DNase I staining relative to the controls, suggesting a reduction in F-actin content in these cells (Fig. 6). Similar results were shown to be true for all SCAR− cell lines tested (nine strains; data not shown); the same was also true for all of the carB −/scrA − strains observed (data not shown). The SCAR− cells contained an F-actin–rich cortex and pseudopods, as did the controls, but the staining of these structures was considerably less intense than that of the WT controls. Oregon Green 488-DNase I, the G-actin–specific stain, showed mainly cytoplasmic staining in both the mutant and wild-type cells. The image analysis software, NIH Image, was subsequently used to analyze the ratio of F-actin to G-actin in the cells in Fig. 6. The results are shown in Table I, and indicate a greater than tenfold difference in relative F-actin and G-actin levels in the two cell types.
Figure 6.
Vegetative SCAR− cells have reduced F-actin and increased G-actin staining. F-actin stain is TRITC-phalloidin, and G-actin staining is Oregon Green 488-DNaseI. Images are three equivalent thickness confocal sections from the middle of the cells collapsed into one image. Bar, 20 μm.
Table I.
Analysis of F-actin and G-actin Ratios in SCAR+ and SCAR− Cells
| SCAR+ cells | SCAR− cells | |||
|---|---|---|---|---|
| Average area (pixels2) | 1866 ± 421 | 1110 ± 211 | ||
| Average F-actin content (pixels/unit area) | 420 ± 75 | 159 ± 65 | ||
| Average G-actin content (pixels/unit area) | 107 ± 63 | 522 ± 126 | ||
| F-actin/G-actin | 3.9 | 0.3 |
Measurements represent analysis of 25–30 cells from each panel of Fig. 6. Individual measurements were taken using NIH Image (1.61) and analyzed using Microsoft Excel.
To determine if SCAR gene disruption affected the actin cytoskeleton during chemotactic movement, mutant and WT cells were deprived of nutrient media to induce aggregation in a submerged culture format. The SCAR− cells showed a dramatically different cell morphology and F-actin–staining pattern during the aggregation stage than did the WT controls. As previously reported, WT cells are roughly oblong-shaped during chemotaxis, with a strong concentration of F-actin at the leading edge and a second, less intense area of F-actin staining at the trailing edge of the cell (Fukui et al., 1987; Fig. 7 a). SCAR−cells show one to three thin pseudopod-like extensions projecting from the cell body with only a weak concentration of F-actin at the distal tip of each (Fig. 7 b). While SCAR− cells did not have a WT morphology, they became polarized, roughly orienting towards the aggregation center and forming visible streams (Fig. 7, inserts). Though the streams were not as tightly organized as those of WT cells, their presence suggests the possibility of chemotactic movement despite abnormal F-actin organization.
Figure 7.
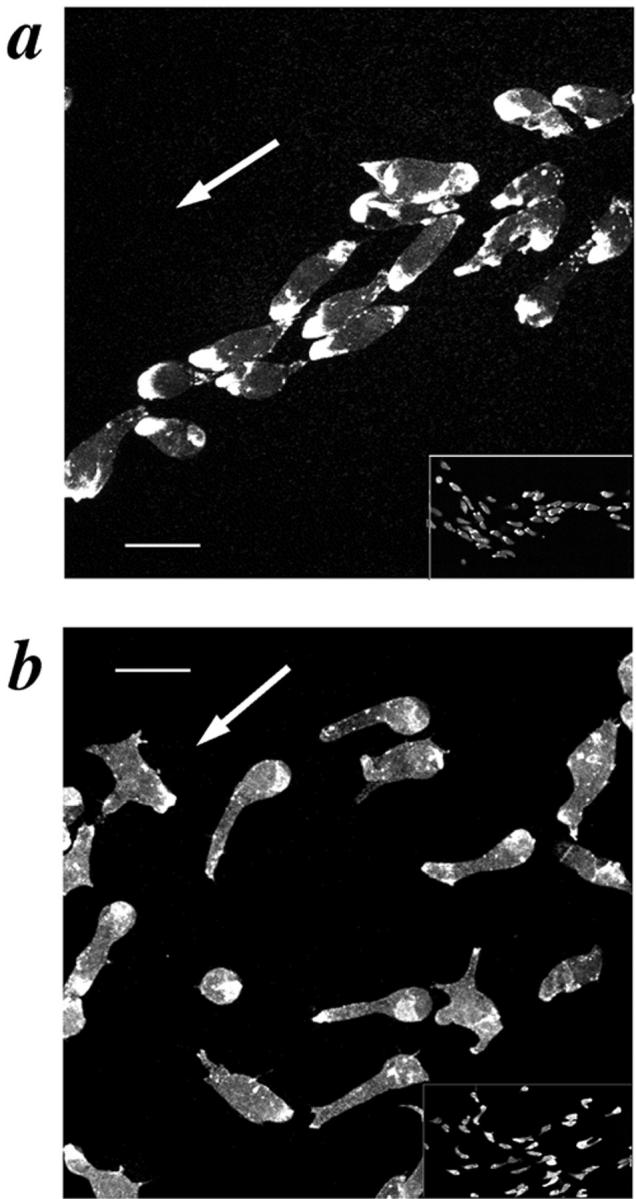
Aggregating SCAR− cells have aberrant cell morphology and F-actin staining. Aggregating cells were prepared by the submerged culture aggregation method and stained with TRITC-phalloidin. (a) HPS400/SCAR+, (b) HPS400/SCAR− cells. The white arrow represents the direction of movement; Bar, 10 μm. Inserts in a and b represent low-magnification views to more clearly compare the aggregation streams produced by the two cell lines.
Discussion
Isolation of SCAR as a Suppressor of the carB Null Strain
Our genetic screen began with the assumption that the cAR2 signaling pathway is attenuated by negative regulators, and that these negative regulators could be eliminated by mutation. Since the carB null strain has a strong phenotype, bypass suppressors were readily identified as clones containing more advanced developmental structures than the parental strain. To facilitate cloning, the disrupted gene in the mutants, we used the REMI insertional mutagenesis technique. This technique has been used successfully to identify suppressors of other developmental mutants in Dictyostelium, and our screen confirms the use of this approach to find genetic suppressors (Shaulsky et al., 1996).
Suppressing a receptor null mutant phenotype with another null mutation entails certain assumptions about the signaling pathways involved. The key assumption is that certain components of the signaling pathway can have a weak endogenous activity that can be enhanced by removing a constraining factor through mutation. In other genetic screens, this assumption has proven justified. For example, in the sevenless signaling pathway, a weak loss-of-function receptor mutation can be suppressed by a loss-of-function mutation in a rasGAP or in an Ets-type transcription factor, yan (Gaul et al., 1992; Lai and Rubin, 1992). In G protein–coupled pathways, SST2, the regulator of G-protein–signaling protein in yeast can be mutated to suppress loss-of-function mutations, partially in the mating pheromone receptor STE2 (Jenness et al., 1987). In the case of the cAR2 pathway, we assumed that either the heterotrimeric G protein or some other downstream component's weak endogenous activity could be enhanced by a new suppressor mutation, or that an alternate receptor such as cAR4 (that cannot normally provide the necessary signal inputs to complete development in the carB null background) could provide enough signal in the presence of a suppressor mutation, to rescue the morphological block of the carB null strain.
Isolation of SCAR as a suppressor of the carB null mutation suggests that it serves as a negative regulator of the cAR2 pathway. Based on SCAR's homology to other regulators of the actin cytoskeleton, we hypothesize that the cAR2 pathway, as in the case of cAR1, includes inputs to actin cytoskeletal reorganization, and that SCAR is somehow acting to modify these inputs. The fact that a functional cAR2 receptor in the SCAR− background leads to the formation of multiple tips provides strong genetic evidence that cAR2 signaling impinges on the same pathway that SCAR affects; however, a direct connection has not been proven. Understanding the exact biochemical nature of the interactions between cAR2, SCAR, and the other components in this pathway will require significant further investigation.
Tip Formation, Signal Transduction, and the Actin Cytoskeleton
Since multicellular morphogenesis can be described in its most basic form as a group of cells undergoing simultaneous cell shape or motility changes, it is not surprising that the actin cytoskeleton has been shown to play an important role in this process. Dictyostelium tip formation is a simple and potentially powerful model of this type of morphogenesis. A role for the actin cytoskeleton in Dictyostelium tip formation is supported by the existence of several mutants in known components of the actin cytoskeleton and the actomyosin complex that affect this process. Null mutations in myosin II heavy chain, or in its associated regulatory light chain block morphogenesis before tip formation (DeLozanne and Spudich, 1987; Chen et al., 1994). Another mutant that deranges development around the time of tip formation is the double mutant in two F-actin cross-linking proteins, alpha-actinin and ABP-120 (Rivero et al., 1996). A third mutant that may be particularly relevant to SCAR is a double knockout of both isoforms of Dictyostelium profilin. These cells have increased F-actin content, giant cells (due to a cytokinesis defect), and arrest before tip formation during development (Haugwitz et al., 1994). Considering the possible binding sites for profilin in the SCAR polyproline repeat region, and the nearly opposite phenotypes for the null mutants, a triple mutant may be very informative. Construction and analysis of such a mutant is underway, and preliminary data suggest at least a genetic interaction between SCAR and profilin (Bear et al., manuscript in preparation).
Besides the cAR2 receptor, other known components of signal transduction cascades also participate in regulating tip formation in Dictyostelium. Overexpression of an activated form of the rasD gene leads to a multiple tip phenotype (Reymond et al., 1986). A double knockout of two isoforms of phosphoinositide 3-kinase also leads to multiple tip phenotype (Zhou et al., 1995). A third strain with a multiple tip phenotype is one in which the tyrosine phosphatase, PTP1, is overexpressed (Howard et al., 1992). This strain also shows altered kinetics of cell shape changes upon return to rich growth medium (Howard et al., 1993). Whether or not these mutations exert their effects on tip formation through changes in the cytoskeleton and potentially through SCAR is not known, but evidence from other systems suggests that proteins of these types could be affecting the actin cytoskeleton (Zigmond, 1996; Retta et al., 1996).
SCAR is the First Described Member of a Conserved WASP-related Protein Family
The SCAR protein appears to define a new family of WASP-related actin cytoskeleton regulators that has been conserved over a significant fraction of eukaryotic evolution. Although SCAR shares several domains in common with WASP (Fig. 4 b), it is clearly not the Dictyostelium homologue of WASP, since a sequence bearing much closer homology to WASP was recently deposited in the Tsukuba cDNA-sequencing project database, and was previously identified by Chang Chung and Richard Firtel (University of California, San Diego) as a protein that interacts with human Cdc42 (personal communication). The existence of other sequences in the databases that bear a much closer relationship to SCAR than to WASP further supports classifying these two families of proteins as distinct entities. The human and nematode SCAR genes are completely sequenced, and based on the similarities throughout their entire lengths, appear to encode homologues of the Dictyostelium SCAR gene (Fig. 4 a). The clones found in the EST databases are not completely sequenced, and may encode homologous proteins or proteins that share some, but not all of SCAR's domains. The human EST clones do not appear to be splice variants of a single gene based on their divergence of nucleotide sequence along the entire length of the clones (data not shown). Several other human EST clones not shown in Fig. 4 a have areas of identical nucleotide sequence with the three shown, and only diverge over short segments. These clones may represent other human SCAR genes, or possibly splice variants of the HsSCAR genes shown. Also, the EST clone named HsSCAR4 may in fact be the 3′ region of the HsSCAR2 or HsSCAR3 gene. Further analysis will be required to completely describe the human SCAR gene family. The C. elegans genome appears to contain only one SCAR-like gene in the regions sequenced thus far, and the S. cerevisiae genome, which does contain a gene related to WASP (bee1; Li, 1997), does not appear to contain a close homologue of SCAR.
Possible Function of the Domains Found in SCAR
Since Dictyostelium SCAR is the first described member of this family of proteins, virtually nothing is known about its function. The domain structure of the SCAR protein and its similarity to WASP, however, suggest that it may regulate the actin cytoskeleton by binding to the components of signal transduction cascades or structural elements of the cytoskeleton. The SHD region of SCAR is highly conserved, but contains no obvious structural motifs to give a clue as to its function. The analogous NH2-terminal region of WASP contains a CRIB/GBD motif known to interact with the small GTPases Cdc42 and Rac (Symons et al., 1996). The NH2-terminal regions of Bni1p, Bnr1p, and p140mDia, members of the formin/diaphanous family, also contain a binding site for small GTPases (Evangelista et al., 1997; Imamura et al., 1997; Watanabe et al., 1997; Fig. 4 b). Perhaps by analogy the conserved SHD represents a binding site for a small GTPase, which in turn may regulate SCAR function. WASP also contains a Plekstrin homology domain in its NH2 terminus that serves as a binding site for the inositol phospholipid PIP2 (Miki et al., 1996). Perhaps the SHD is a novel binding site for another species of inositol phospholipid or inositol polyphosphate. Several of these molecules have been proposed to be second messengers in a variety of cellular processes, including actin cytoskeleton rearrangement (reviewed in Fukuda and Mikoshiba, 1997). Establishing the function of the SHD domain is a goal presently under investigation.
Between the SHD and the polyproline repeat region, all SCAR proteins contain a region rich in basic residues. A similar patch of basic residues is seen in WASP just NH2-terminal of the GBD/CRIB motif. These basic residues may be an intramolecular binding site for the acidic region of residues at the extreme COOH terminus. In the case of N-WASP, fusion proteins containing the basic region, the CRIB/GBD motif, and the acidic region, respectively, have been shown to interact in in vitro binding assays (Miki et al., 1998a ). This interaction can be disrupted by adding GTP-bound Cdc42. These data support a model in which binding an activated GTPase to the CRIB/GBD motif in the NH2-terminal region of N-WASP releases an intramolecular inhibitory interaction, and exposes other domains of the molecule such as the WH2 domain. It is tempting to speculate that SCAR may undergo a similar reaction when the appropriate molecule binds to its SHD region.
SCAR and WASP both have a region of polyproline repeats in the central part of their respective sequences. The prolines in WASP and SCAR form a specific type of polyproline repeat, fitting the consensus (XPPPPP, X = G, A, L, S) defined by Purich and Southwick as an ABM-2–type repeat (1997). Repeats of this type are found in many actin cytoskeleton–associated proteins such as the actin monomer–binding protein profilin (Southwick and Purich, 1997), as well as those containing WW protein interaction domains (Ermekova et al., 1997) and some that contain SH3 domains. Many of the latter are known to be involved in cell signaling and/or cytoskeletal regulation (Pawson and Scott, 1997). While there are no reports of WW motif–containing proteins interacting with WASP, evidence exists for WASP-like proteins interacting with profilin and with a number of SH3 domain–containing proteins.
Although a physical connection between profilin and WASP has yet to be established, several other ABM-2 repeat–containing proteins have been shown to associate physically with profilin via their polyproline repeats. These include the proteins VASP and the VASP/ena family member mena, as well as the diaphanous homologues Bni1p, p140mDia, and cdc12p (Reinhard et al., 1995; Gertler et al., 1996; Imamura et al., 1997; Watanabe et al., 1997; Chang et al., 1997). The functional nature of profilin's interaction with ABM-2 repeat containing proteins is demonstrated by the synthetic lethality, in fission yeast, of the profilin mutation (cdc3) in combination with a mutation in a member of the formin family (cdc12; Chang et al., 1997).
SH3 domain-containing proteins have also been shown to interact with proline-rich sites that overlap with ABM-2 repeats (Finan et al., 1996; Bedford et al., 1997). For these binding domains, the residues surrounding the prolines (either on the NH2-terminal or COOH-terminal side) strongly determine the binding specificity. Both WASP and N-WASP contain consensus SH3 binding sites, and have been demonstrated to interact with the SH3 domains of c-Src, p85α, PLCγ, Nck, and Grb2 by in vitro binding experiments (Finan et al., 1996; Miki et al., 1996). The ABM-2 repeats found in SCAR and all the SCAR-like sequences do not fit the SH3 binding consensus; however, this fact does not rule out the binding of an atypical SH3 domain (like the Abl tyrosine kinase type) to SCAR in its polyproline repeat region. Further experimentation will be required to determine if the polyproline repeats in SCAR are binding sites for any or all of these ligands.
The COOH-terminal region of the SCAR protein suggests that it is most closely related to the WASP protein family. The domain immediately COOH-terminal of the ABM-2 repeats in both SCAR and WASP is a so-called WH2 domain (Symons et al., 1996). This motif was first found in the yeast actin-binding protein verprolin. When the verprolin gene in yeast is disrupted by homologous recombination, severe defects in cell polarity, organization of the actin patches, and growth are seen (Donnelly et al., 1993). The WH2 domains found in verprolin and N-WASP were both recently shown to bind directly to actin, mostly likely in its monomeric form (Vaduva et al., 1997; Miki et al., 1998b). Several other uncharacterized sequences in the databases contain WH2 domains, but most of these fit into either the WASP, SCAR, or verprolin families. Interestingly, nearly all sequences containing WH2 domains also contain ABM-2 polyproline repeats, suggesting a close tie between their respective roles in protein function. While to date we have no data for a direct interaction between SCAR and actin, in vitro experiments to address that question are presently underway.
A short region distal of the WH2 domain seems to define a conserved motif specific for SCAR sequences (SCAR-motif; Fig. 4 b). The corresponding region of WASP is different, and was found to be homologous to a region of ADF/cofilin (Miki et al., 1996; WASP-motif; Fig. 4 b). The SCAR-motif and the cofilin-homologous region or WASP-motif, however, are both anchored by two conserved basic residues. The verprolin family also appears to contain a region of extended homology downstream of its WH2 domain; however, this region is quite distinct from the SCAR and WASP motifs (verprolin motif; Fig. 4 b). In WASP, the functional importance of the two basic residues in the WASP motif is suggested by the existence of a missense mutation (R478E) that causes a severe form of WAS in patients (Kolluri et al., 1995). The importance of this region was further confirmed by Miki et al. when they found that a fusion protein containing the WH2 domain and the cofilin-homologous region from N-WASP had very strong actin-depolymerizing activity (1998b). How this relates to WASP or SCAR function in vivo remains to be determined, but such data suggest that they may exert their effects on the actin cytoskeleton by changing the polymerization state of actin.
The COOH-terminal region of the protein also seems to define a conserved structural domain shared between SCAR and WASP. It consists of a highly acidic region in both WASP and N-WASP (and Dictyostelium WASP; Saxe, unpublished observations) as well as all of the SCAR sequences, and is anchored by an invariant tryptophan residue within one or two residues of the end of the protein. The existence of both the acidic and basic regions in analogous positions in the SCAR sequence suggests that a similar intramolecular inhibition might be occurring in SCAR that has been hypothesized for N-WASP. Further experimentation will be required to confirm such a model.
The data presented in this paper on cell morphology and actin distribution during both vegetative growth and chemotaxis in the SCAR− strain is consistent with the idea that the SCAR protein plays a role in regulating actin polymerization. In vegetative cells the lower amount of F-actin suggests that SCAR may act to enhance actin polymerization. Whether this effect is direct or indirect remains to be elucidated, but SCAR's potential binding sites for both profilin, a protein known to be involved in the polymerization of actin (Pantaloni and Carlier, 1993), and actin itself suggests that SCAR's effects on polymerization may be fairly direct. The exact nature of SCAR's effect(s) on actin polymerization are currently being pursued through in vivo and in vitro assays. The aberrant cell morphology and actin staining seen during aggregation in the SCAR− cells suggests that this gene product may also participate in the signal transduction pathway from cell surface receptors to the reorganization of the actin cytoskeleton. This result supports the hypothesis that SCAR was identified as a suppressor of the carB null strain because it participates in a pathway that connects the cAR2 receptor to the actin cytoskeleton. The ultimate result of such a signaling cascade is multicellular morphogenesis regulation.
Acknowledgments
The authors would like to thank Sonya Greene, John Crowe, Cheryl Jones, and all the members of the Saxe lab for technical assistance and support. We also thank Laura Fox and Win Sale for assistance with videomicroscopy, Win Sale and Art English for assistance in using NIH Image to quantitate F-actin and G-actin levels, and Danny Rouk for assistance with graphics. We would also especially thank Steve Alexander for providing the finger stage cDNA library. For communicating results before publication, we thank Richard Firtel and Chang Chung.
This work was supported by National Institutes of Health grant GM45705 to C.L. Saxe.
Abbreviations used in this paper
- ABM-2
actin-based motility type 2 repeat
- DB
developmental buffer
- EST
expressed sequence tag
- REMI
restriction enzyme–mediated integration
- SCAR
suppressor of cAR
- SH3
Src homology 3 domain
- SHD
SCAR Homology domain
- WASP
Wiskott-Aldrich syndrome protein
- WH2
WASP homology 2 domain
- WT
wild-type
Footnotes
The present address of James E. Bear is Center for Cancer Research and Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139. The present address of John F. Rawls is Department of Genetics, Washington University, St. Louis, Missouri 63110.
Address all correspondence to Charles L. Saxe III, Department of Cell Biology, Emory University School of Medicine, 1648 Pierce Drive, Atlanta, GA 30322. Tel.: 404-727-6248; Fax: 404-727-6256; E-mail: karl@cellbio.emory.edu
References
- Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideumcytokinesis mutants by restriction enzyme-mediated integration of the Blasticidin S resistence marker. Biochem Biophys Res Comm. 1994;205:1808–1814. doi: 10.1006/bbrc.1994.2880. [DOI] [PubMed] [Google Scholar]
- Aspenström P, Lindberg U, Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr Biol. 1996;6:70–75. doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Inouye CJ, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. . Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Chan DC, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO (Eur Mol Biol Organ) J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks M, Kay RR. Combinatorial control of cell differentiation by cAMP and DIF-1 during development of Dictyostelium discoideum. . Development. 1990;110:977–984. doi: 10.1242/dev.110.3.977. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Devreotes PN. Molecular insights into eukaryotic chemotaxis. FASEB J. 1991;5:3078–3085. [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Ostrow BD, Tafuri SR, Chisholm RL. Targeted disruption of the DictyosteliumRMLC gene produces cells defective in cytokinesis and development. J Cell Biol. 1994;127:1933–1944. doi: 10.1083/jcb.127.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-Y, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. . Trends Genet. 1996;12:52–57. doi: 10.1016/0168-9525(96)81400-4. [DOI] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyosteliummyosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Derry JMJ, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich Syndrome. Cell. 1994;78:635–644. [Google Scholar]
- Devreotes PN. G Protein–linked signaling pathways control the developmental program of Dictyostelium. . Neuron. 1994;12:235–241. doi: 10.1016/0896-6273(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Dharmawardhane S, Warren V, Hall AE, Condeelis J. Changes in the association of actin-binding proteins with the actin cytoskeleton during chemotactic stimulation of Dictyostelium discoideum. . Cell Motil Cytoskelet. 1989;13:57–63. doi: 10.1002/cm.970130107. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha Subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly SFH, Pocklington MJ, Pallotta D, Orr E. A proline-rich protein, verprolin, involved in cytoskeletal organization and cellular growth in the yeast Saccharomyces cerevisiae. . Mol Microbiol. 1993;10:585–596. doi: 10.1111/j.1365-2958.1993.tb00930.x. [DOI] [PubMed] [Google Scholar]
- Ermekova KS, Zambrano N, Linn H, Minopolis G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homologue of Drosophila enabled. . J Biol Chem. 1997;272:32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Finan PM, Soames CJ, Wilson L, Nelson DL, Stewart DM, Truong O, Hsuan JJ, Kellie S. Identification of regions of the Wiskott-Aldrich Syndrome protein responsible for Association with selected Src homology 3 domains. J Biol Chem. 1996;271:26291–26295. doi: 10.1074/jbc.271.42.26291. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Mikoshiba K. The function of inositol high polyphosphate binding proteins. Bioessays. 1997;19:593–603. doi: 10.1002/bies.950190710. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Yumura S, Yumura TK. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and the their changes during chemotaxis. Methods Cell Biol. 1987;28:347–356. doi: 10.1016/s0091-679x(08)61655-6. [DOI] [PubMed] [Google Scholar]
- Gallaego MD, Santamaria M, Peña J, Molina IJ. Defective actin reorganization and polymerization of Wiskott-Aldrich T cells in response to CD3-mediated stimulation. Blood. 1997;90:3089–3097. [PubMed] [Google Scholar]
- Gaul U, Mardon G, Rubin GM. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Ginsburg GT, Gollop R, Yu Y, Louis JM, Saxe CL, Kimmel AR. The regulation of Dictyosteliumdevelopment by transmembrane signaling. J Euk Microbiol. 1995;42:200–205. doi: 10.1111/j.1550-7408.1995.tb01565.x. [DOI] [PubMed] [Google Scholar]
- Haugland RP, You W, Paragas VB, Wells KS, DuBose DA. Simultaneous visualization of G- and F-actin in endothelial cells. J Histochem Cytochem. 1994;42:345–350. doi: 10.1177/42.3.8308251. [DOI] [PubMed] [Google Scholar]
- Haugwitz M, Noegel AA, Karakesisoglou J, Schleicher M. Dictyosteliumamoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell. 1994;79:303–314. doi: 10.1016/0092-8674(94)90199-6. [DOI] [PubMed] [Google Scholar]
- Howard PK, Sefton BM, Firtel RA. Analysis of a spatially regulated phosphotyrosine phosphatase identifies tyrosine phosphorylation as a key regulator pathway in Dictyostelium. . Cell. 1992;71:637–647. doi: 10.1016/0092-8674(92)90597-6. [DOI] [PubMed] [Google Scholar]
- Howard PK, Sefton BM, Firtel RA. Tyrosine phosphorylation of actin in Dictyosteliumassociated with cell-shape changes. Science. 1993;259:241–244. doi: 10.1126/science.7678470. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets fo the Rho family of small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness DD, Goldman BS, Hartwell LH. Saccharomyces cerevisiaemutants unresponsive to alpha-factor binding and extragenic suppression. Mol Cell Biol. 1987;7:1311–1319. doi: 10.1128/mcb.7.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney D, Cairns L, Remold-O'Donnell E, Peterson J, Rosen FS, Parkman R. Morphological abnormalities in the lymphocytes of patients with the Wiskott-Aldrich syndrome. Blood. 1986;68:1329–1332. [PubMed] [Google Scholar]
- Kirchhausen T, Rosen FS. Disease mechanism: unravelling Wiskott-Aldrich syndrome. Curr Biol. 1996;6:676–678. doi: 10.1016/s0960-9822(09)00447-3. [DOI] [PubMed] [Google Scholar]
- Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegansnervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- Kolluri R, Shehabeldin A, Peacocke M, Lamhonwah A-M, Teichert-Kuliszewska K, Weissman S, Siminovitch KA. Identification of WASP mutations in patients with Wiskott-Aldrich syndrome and isolated thrombocytopenia reveals allelic heterogeneity at the WAS locus. Human Mol Genet. 1995;4:1119–1126. doi: 10.1093/hmg/4.7.1119. [DOI] [PubMed] [Google Scholar]
- Kolluri R, Tolias KF, Carpenter CL, Rosen FS, Kirchhausen T. Direct interaction of the Wiskott-Aldrich syndrome protein with the GTPase Cdc42. Proc Natl Acad Sci USA. 1996;93:5615–5618. doi: 10.1073/pnas.93.11.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyosteliumby restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z-C, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yangene, an ETS domain protein. Cell. 1992;70:609–620. doi: 10.1016/0092-8674(92)90430-k. [DOI] [PubMed] [Google Scholar]
- Larochelle DA, Vithalani KK, De Lozanne A. A novel member of the rho family of small GTP-binding is specifically required for cytokinesis. J Cell Biol. 1996;133:1321–1330. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical acin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeleton rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO (Eur Mol Biol Organ) J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998a;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Miki H, Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem Biophys Res Commun. 1998b;243:73–78. doi: 10.1006/bbrc.1997.8064. [DOI] [PubMed] [Google Scholar]
- Molina IJ, Kenney DM, Rosen FS, Remold-O'Donnell E. T cell lines characterize events in the pathogensis of the Wiskott-Aldrich syndrome. J Exp Med. 1992;176:867–874. doi: 10.1084/jem.176.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe DJ, Loebbert R, Bric E, Whitton T, Prawitt D, Vu D, Buckler A, Winterpacht A, Zabel B, Houseman DE. Systematic screening of an arrayed cDNA library by PCR. Proc Natl Acad Sci USA. 1995;92:2209–2213. doi: 10.1073/pnas.92.6.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs HD, Slichter SJ, Harker LA, Von Behrens WE, Clark RA, Wedgewood RJ. The Wiskott-Aldrich syndrome: studies of lymphocytes, granulocytes, and platelets. Blood. 1980;55:243–252. [PubMed] [Google Scholar]
- Pantaloni D, Carlier M-F. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. . Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Purich DL, Southwick FS. ABM-1 and ABM-2 homology sequences: consensus docking sites for actin-based motility defined by oligoproline regions in listeria ActA surface protein and human VASP. Biochem Biophys Res Comm. 1997;231:686–691. doi: 10.1006/bbrc.1997.6158. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO (Eur Mol Biol Organ) J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retta RF, Barry ST, Critchley DR, Defilippi P, Silengo L, Tarone G. Focal adhesions and stess fiber formation is regulated by tyrosine phosphatase activity. Exp Cell Res. 1996;229:307–317. doi: 10.1006/excr.1996.0376. [DOI] [PubMed] [Google Scholar]
- Reymond CD, Gomer RH, Nellen W, Theibert A, Devreotes PN, Firtel RA. Phenotypic changes induced by a mutated ras gene during the development of Dictyosteliumtransformants. Nature. 1986;323:340–343. doi: 10.1038/323340a0. [DOI] [PubMed] [Google Scholar]
- Rivero F, Köppel B, Perancino B, Bozzaro S, Siegert F, Weijer CJ, Shleicher M, Albrecht R, Noegel AA. The role of cortical cytoskeleton: F-actin crosslinking proteins protect against osmotic stress, ensure cell size, cell shape and motility, and contribute to phagocytosis and development. J Cell Sci. 1996;109:2679–2691. doi: 10.1242/jcs.109.11.2679. [DOI] [PubMed] [Google Scholar]
- Saxe CL, III, Ginsburg GT, Louis JM, Johnson R, Devreotes PN, Kimmel AR. CAR2, a prestalk cAMP receptor required for normal tip formation and late development of Dictyostelium discoideum. . Genes Dev. 1993;7:262–272. doi: 10.1101/gad.7.2.262. [DOI] [PubMed] [Google Scholar]
- Saxe CL, III, Yu Y, Jones CJ, Bauman A, Haynes C. The cAMP receptor subtype cAR2 is restricted to a subset of prestalk cells during Dictyosteliumdevelopment and displays unexpected DIF-1 responsiveness. Dev Biol. 1996;174:202–213. doi: 10.1006/dbio.1996.0066. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Escalante R, Loomis WF. Developmental signal transduction pathways uncovered by genetic suppressors. Proc Natl Acad Sci USA. 1996;93:15260–15265. doi: 10.1073/pnas.93.26.15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Vaduva G, Martin NC, Hopper AK. Actin-binding Verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J Cell Biol. 1997;139:1821–1833. doi: 10.1083/jcb.139.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jochusch BM, Narumiya S. p140mDia, a mammalian homologue of Drosophila diaphanous, is a target protein for Rho small GTPase and is ligand for profilin. EMBO (Eur Mol Biol Organ) J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Takegawa K, Emr SD, Firtel RA. A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vsp34p PI 3-kinase homologues during growth and development. Mol Cell Biol. 1995;15:5645–5656. doi: 10.1128/mcb.15.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH. Signal transduction and actin filament organization. Curr Opin Cell Biol. 1996;8:66–73. doi: 10.1016/s0955-0674(96)80050-0. [DOI] [PubMed] [Google Scholar]