Abstract
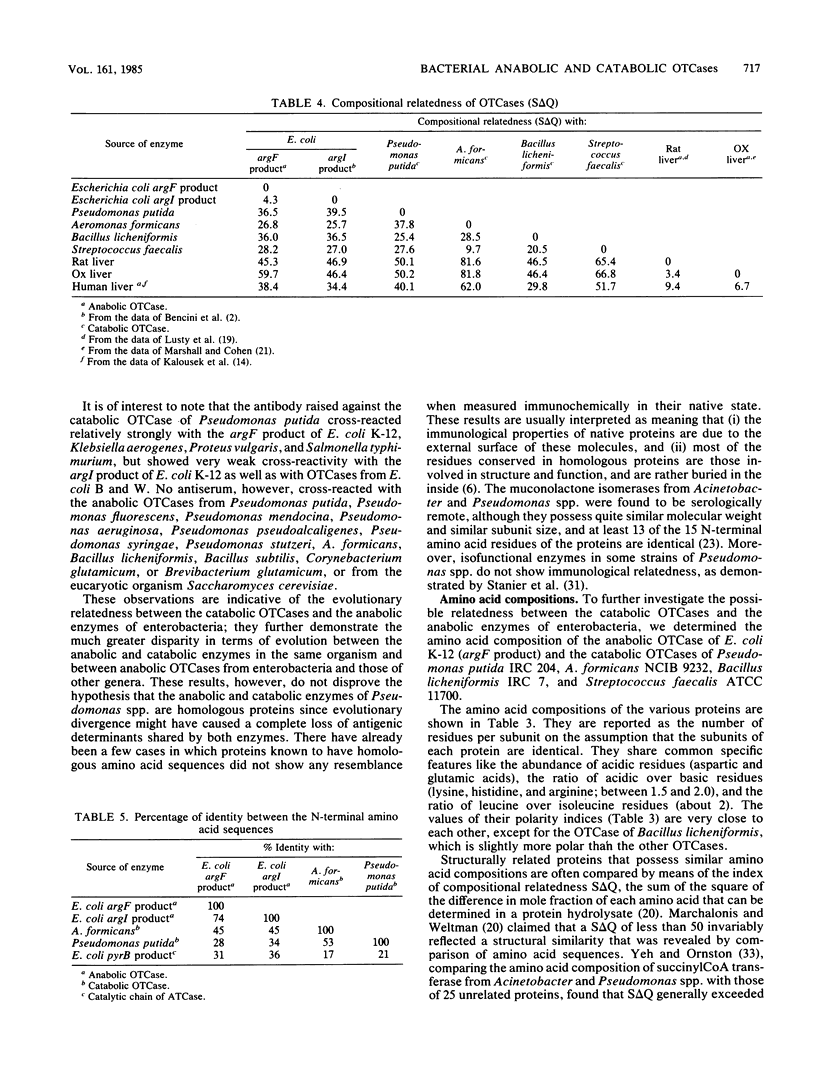
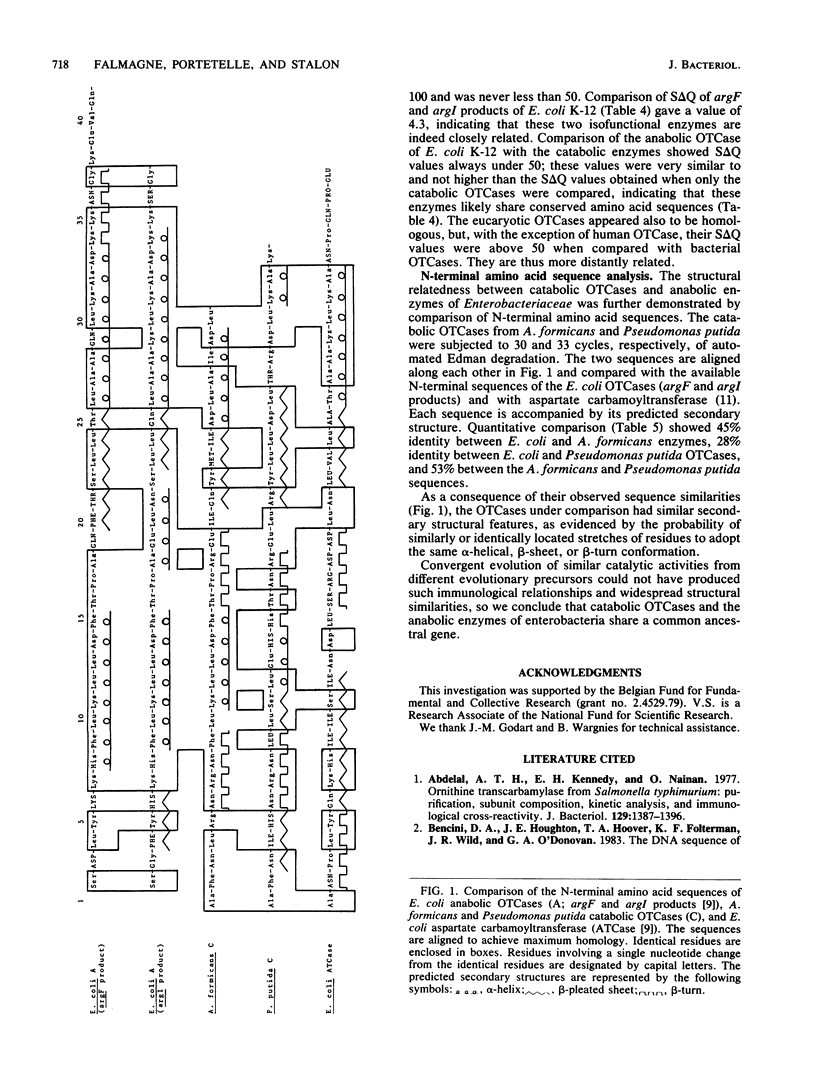
Purified catabolic ornithine carbamoyltransferase of Pseudomonas putida and anabolic ornithine carbamoyltransferase (argF product) of Escherichia coli K-12 were used to prepare antisera. The two specific antisera gave heterologous cross-reactions of various intensities with bacterial catabolic ornithine carbamoyltransferases formed by Pseudomonas and representative organisms of other bacterial genera. The immunological cross-reactivity observed only between the catabolic ornithine carbamoyltransferases and the anabolic enzymes of enterobacteria suggests that these proteins share some structural similarities. Indeed, the amino acid composition of the anabolic ornithine carbamoyltransferase of E. coli K-12 (argF and argI products) closely resembles the amino acid compositions of the catabolic enzymes of Pseudomonas putida, Aeromonas formicans, Streptococcus faecalis, and Bacillus licheniformis. Comparison of the N-terminal amino acid sequence of the E. coli anabolic ornithine carbamoyltransferase with that of the A. formicans and Pseudomonas putida catabolic enzymes shows, respectively, 45 and 28% identity between the compared positions; the A. formicans sequence reveals 53% identity with the Pseudomonas putida sequence. These results favor the conclusion that anabolic ornithine carbamoyltransferases of enterobacteria and catabolic ornithine carbamoyltransferases derive from a common ancestral gene.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdelal A. T., Kennedy E. H., Nainan O. Ornithine transcarbamylase from Salmonella typhimurium: purification, subunit composition, kinetic analysis, and immunological cross-reactivity. J Bacteriol. 1977 Mar;129(3):1387–1396. doi: 10.1128/jb.129.3.1387-1396.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K., Lauwers N., Stalon V., Wiame J. M. Oxygen and nitrate in utilization by Bacillus licheniformis of the arginase and arginine deiminase routes of arginine catabolism and other factors affecting their syntheses. J Bacteriol. 1978 Sep;135(3):920–927. doi: 10.1128/jb.135.3.920-927.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck C., Portetelle D., Burny A., Zavada J. Topographical analysis by monoclonal antibodies of BLV-gp51 epitopes involved in viral functions. Virology. 1982 Oct 30;122(2):353–362. doi: 10.1016/0042-6822(82)90235-5. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion A. B., Soderberg K. L., Wilson A. C. Immunological comparison of azurins of known amino acid sequence. Dependence of cross-reactivity upon sequence resemblance. J Mol Evol. 1975 Sep 8;5(4):291–305. doi: 10.1007/BF01732216. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Beta-turns in proteins. J Mol Biol. 1977 Sep 15;115(2):135–175. doi: 10.1016/0022-2836(77)90094-8. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Crabeel M., Charlier D., Cunin R., Glansdorff N. Cloning and endonuclease restriction analysis of argF and of the control region of the argECBH bipolar operon in Escherichia coli. Gene. 1979 Mar;5(3):207–231. doi: 10.1016/0378-1119(79)90079-9. [DOI] [PubMed] [Google Scholar]
- Gigot D., Glansdorff N., Legrain C., Piérard A., Stalon V., Konigsberg W., Caplier I., Strosberg A. D., Hervé G. Comparison of the N-terminal sequences of aspartate and ornithine carbamoyltransferases of Escherichia coli. FEBS Lett. 1977 Sep 1;81(1):28–32. doi: 10.1016/0014-5793(77)80920-4. [DOI] [PubMed] [Google Scholar]
- Halleux P., Legrain C., Stalon V., Piérard A., Wiame J. M. Regulation of the catabolic ornithine carbamoyltransferase of Pseudomonas fluorescens. A study of the quaternary structure. Eur J Biochem. 1972 Dec 4;31(2):386–393. doi: 10.1111/j.1432-1033.1972.tb02545.x. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. Direct microsequence analysis of polypeptides using an improved sequenator, a nonprotein carrier (polybrene), and high pressure liquid chromatography. Biochemistry. 1978 May 30;17(11):2124–2133. doi: 10.1021/bi00604a016. [DOI] [PubMed] [Google Scholar]
- Kalousek F., François B., Rosenberg L. E. Isolation and characterization of ornithine transcarbamylase from normal human liver. J Biol Chem. 1978 Jun 10;253(11):3939–3944. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Legrain C., Halleux P., Stalon V., Glansdorff N. The dual genetic control of ornithine carbamolytransferase in Escherichia coli. A case of bacterial hybrid enzymes. Eur J Biochem. 1972 May;27(1):93–102. doi: 10.1111/j.1432-1033.1972.tb01814.x. [DOI] [PubMed] [Google Scholar]
- Legrain C., Stalon V., Noullez J. P., Mercenier A., Simon J. P., Broman K., Wiame J. M. Structure and function of ornithine carbamoyltransferases. Eur J Biochem. 1977 Nov 1;80(2):401–409. doi: 10.1111/j.1432-1033.1977.tb11895.x. [DOI] [PubMed] [Google Scholar]
- Legrain C., Stalon V. Ornithine carbamoyltransferase from Escherichia coli W. Purification, structure and steady-state kinetic analysis. Eur J Biochem. 1976 Mar 16;63(1):289–301. doi: 10.1111/j.1432-1033.1976.tb10230.x. [DOI] [PubMed] [Google Scholar]
- Lusty C. J., Jilka R. L., Nietsch E. H. Ornithine transcarbamylase of rat liver. Kinetic, physical, and chemical properties. J Biol Chem. 1979 Oct 25;254(20):10030–10036. [PubMed] [Google Scholar]
- Marshall M., Cohen P. P. Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. I. Isolation and subunit structure. J Biol Chem. 1972 Mar 25;247(6):1641–1653. [PubMed] [Google Scholar]
- Mercenier A., Simon J. P., Vander Wauven C., Haas D., Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol. 1980 Oct;144(1):159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Meagher R. B., Ornston L. N. Relationships among enzymes of the beta-ketoadipate pathway. IV. Muconolactone isomerase from Acinetobacter calcoaceticus and Pseudomonas putida. J Biol Chem. 1974 Dec 10;249(23):7410–7419. [PubMed] [Google Scholar]
- Penke B., Ferenczi R., Kovács K. A new acid hydrolysis method for determining tryptophan in peptides and proteins. Anal Biochem. 1974 Jul;60(1):45–50. doi: 10.1016/0003-2697(74)90129-8. [DOI] [PubMed] [Google Scholar]
- Stalon V., Legrain C., Wiame J. M. Anabolic ornithine carbamolytransferase of Pseudomonas. The bases of its functional specialization. Eur J Biochem. 1977 Apr 1;74(2):319–327. doi: 10.1111/j.1432-1033.1977.tb11396.x. [DOI] [PubMed] [Google Scholar]
- Stalon V., Mercenier A. L-arginine utilization by Pseudomonas species. J Gen Microbiol. 1984 Jan;130(1):69–76. doi: 10.1099/00221287-130-1-69. [DOI] [PubMed] [Google Scholar]
- Stalon V., Ramos F., Piérard A., Wiame J. M. Regulation of the catabolic ornithine carbamoyltransferase of Pseudomonas fluorescens. A comparison with the anabolic transferase and with a mutationally modified catabolic transferase. Eur J Biochem. 1972 Aug 18;29(1):25–35. doi: 10.1111/j.1432-1033.1972.tb01953.x. [DOI] [PubMed] [Google Scholar]
- Stalon V., Ramos F., Piérard A., Wiame J. M. The occurrence of a catabolic and an anabolic ornithine carbamoyltransferase in Pseudomonas. Biochim Biophys Acta. 1967 May 16;139(1):91–97. doi: 10.1016/0005-2744(67)90115-5. [DOI] [PubMed] [Google Scholar]
- Stalon V., Simon J. P., Mercenier A. Enzymes of arginine utilization and their formation in Aeromonas formicans NCIB 9232. Arch Microbiol. 1982 Dec 3;133(4):295–299. doi: 10.1007/BF00521293. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Wachter D., Gasser C., Wilson A. C. Comparative immunological studies of two Pseudomonas enzymes. J Bacteriol. 1970 May;102(2):351–362. doi: 10.1128/jb.102.2.351-362.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargnies B., Lauwers N., Stalon V. Structure and properties of the putrescine carbamoyltransferase of Streptococcus faecalis. Eur J Biochem. 1979 Nov 1;101(1):143–152. doi: 10.1111/j.1432-1033.1979.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Yeh W. K., Ornston L. N. Evolutionarily homologous alpha 2 beta 2 oligomeric structures in beta-ketoadipate succinyl-CoA transferases from Acinetobacter calcoaceticus and Pseudomonas putida. J Biol Chem. 1981 Feb 25;256(4):1565–1569. [PubMed] [Google Scholar]



