SUMMARY
To investigate the disposal of non-glycosylated BiP substrates, we used a non-secreted κ LC, which exists in a partially (ox1) and completely (ox2) oxidized state. The ox2 form is partially reduced in order to be degraded and only the ox1 form is ubiquitinated and associates with both Herp, and Derlin-1. Herp is in a complex with ubiquitinated proteins and with the 26S proteasome, suggesting that it plays a role in linking substrates with the proteasome. Over-expressed Herp also interacts with two other BiP substrates but not with two calnexin substrates. Either expression of p97 or Hrd1 mutants, which are in a complex with Herp and Derlin-1, or reducing Herp levels inhibited the degradation of the BiP substrates, whereas the latter had no effect on the degradation of the calnexin substrates. This suggests that there is some distinction in the pathways used to dispose of these two types of ERAD substrates.
INTRODUCTION
The biosynthesis of proteins in the endoplasmic reticulum (ER) is tightly monitored by a mechanism termed ER quality control to assure that only properly folded and assembled proteins reach their final destination. Those proteins that fail to mature properly are identified and retrotranslocated for degradation by the 26S proteasome via a process known as ER associated degradation (ERAD) (Ellgaard et al., 1999). The ERAD process begins with the recognition of a substrate as being misfolded or unfolded. Next, the substrate is transported across the ER membrane through a proteinaceous channel called the retrotranslocon, which shares some properties with the translocon. Once in the cytosol, the substrate is polyubiquitinated and degraded by the 26S proteasome (Meusser et al., 2005). In S. cerevisiae, there are three major ERAD sub-pathways defined by which region of the protein is misfolded (i.e., luminal, transmembrane, or cytosolic). These pathways utilize distinct ubiquitin-ligase complexes and vary in their reliance on Der1p and Usa1p (Carvalho et al., 2006; Denic et al., 2006). Proteins with misfolded ER-luminal domains use the ERAD-L pathway, in which the Hrd1p/Hrd3p ubiquitin ligase forms a near stoichiometric membrane core complex by binding to Der1p via the linker protein Usa1p. Whereas proteins with misfolded transmembrane or cytosolic domains are not dependent on either Der1p or Usa1p. Der1p possesses four transmembrane domains and is thought to constitute part of the channel for retrotranslocation of this group of substrates (Hiller et al., 1996; Knop et al., 1996). All three ERAD sub-pathways converge at the Cdc48p ATPase complex, which uses the energy of hydrolysis to extract the substrate (Rabinovich et al., 2002). Mammalian equivalents of a number of these proteins including three different Der1p homologues, Derlin-1, Derlin-2 and Derlin-3, have been identified (Ye et al., 2004)(Lilley and Ploegh, 2004)(Oda et al., 2006), although it is still unclear if distinct ERAD sub-pathways exist in mammalian cells.
Recent progress has been made in elucidating ERAD pathways for misfolded glycoproteins in mammalian cells. Most glycoproteins interact with the calnexin/calreticulin chaperone family during folding. These chaperones monitor the cyclic processing of N-linked glycans that occurs on unfolded glycoproteins (Hebert et al., 1995). If mannosidases trim the glycan too far, the protein is no longer a substrate for reglucosylation and instead is recognized by an ER-degradation enhancing alpha-mannosidase-like protein (EDEM), which targets the protein for retrotranslocation and degradation (Oda et al., 2003)(Molinari et al., 2003). The misfolded α1-antitrypsin NullHong Kong (AAT NHK) glycoprotein is a substrate of calnexin/calreticulin (Cabral et al., 2001) and requires either Derlin-2 or -3 for its degradation but its turnover is not affected when Derlin-1 levels are reduced (Oda et al., 2006).
It is much less clear how unfolded, non-glycosylated proteins that utilize BiP are recognized and targeted for degradation. Studies in yeast demonstrate that BiP is required for the retrotranslocation of a number of ERAD substrates (Plemper et al., 1997)(Brodsky et al., 1999). In mammalian cells, it has been reported that the BiP binding domain controls the rate of degradation of some Ig LC mutants (Skowronek et al., 1998). To elucidate the ERAD pathway for non-glycosylated BiP substrates, we used three different BiP binding proteins; a non-secreted Ig κ LC, a mutant Ig λ LC, and a truncated Ig γ HC. We find that all three BiP substrates interact with a subset of ERAD components, which includes ones that are not used to dispose of glycosylated AAT mutants.
RESULTS
Non-secreted κ LC in P3U.1 (γ−, κ+) cells are degraded by the 26S proteasome
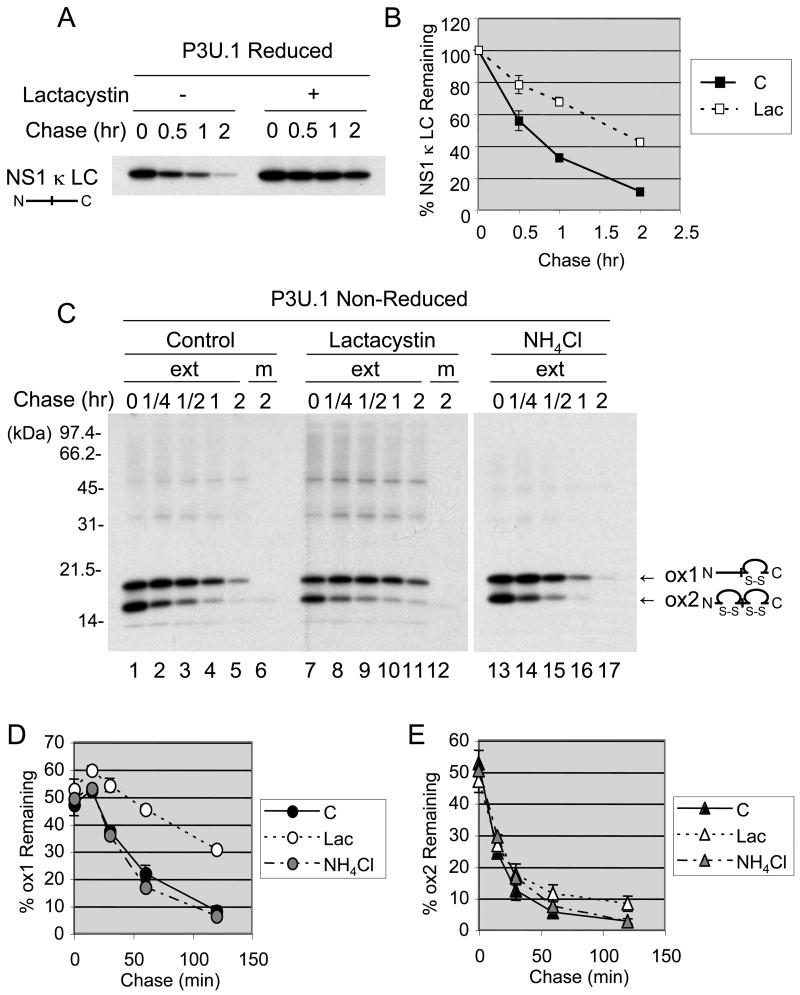
The non-glycosylated, non-secreted κ LC produced in the P3U.1 plasmacytoma line binds to BiP (Ma et al., 1990) and is degraded by the 26S proteasome (Chillaron and Haas, 2000), which strongly indicates the existence of an ERAD pathway for this group of proteins. Thus, in order to better understand this ERAD pathway, we first examined the half-life of this LC, which is also referred to as NS1 κ LC, in the presence and absence of lactacystin. Examination of the metabolically labeled LC under reducing conditions demonstrated that lactacystin significantly inhibited degradation of the unassembled κ LC, confirming that it is an ERAD substrate (Fig. 1A and 1B). The NS1 κ LC is composed of two Ig domains and each domain has a pair of cysteine residues, which form an intrachain disulfide bond. In addition, a penultimate cysteine forms the covalent interaction with Ig HC. NS1 κ LC exists in both a partially (ox1) and fully (ox2) oxidized form, which can be distinguished on non-reducing SDS gels (Knittler et al., 1995). When the immunoprecipitated LC were examined under non-reducing conditions, we found that the level of the ox1 form increased initially at 15 min (Fig. 1C and 1D), both in the presence and absence of lactacystin. The ox1 form then decreased gradually in the absence of lactacystin but was stabilized in its presence. Conversely, the level of the ox2 form decreased rapidly under both conditions (Fig. 1C and 1E). It should be noted that neither form of the NS1 κ LC was secreted into the medium during the 2 hours of chase (Fig. 1C, lanes 6 and 12). Incubation of cells with NH4Cl, an inhibitor of lysosomal degradation, did not affect the turn-over of either form of the κ LC (Fig. 1C, D, and E), indicating that the ox2 form was not transported to the lysosomes for degradation. Thus, it appears that both the ox1 and ox2 intermediates are ultimately degraded by the 26S proteasome. The fact that the ox1 form is preferentially stabilized by lactacystin suggests that the ox2 form may need to be partially reduced to the ox1 form in order to be degraded, and that this step occurs somewhere prior to the lactacystin-mediated inhibition of the proteasome. Interestingly, we saw no evidence of the accumulation of a fully reduced form of LC when degradation was inhibited.
Figure 1. Non-secreted κ LC in P3U.1 (γ−, κ +) are degraded by the 26S proteasome.
(A) P3U.1 cells were pre-incubated in the presence or absence of 10 μM lactacystin, labeled for 15 min and chased for the indicated times in the presence or absence of lactacystin. NS1 κ LC were immunoprecipitated from cell extracts and subjected to SDS-PAGE under reducing conditions, followed by autoradiography. (B) The intensity of the κ LC band in each lane was expressed as a percent of the signal present at time=0 (100%) and plotted. The mean values and standard errors were calculated from two independent experiments. (C) Cells were prepared as in A except that labeling period was 2 min and the chased was performed alone, with lactacystin, or with NH4Cl as indicated. NS1 κ LC were immunoprecipitated from cell extracts (ext) or medium (m) and subjected to SDS-PAGE under non-reducing conditions, followed by autoradiography. Two different folding forms of the NS1 κ LC (ox1; a partially oxidized form and ox2; a completely oxidized form) are indicated. The ox1 (D) and ox2 (E) levels of κ LC in each lane in (C) were quantified and expressed as a percent of total amount of LC present at t=0. The mean values and standard errors were calculated from two independent experiments.
The partially oxidized form of non-secreted κ LC in P3U.1 cells is ubiquitinated
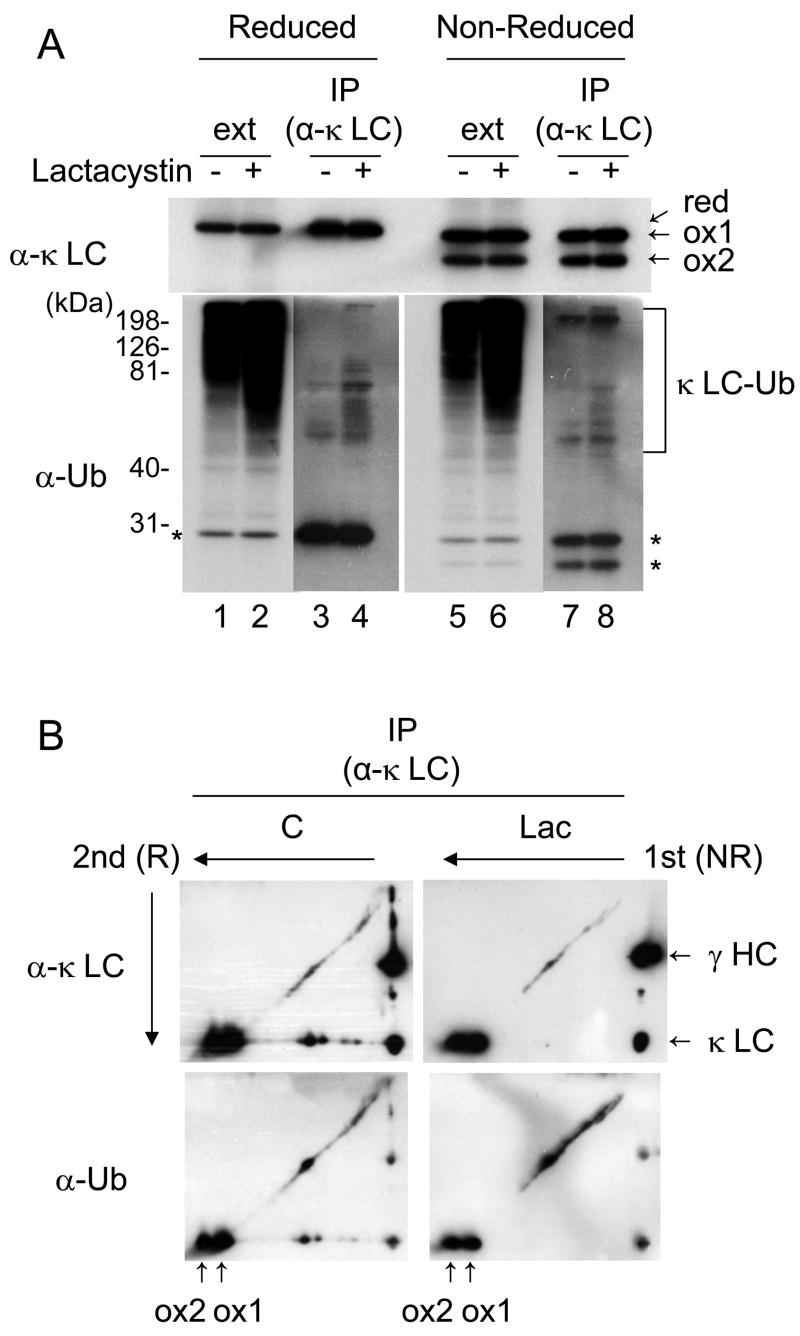
We next determined if the NS1 κ LC was ubiquitinated in P3U.1 cells. Cell lysates were prepared from control and lactacystin-treated cells and LC were immunoprecipitated. These samples were then resolved under reducing conditions as well as non-reducing conditions in order to detect both forms of the κ LC (Fig. 2A). Immunoblot analysis using a monoclonal antibody that recognizes ubiquitinated proteins revealed the accumulation of ubiquitin-conjugated molecules in the whole cell extract after lactacystin treatment (Fig. 2A lane 1, 2, 5 and 6). When the immunoprecipitated κ LC were similarly analyzed, we detected slower migrating forms of the NS1 κ LC, which increased with lactacystin treatment and were recognized by an anti-ubiquitinated proteins antibody, demonstrating that they represented ubiquitinylated forms κ LC (Fig. 2A lane 3, 4, 7 and 8). The same immunoprecipitated samples were next analyzed by 2-dimensional SDS-PAGE (1st dimension under non-reducing conditions and 2nd under reducing conditions) followed by immunoblotting with either a polyclonal goat anti-κ LC or the monoclonal antibody that recognizes ubiquitinated proteins. Examination of the filters with the anti-κ LC antibody illuminated two spots on the far left that corresponded to the ox1 and ox2 forms of the LC, as well as a diagonal that emanated from the ox1 but not the ox2 form (Fig. 2B). This diagonal was also recognized by the anti-ubiquitinated proteins antibody, demonstrating that only the ox1 form is a substrate for ubiquitination. Unfortunately, the secondary antibody (goat anti-mouse Ig) used to detect the mouse monoclonal anti-ubiquitinated proteins antibody also recognizes the mouse κ LC produce by this plasmacytoma line and results in an additional complication for interpreting the data, since the unmodified LC are also detected (Fig. 2A lower panel and 2B lower panel). Nonetheless, there is no evidence of poly-ubiquinated forms trailing behind the ox2 form of the NS1 κ LC in these 2-D blots. Together with the data in Fig.1, this strongly suggests that the ox2 form is converted to the ox1 form in order to be retro-translocated, ubiquitinated, and degraded.
Figure 2. The partially oxidized form of non-secreted κ LC in P3U.1 cells is ubiquitinated.
(A) Cell lysates from P3U.1 cells treated in the presence or absence of lactacystin for 6 hr were immunoprecipitated with anti-κ LC antibody, and SDS-PAGE separation was performed under reducing or non-reducing conditions, followed by immunoblot analysis. The three different folding forms of the NS1 κ LC were detected (red, a completely reduced form, ox1 and ox2) in the top panel. In the bottom panel the ubiquitinated forms of LC are shown and the unmodified NS1 LC, which are also detected by the secondary antibody (goat anti-mouse Ig), are indicated with asterisks. (B) The same immunoprecipitated samples as in (A) were analyzed by 2-dimentional SDS-PAGE, followed by immunoblotting with the indicated antibodies.
NS1 κ LC interacts with Herp and Derlin-1
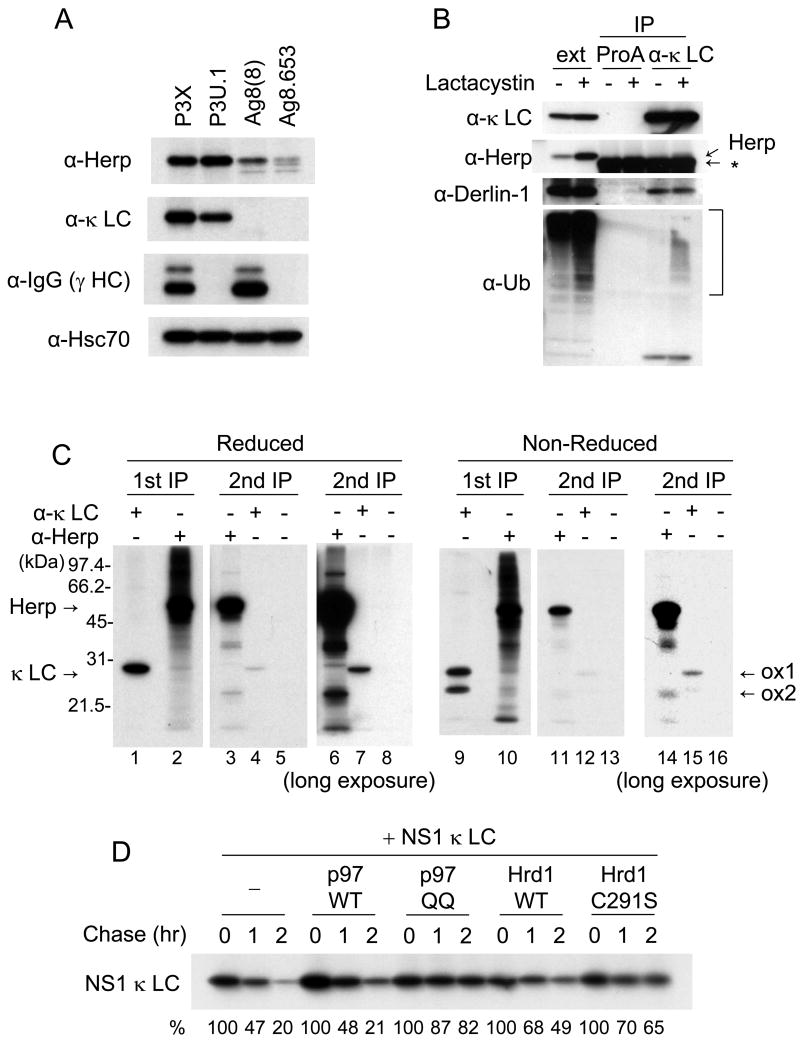
Since our data revealed that the NS1 κ LC is ubiquitinated and degraded by the proteasome, we wished to determine how it was extracted from the ER lumen. Recently the degradation of a glycosylated calnexin substrate was shown to be dependent on Derlin-2 and -3 expression but on not Derlin-1 (Oda et al., 2006). Derlin-1 is in a complex with Herp, the AAA ATPase Cdc48/p97, and the ubiquitin ligase Hrd1 (Schulze et al., 2005). As a first step toward determining if the degradation of LC might require this protein complex, we compared the expression of Herp protein in four related plasmacytoma lines with different Ig expression patterns; P3X (γ+, κ+), P3U.1 (γ−, κ+), Ag8(8) (γ+, κ−), and Ag8.653 (γ−, κ−) (Fig. 3A). Herp was highly expressed in P3X and P3U.1 cells, with Ag8(8) cells expressing much lower levels, and the Ig− Ag8.563 cells showing only minimally detectable levels of Herp (Fig. 3A). This pattern appeared to correlate better with κ LC expression and not simply with the accumulation of unfolded proteins, as the expression of Herp was fairly low in Ag8(8) cells, which accumulate large quantities of incompletely folded HC that are turned over quite slowly (Lee et al., 1999). In addition, although the κ LC level in P3U.1 was slightly lower than that in P3X, Herp levels were actually somewhat higher in P3U.1 cells (Fig. 3A). This might reflect the fact that only a portion of the LC fail to assembled and are degraded in P3X cells, which secrete IgG κ, whereas all of the LC in P3U.1 cells are retained and degraded (data not shown). Therefore, we concentrated on the P3U.1 cells for the remaining experiments.
Figure 3. The partially oxidized form of non-secreted κ LC, which is a BiP substrate, interacts with Herp and Derlin-1.
(A) Whole cell extracts were prepared from four related plasmacytoma cell lines; P3X (γ+, κ+), P3U.1 (γ−, κ+), Ag8(8) (γ+, κ−) and Ag8.653 (γ−, κ−). Proteins were subjected to SDS-PAGE followed by immunoblot analysis using antibodies as indicated. Hsc70 was detected as a loading control. (B) P3U.1 cells treated with or without lactacystin for 6h were lysed in the presence of apyrase. After removing a portion (ext), NS1 κ LC were immunoprecipitated with anti-κ LC or Protein A Sepharose alone as negative control and subjected to immunoblot analysis as indicated. A non-specific band detected with the anti-Herp antiserum is indicated with an asterisk. (C) P3U.1 cells were labeled with 35S-methionine/cysteine in the presence of lactacystin for 6 hr and lysed as in (B) and extracts were immunoprecipitated with anti-Herp or anti-κ LC antisera. The proteins precipitated with anti-Herp antiserum was further subjected to second immunoprecipitation by using anti-Herp (H), anti-κ LC antisera or Protein A Sepharose alone. Samples were subjected to SDS-PAGE under reducing or non-reducing conditions, followed by autoradiography. (D) NS κ LC were transiently expressed in 293T cells together with an empty vector control, p97 wild type, p97 QQ, Hrd1 wild type or Hrd1 C291S. At 24 hr post-transfection, cells were labeled with 35S-methionine/cysteine for 15 min and chased for the indicated times. LC were immunoprecipitated and subjected to SDS-PAGE, followed by autoradiography. The κ LC band in each lane was quantified and expressed as a percent of the signal present at t=0. The values are shown under each lane.
To determine if the NS1 κ LC interacts with either Derlin-1 or Herp, we incubated cells with or without lactacystin, isolated LC and performed western blot analyses to identify associated proteins. Readily detectable amounts of Derlin-1 co-precipitated with the κ LC (Fig. 3B), suggesting that this non-glycosylated BiP substrate might be retro-translocated through a channel containing Derlin-1 before being degraded in cytosol. In addition to Derlin-1, it appeared that a band recognized by the anti-Herp antiserum was co-precipitated with the κ LC, but the presence of a non-specific band in the same region made it difficult to determine this with any certainty. Thus, to further confirm the interaction of the NS1 κ LC with Herp, a two-step immunoprecipitation was carried out on metabolically labeled cell extracts from lactacystin-treated P3U.1 cells (Fig. 3C). Although we were unable to detect any Herp protein in the anti-κ precipitated material, immunoprecipitation with the anti-Herp antibody revealed a faint band that co-migrated with the κ LC (Fig. 3C lane 2). To determine if indeed this was the NS1 κ LC, a parallel sample was first immunoprecipitated with anti-Herp and then boiled in the presence of SDS to release bound proteins from the beads. The eluted material was diluted and then divided into three portions, which were subjected to a second immunoprecipitation with either anti-Herp, anti-κ, or Protein A-Sepharose alone (Fig. 3C lane 3 – 8). We found that indeed the NS1 κ LC could be immunoprecipitated from the material that was bound to Herp (lanes 4 and 7), indicating that NS1 κ LC and Herp interact physically in vivo. When the second immunoprecipitation was done with the anti-Herp antibody, Herp was re-precipitated but not the κ LC band (lanes 3 and 6), because the interaction between these two proteins was disrupted by boiling in the presence of SDS as expected. Next, to identify which form of the NS1 κ LC binds to Herp, the same two-step immunoprecipitation was performed under non-reducing conditions (Fig. 3C lane 9 – 16). We found that the ox1 form of NS1 κ LC was preferentially co-precipitated with Herp (Fig. 3C lane 12 and 15), indicating that the partially folded form that binds to BiP (Knittler et al., 1995) and is ubiquitinated, also interacts with Herp.
The degradation of NS1 κ LC requires the activity of p97 and Hrd1
Because both Herp and Derlin-1 form a complex with p97 and Hrd1, we next examined whether either of these proteins were also involved in the degradation pathway of NS1 κ LC. To do so, we co-transfected 293T cells with NS1 κ LC along with either wild-type p97, a dominant-negative p97 mutant (QQ) (Ye et al., 2001), wild-type Hrd1, or a dominant negative Hrd1 mutant (C291S) (Nadav et al., 2003), and pulse-chase experiments were conducted to detect changes in the half-life of the LC. We found that the p97 QQ mutant dramatically inhibited the degradation of NS1 κ LC, whereas over-expression of wild type p97 did not affect its half-life (Fig. 3D). Over-expression of both wild-type and mutant Hrd1 inhibited the degradation of NS1 κ LC, although the effects with the mutant were somewhat greater. The ability of wild-type Hrd1 to also inhibit degradation is consistent with previous studies where the over-expression of ubiquitination enzymes actually inhibits ERAD instead of increasing protein turnover (Kikkert et al., 2004) (Shen et al., 2007). Together these results strongly implicate the Derlin-1, Herp, p97, Hrd1 complex in the disposal of this non-glycosylated BiP substrate.
Over-expressed Herp-FLAG interacts with several BiP substrates but not with a well characterized ERAD substrate that uses the calnexin/calreticulin pathway
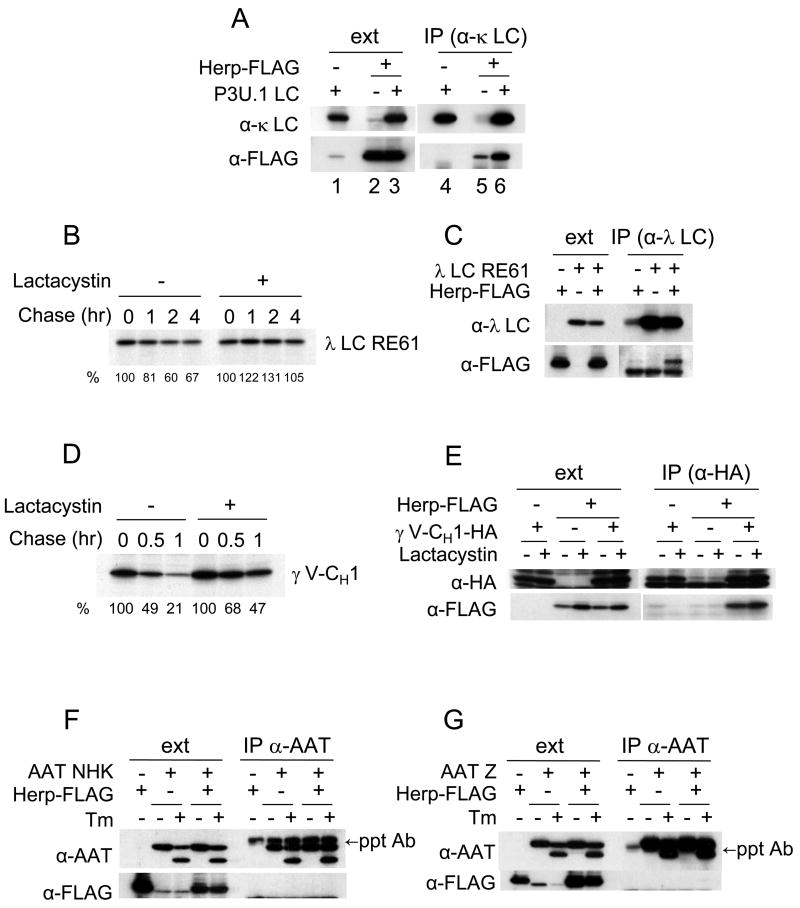
To determine if Herp binds to other BiP-binding, ERAD substrates, we employed a transient expression system. A FLAG-tagged version of Herp (Herp-FLAG) was co-expressed in 293T cells along with several different BiP substrates, since the endogenous level of Herp is very low in this cell line. First, the interaction of Herp-FLAG and NS1 κ LC was tested to ensure that the interaction we observed with endogenous proteins (Fig. 3C) could be reproduced. Although a small amount of Herp appeared to be precipitated with anti-κ in the absence of LC expression, the amount of Herp-FLAG that was isolated increased significantly in the presence of the NS1 κ LC (Fig. 4A lane 5 and 6), indicating that Herp-FLAG binds to the NS1 κ LC in this system as expected. Next, we examined two other non-secreted BiP binding proteins (i.e., a non-secreted λ LC mutant (RE61) and a truncated, HA-tagged γ HC (HA-γ V-CH1)). We found that the turnover of both proteins was significantly diminished by treating with lactacystin (Fig. 4B and 4D), indicating that they were degraded by 26S proteasome via the ERAD pathway. To investigate if these substrates could also interact with Herp, 293T cells were transfected with either the λ LC RE61 mutant (Fig. 4C) or the truncated HC (Fig. 4E) alone or with Herp-FLAG. Both RE61 LC (Fig. 4C) and γ V-CH1 (Fig. 4E) were co-precipitated with Herp, demonstrating that Herp interacts with a number of non-glycosylated ERAD substrates that bind to BiP.
Figure 4. Over-expressed Herp-FLAG interacts with the BiP substrates, i.e., non-secreted λ LC mutant and unassembled Ig γ HC mutant, but not with the calnexin/calreticulin substrates, i.e., α1-antitrypsin variants.
NS1 κ LC expressed in P3U.1 cells (A), RE61 λ LC and BiP (C) or HA-γ V-CH1 (E) were transiently co-expressed with or without Herp-FLAG in 293T cells. At 24 hr post-transfection, cell extracts were prepared from cells treated with or without lactacystin and immunoprecipitated with anti-κ LC antiserum (A), anti-λ LC antiserum (C) or anti-HA antibody (E). Cell extracts and precipitated samples were subjected to immunoblot analyses as indicated. Non-secreted λ LC RE61 (B) or HA-γ V-CH1 (D) were transiently expressed in 293T cells. At 24 hr post-transfection, cells were labeled with 35S-methionine/cysteine for 15 min and chased for the indicated times in the presence or absence of lactacystin. Immunoprecipitated samples from cell extracts were subjected to SDS-PAGE, followed by autoradiography. The signals for λ LC RE61 (B) or HA-γ V-CH1 (D) were quantified as expressed as a percent of that present at t=0. The values are shown at the bottom of each lane. The α1-antitrypsin (AAT) NHK variant (F) or Z variant (G) was transiently co-expressed with or without Herp-FLAG in 293T cells. At 24 hr post-transfection, cell extracts were prepared after treatment with or without tunicamycin for 3 hr and subjected to immunoprecipitation with anti-α1-antitrypsin antiserum. Cell extracts and precipitated samples were subjected to immunoblot analysis as indicated.
We next determined whether Herp could interact with two well-characterized non-secreted glycoproteins that are substrates of calnexin/calreticulin; the α-antitrypsin NHK variant (Fig. 4F) and the Z variant (Fig. 4G). Cells were transfected with either the NHK or Z variant alone or with Herp-FLAG and immunoprecipitated with anti-α1-antitrypsin. Although Herp was expressed at reasonably high levels in these cells, we were unable to detect any association of Herp with either of the α1-antitrypsin variants. Transfected cells were also treated with tunicamycin to determine if Herp would bind to either of these proteins in the absence of glycosylation, but again, there was no evidence of Herp binding (Fig. 4F, G). Of note, unlike some glycoproteins, the lack of N-linked glycans does not appreciably convert these calnexin substrates into BiP substrates (data not shown), which is in keeping with published data on the NHK-QQQ mutant that cannot be glycosylated but still binds to EDEM (Oda et al., 2006). These data suggest that there is some specificity in terms of which ERAD substrates Herp can interact with.
Herp interacts with 26S proteasome and ubiquitinated substrates
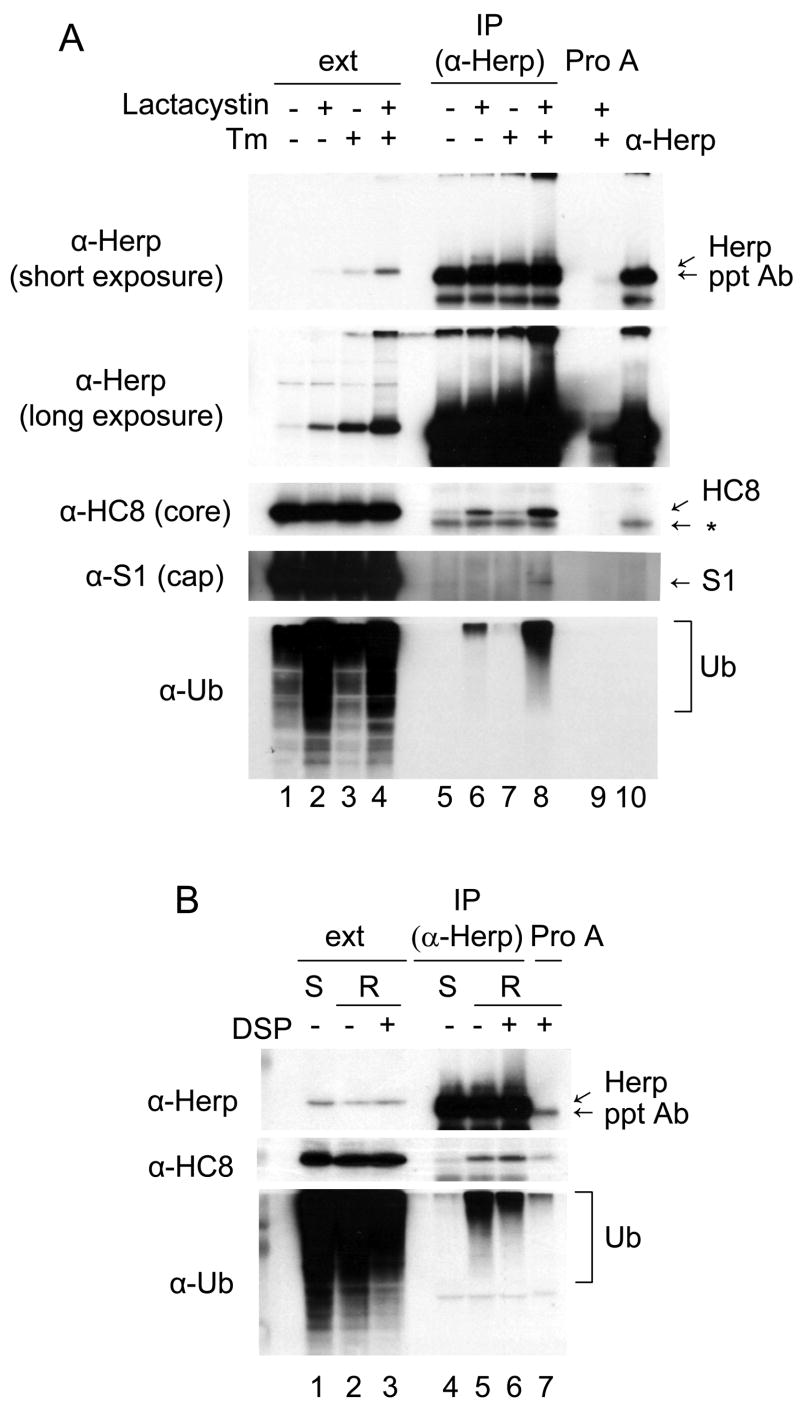
To further investigate the general involvement of Herp in proteasomal degradation, we used control and UPR-activated NIH3T3 cells, because there are commercial antibodies that recognize murine proteasomal subunits, Herp is significantly up-regulated by ER stress in these cells, and our analyses would not be further complicated by the presence of endogenous Ig subunits. NIH3T3 cells were treated with the indicated combinations of lactacystin and tunicamycin and cell lysates were prepared. An aliquot of the cell extract (ext) was removed for direct loading on the gels, and the remainder was immunoprecipitated with the anti-Herp antibody. As expected, the Herp protein level increased in response to tunicamycin treatment (Kokame et al., 2000; Ma and Hendershot, 2004; van Laar et al., 2000; Yamamoto et al., 2004), and in each case, it was further elevated in the presence of lactacystin (Fig. 5A lanes 1–4), which is in keeping with studies showing that Herp is rapidly degraded by the 26S proteasome (Sai et al., 2003). When Herp was isolated from cells that were treated with lactasystin, we found that both a core (H8) and cap (S1) subunit of the 26S proteasome were co-precipitated with Herp, as were ubiquitinated proteins (Fig. 5A lanes 6 and 8). Only very small amounts of the HC8 subunit and Ub proteins could be detected in the absence of lactasystin treatment (Fig. 5A, lanes 5 and 7). In order to exclude the possibility that the smear of Herp-associated bands detected by the anti-ubiquitinated proteins antibody represented ubiquitinated Herp, cell extracts were prepared from tunicamycin and lactacystin treated cells with two different lysing buffers for immunoprecipitation. One contained 0.6% SDS to disrupt protein-protein interactions (Fig. 5B, lane1), while the other was RIPA buffer (Fig. 5B lane 2 and 3). The cleavable cross-linker, DSP was included in one set of cells that were lysed with RIPA buffer. Although HC8 and ubiquitinated protein were co-precipitated with Herp protein in RIPA lysates regardless of whether DSP was present (Fig. 5B lane 5 and 6), these interactions were not detected in the cell extract containing SDS (Fig. 5B lane 4). These results demonstrate that Herp interacts with the 26S proteasome and strongly suggest that it is associated ubiquitinated substrates.
Figure 5. Herp interacts with the 26S proteasome and ubiquitinated substrates.
(A) NIH3T3 cells were treated as indicated, crosslinked with DSP and lysed. The clarified extracts were immunoprecipitated with anti-Herp antiserum or Protein A-Sepharose as a control. Total cell extracts, precipitated samples, and the anti-Herp antiserum alone were subjected to immunoblot analysis. A non-specific band is indicated with an asterisk. (B) NIH3T3 cells treated with tunicamycin for 24 hr and lactacystin for 6 hr were lysed in SDS buffer (S) or in RIPA buffer (R) with or without DSP. Lysates were immunoprecipitated with anti-Herp antiserum or Protein A-Sepharose as a control. Cell extracts and precipitated samples were subjected to immunoblot analyses as indicated.
Herp is required for the degradation of non-glycosylated BiP substrates, but not for glycosylated calnexin/calreticulin substrates
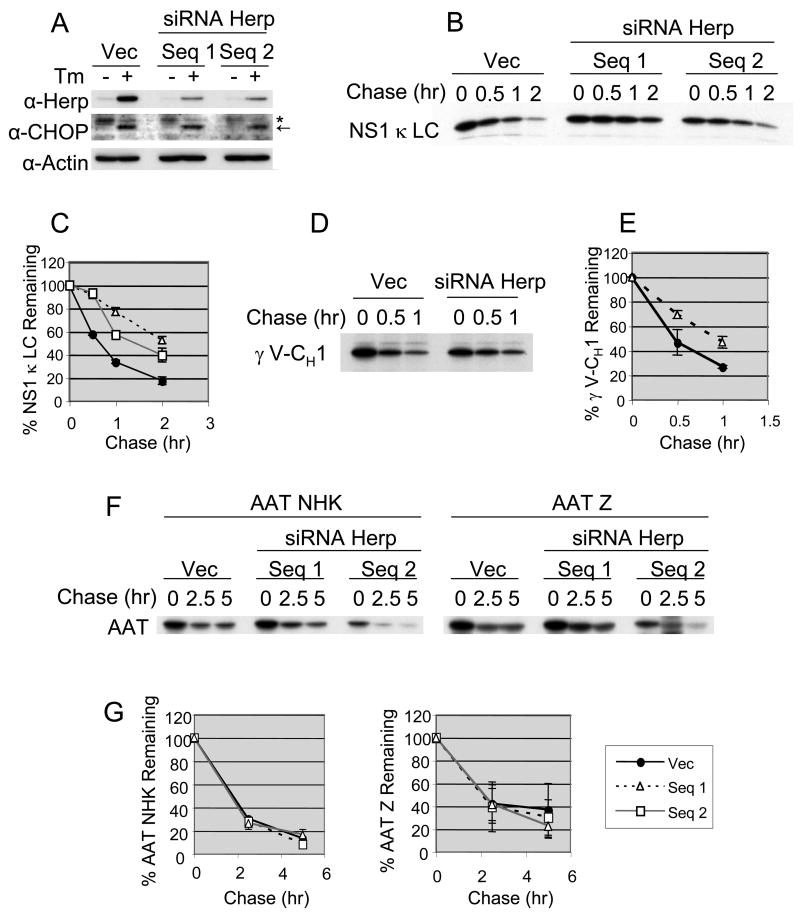
Since we found that Herp interacts with several BiP bound proteins that are degraded by the proteasome and is associated with both ubiquitinated substrates and with components of proteasome, we next tested if Herp was required for the degradation of the NS1 κ LC. In order to examine this, we first attempted to lower Herp expression in the P3U.1 cells with siRNA. Although lines could be obtained that initially had decreased levels of Herp, we were unable to maintain these for any period of time. Thus, we established several NIH3T3 cell lines that stably expressed two different doxycycline-inducible, Herp-specific siRNAs. Since the steady-state level of Herp is normally quite low but is significantly up-regulated under ER stress conditions, we first examined Herp protein levels with and without tunicamycin treatment in these NIH3T3 cell lines. After treating with doxycycline for 24 hrs, we found that both of the lines expressing the siRNAs had significantly reduced levels of Herp as compared to control cells under both conditions, although this was much more obvious in the tunicamycin treated cells (Figure 6A). In all three lines, the cells still responded to ER stress by inducing CHOP expression, which is regulated by the same UPR pathways that control Herp expression (Ma and Hendershot, 2004), arguing that only this target is affected. To study the effect of reducing Herp levels on the stability of ERAD substrates, we transfected these cell lines with the NS1 κ LC, the truncated HC, γ V-CH1, or the two AAT variants and determined their half-lives. In the cell lines expressing Herp siRNA, the turn-over of the NS1 κ LC (Fig. 6B and 6C) and the truncated HC (Fig. 6D and 6E) were reduced compared to that observed in the control cells. The cells expressing sequence #1 had the lowest Herp levels and the half-life of the LC was most dramatically affected in these cells. Conversely, when both of the AAT variants were similarly examined, the reduction of Herp expression did not inhibit their turn-over (Fig. 6F and 6G). These data demonstrate that Herp is required for the efficient degradation of the NS1 κ LC and the truncated HC, but not of the AAT variants.
Figure 6. siRNA mediated repression of Herp leads to the stabilization of non-secreted κ LC, but not of α1-antitrypsin variants.
(A) NIH3T3 cell lines with doxycyclin-inducible siRNA for Herp were created. Vec indicates control line with empty vector. Cells were treated with doxycyclin in the presence or absence of tunicamycin for 24 hr and subjected to immunoblot analysis as indicated. CHOP is indicated with an arrow and a non-specific band recognized by this antiserum with an asterisk. These NIH3T3 cell lines were transfected with vectors encoding NS1 κ LC (B), HA-γ V-CH1 (D) or AAT variants (F). Cells were treated with doxycyclin for 24 hr after transfection to induce shRNA expression, followed by metabolic labeling with 35S-methionine/cysteine for 15 min and chased for the indicated times. κ LC, HA-γ V-CH1, or AAT variants were immunoprecipitated from the appropriate cell extracts and subjected to SDS-PAGE followed by autoradiography. The signals for κ LC (C), γ V-CH1 (E) or the two AAT variants (G) were quantified and plotted. A closed circle, an opened triangle and an opened square show the empty vector, the siRNA Herp sequence #1 and the sequence #2 respectively. The mean values and standard errors were calculated from more than two independent experiments.
DISCUSSION
In this study, we examined the ERAD pathway used for the disposal of three soluble, non-glycosylated BiP substrates that are not secreted. The degradation of these proteins was inhibited by lactacystin, demonstrating that they are degraded by the proteasome and can be considered bona fide ERAD substrates. All three proteins were shown to interact with Herp. Although Herp is an ER-localized membrane protein, topological predictions and protease digestion studies suggest that it is almost entirely cytosolically disposed (Kokame et al., 2000). If indeed this is the case, the interaction of Herp with these ERAD substrates is either indirect or must occur after the ERAD substrate enters or emerges from the retro-translocon. In focusing on the NS1 κ LC, we found that it was ubiquitinated and interacted with Derlin-1, which is one of three Derlin family members that are thought to be retro-translocon components (Lilley and Ploegh, 2004)(Ye et al., 2004).
Immunoprecipitation of Herp revealed that, in addition to associating with ubiquitinated substrates, Herp also bound components of the 26S proteasome and that this interaction was increased when degradation was inhibited. We interpret these findings to suggest that Herp may serve as a bridge to direct partially or fully translocated proteins to either ubiquitin ligases and/or the proteasome for degradation. While it is formally possible that Herp’s association with the proteasome is indirect and occurs through its interaction with ubiquitinated substrates, the fact that reducing Herp levels slowed the turnover of the NS1 κ LC and the HC mutant argue that it is playing a direct role in assisting degradation. We do not currently know which region of Herp is involved in interaction with the proteasome, but the presence of an Ubl domain at its N-terminus is a good candidate for this interaction. Consistent with previous studies which demonstrated that Herp interacts with the Hrd1 ubiquitin ligase directly and the p97 ATPase complex indirectly (Schulze et al., 2005), we detected both proteins in association with Herp (data not shown). In addition, we found that dominant negative mutants of both p97 and Hrd1 inhibited the degradation of NS1 κ LC. These data indicate that p97, Hrd1, Derlin-1 and Herp are all involved in LC degradation (Figure 7).
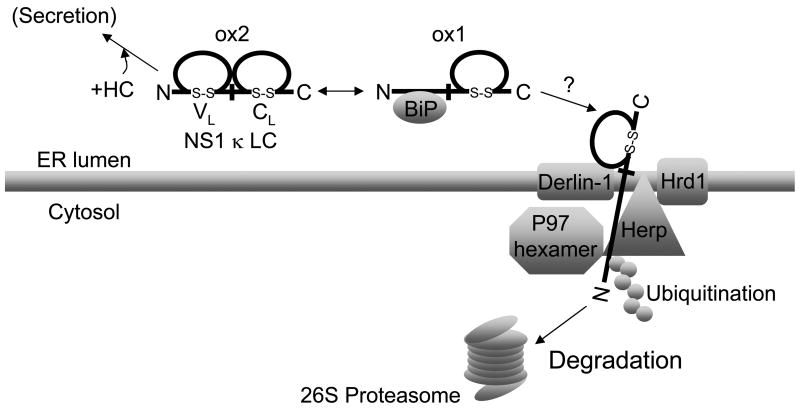
Figure 7. Model for degradation of NS1 κ LC.
The NS1 κ LC exists in equilibrium between a partially oxidized form (ox1) which binds to BiP via its N-terminal VL domain and a completely oxidized form (ox2) that does not bind to BiP. Both forms are degraded by the proteasome, but the ox2 form must first be partially reduced to ox1. The BiP-bound ox1 form of NS1 κ LC is recruited to a retro-translocon composed of Derlin-1, Herp, Hrd1, and p97, where it is then at least partially retrotranslocated and ubiquitinated. The LC is then either pulled through the translocon with the disulfide bond in the CL domain intact, or the proteasome begins to degrade this substrate before it is fully reduced. Herp might play a role in either linking substrate with the proteasome, in enhancing the ubiquitination of substrates by interacting with the Hrd1 ubiquitin ligase, or both.
The NS1 κ LC provides some insights as to how soluble, disulfide bond containing proteins are extracted from the ER (Figure 7). This LC exists in both a completely oxidized and partially oxidized form. Our data reveal that inhibition of degradation stablizes the partially oxidized form (ox1) but not the fully oxidized form (ox2) and that only the ox1 form is ubiquitinated. This strongly implies that the completely folded form (ox2) needs to be partially reduced for retrotranslocation. In keeping with this, Herp binds exclusively to the ox1 form. Importantly, since BiP binds only to the unfolded or partially folded form of NS1 κ LC, we hypothesize that BiP can transport the substrate to the Derlin-1/Herp complex for degradation. Somewhat surprisingly, we were unable to detect any fully reduced LC under any conditions. Our previous studies revealed that the N-terminal VL domain of this LC is the BiP binding site (Skowronek et al., 1998) and that the CL domain folds very stably (Hellman et al., 1999). Thus, it is reasonable to speculate that the unfolded VL domain is threaded into the retrotranslocon, where is encounters p97, Herp, and Hrd1. The folded CL domain may either represent a kinetic barrier for unfolding or for extraction. In the latter case, it would be pulled through the retrotranslocon with its disulfide bond intact. However, we do not currently know whether the stabilized protein is inside the ER or in the cytosol. Further experiments are underway to determine this.
Recently, it was reported that there are distinct retrotranslocation channels for different types of ERAD substrates in yeast (Carvalho et al., 2006)(Denic et al., 2006). Proteins with misfolded luminal domains are retrotranslocated through a channel consisting of ubiquitin ligase Hrd1p, Der1p, a yeast homolog of Derlin-1, and Usa1p. Since Herp was able to substitute for Usa1p in yeast cells (Carvalho et al., 2006), and forms a complex with Derlin-1, Hrd1, and VIMP in mammalian cells (Schulze et al., 2005), it is reasonable to speculate Herp is the mammalian counterpart of Usa1p, and thus serves to link the E3 ligase with the retrotranslocon. We showed here that NS1 κ LC, an ER luminal protein, binds to both Derlin-1 and Herp. However, neither Derlin-1 nor Herp interacted with NHK or PiZ AAT. In keeping with this, lowering Herp levels did not affect the degradation of either of these AAT variants, but it had significant effects on the turnover of BiP substrates. The fact that lowering Herp levels did not compromise the degradation of the AAT variants argues that Herp may not be a component of Derlin-2 or -3 containing retrotranslocons that interact with EDEM (Oda et al., 2006). Together these data suggest that there are distinct retrotranslocation pathways in the mammalian ER and that a Derlin-1/Herp containing pathway is responsible for degrading at least some non-glycosylated BiP substrates. As all of our substrates were soluble proteins, we cannot comment on the role of Herp in disposing of membrane bound proteins that bind BiP.
Evidently p97 and Hrd1 represent common components of retrotranslocons, since dominant negative mutants of these proteins inhibited the degradation of the NS1 κ LC in our study, and it was previously shown that they interact with Derlin-2 and -3 (Oda et al., 2006)(Wilhovsky et al., 2000). The presence of EDEM in the Derlin-2 and -3 containing channel(s) provides a means for targeting glycosylated substrates to these retrotranslocons, however it is not clear how BiP bound substrates are sent to the Derlin-1/Herp containing channel. One likely candidate would be an ER-localized DnaJ protein; a possibility that we are currently investigating. Although our data suggest that there may be a separation of retrotranslocon substrates based on their glycosylation status or the chaperone system used, it is important to note that the number of substrates that have been examined is still quite limited.
In summary, we provide the first characterization of an ERAD pathway for non-glycosylated, soluble BiP substrates. We find that this pathway involves Derlin-1 containing retrotranslocons, and requires Herp, p97, and Hrd1. In addition, our studies provide a better understanding of the disposal of disulfide bond containing proteins and strongly suggest that there is a substrate specificity for mammalian retrotranslocons.
EXPERIMENTAL PROCEDURES
Cell lines and antibodies
Mouse plasmacytoma cell lines P3X63Ag (γ+, κ+) (Kohler and Milstein, 1975), P3U.1 (γ−, κ+) (Kohler et al., 1976), Ag8(8) (γ+, κ−) (Bole et al., 1986), and Ag8.653 (γ−, κ−) (Kearney et al., 1979) were grown in complete RPMI-1640 medium containing 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin-streptomycin, and 55 μM 2-mercaptoethanol (2ME). 293T and NIH3T3 cells were cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/ml penicillin-streptomycin. All cell lines were cultured at 37°C under 5% CO2.
The anti-Herp antibody was raised against recombinant human Herp fused to the amino-terminal glutathione S-transferase-tag (GST-Herp) and expressed in E. coli. The antiserum from an immunized rabbit was affinity purified with GST-Herp covalently coupled to CNBr-activated Sepharose 4 fast flow (Amersham). Polyclonal anti-calnexin (Meunier et al., 2002) antibodies have been described previously. The monoclonal anti-HA (12CA5) antibody was kindly provided by Dr. Al Reynolds (Vanderbilt University, USA), the polyclonal anti-Derlin-1 antiserum was a generous gift from Dr. Yihong Ye (NIDDK, USA), the anti-p97 antibody was kindly provided by Dr. George DeMartino (UTSW, USA), and the anti-Hrd1 antibody was a generous gift from Dr. Emmanuel Wiertz (Leiden University, The Netherlands). All other antibodies were purchased from companies; anti-mouse IgG (Igγ and κ) and anti-mouse IgM (Igμ and λ) (Southern Biotech), anti-actin, anti-Hsc70, and anti-FLAG D-8 (Santa Cruz), anti-ubiquitinated proteins FK2, anti-HC8, and anti-S1 (BIOMOL), and anti-α1 antitrypsin (MP Biomedicals).
Preparation of cell extracts
Cells were lysed in either RIPA buffer [0.15 M NaCl, 10 mM Tris-HCl (pH 7.5), 1% Nonidet P-40 (NP-40), 0.1% SDS, 0.2% deoxycholate] or SDS lysis buffer [50 mM Tris-HCl (pH 8.0), 0.6% SDS] as indicated. For lysis with SDS, samples were incubated at 95°C for 10 min and the SDS buffer was then diluted with a 4-fold volume of buffer containing 10 mM sodium phosphate (pH 7.2), 2 mM EDTA, 0.25 M NaCl, 0.1% NP-40 followed by centrifugation to remove debris. All buffers contained 10 mM N-ethylmaleimide (NEM), 0.25 mM phenylmethylsulphonyl fluoride (PMSF), and a protease inhibitor cocktail (Complete, Roche). When proteins were analyzed under nonreducing conditions, cells were washed with ice-cold PBS containing 10 mM NEM and then lysed with buffer containing NEM and 10 units/ml of apyrase (Sigma). In experiments where protein cross-linking was performed, cells were incubated with the membrane permeable, thiol-cleavable cross-linker dithiobis (succinimidylpropionate) (DSP) before cell lysis as described previously (Meunier et al., 2002).
Metabolic Labeling and Pulse-Chase Experiments
For metabolic labeling, cells (3× 107cells) were incubated in methionine- and cysteine-free RPMI 1640 labeling medium containing 10% dialyzed FBS with 600 μCi of 35S-TransLabel (MP Biomedicals) for 6 h and then lysed in the presence of apyrase. For pulse-chase experiments, cells (1× 106cells) were incubated for 30 min in labeling media and then pulse-labeled with 100 μCi 35S-TransLabel for 2 or 15 min as indicated. The chase was initiated by washing the cells twice with cold PBS and then adding an excess of unlabeled methionine (2 mM) and cysteine (2 mM) to the chase media. Aliquots of cells were removed at the indicated times of chase, and cells were separated from the media and washed once with ice-cold PBS containing 10 mM NEM before lysing in the presence of apyrase. The media and cell lysates were immunoprecipitated as indicated. For proteasome inhibition experiments, 5 μM lactacystin was added 3h prior to the starvation period and included throughout the metabolic labeling and chase. For lysosomal inhibition, 10 mM NH4Cl was added during the 30 min starvation period and throughout the labeling and chase. To determine half-lives of the various proteins under different conditions, the signal for each band was quantified by phosphorimager and the bands present in the chase samples were expressed as a percent of the signal present in the pulse lane (t=0). In the case of non-reduced gels where the ox1 and ox2 form of LC were analyzed, the signal for each band was expressed as a percent of the total LC present (ox1 + ox2) in the pulse sample.
Immunoprecipitation, 2D gel analysis
For immunoprecipitation, antibodies were mixed with the cell lysates made with either RIPA buffer or SDS lysis buffer as indicated, and the immune complexes were recovered with Protein A-Sepharose (Sigma). To identify co-precipitating proteins in immune complexes, the Protein A beads were incubated in 60 μl of 2% SDS elution buffer and boiled for 15 min. The resulting supernatant was diluted to 0.1% SDS and reimmunoprecipitated as indicated. For 2D gel analysis, immunoprecipitated samples were loaded onto a SDS-polyacrylamide gel and separated under non-reducing conditions. Each lane was cut and then incubated with 2x SDS sample buffer containing 2-mercaptoethanol at room temperature for 20 min to reduce the disulfide bonds. The gel slice was then applied to a second SDS-polyacrylamide gel and separated in the second dimension.
Transfection of expression vector
DNAs encoding the NS1 κ LC (Skowronek et al., 1998), a truncated Ig HC comprised of the VH and CH1 domains plus a C-terminal 9 residue epitope from the influenza hemagglutinin (HA-γ V-CH1) (Lee et al., 1999), a nonsecreted λ LC point mutant, RE61, in which arginine61 in the VL domain is mutated to glutamine (Yair Argon, unpublished data), and the α1-antitrypsin NHK and Z variants (Sifers et al., 1988) have been described. A cDNA encoding human Herp was PCR amplified using primers from the published sequence (Kokame et al., 2000; van Laar et al., 2000) and ligated along with a C-terminal FLAG tag into the pcDNA3 vector (Invitrogen). Wild type p97 and an ATP hydrolysis-defective mutant (QQ) were a kind gift from Dr. Yihong Ye (NIDDK, USA) and Dr. Hemmo Meyer (ETH, Switzerland). The pcDNA3/Hrd1 wild type and Hrd1 C291S vectors were generously supplied by Dr. Yuval Reiss (Proteologics, Israel). The recombinant plasmids were introduced into 293T cells using calcium phosphate precipitation (Sambrook and Russell, 2001), or into NIH3T3 cells using FuGENE 6 Transfection Reagent (Roche) and Lipofectamine LTX Reagent (Invitrogen).
Inducible RNA interference
The pSuperior-puro vector (Oligoengine) was used to express shRNA molecules from a tetracycline-inducible H1 promoter. Annealed oligonucleotides (5′-TGTGAAGAATCCCTCCAAA-3′ and 5′-TGTTCAAATTACACTAAGT-3′ for mHerp RNAi) were ligated into pSuperior-puro as directed by the manufacture, to produce pSuperior-puro-Herp. NIH3T3 cells were transfected with pcDNA6/TR (Invitrogen), which expresses the tetracycline repressor protein (tetR) from a CMV promotor and selected in 3 μg/ml blasticidin S HCl (Invitrogen). These cells were then transfected with pSuperior-puro-Herp and individual clones were isolated after selection in 3 μg/ml puromycin. shRNA expression was then induced with 2 μg/ml doxycycline for 24 hours and the level of Herp expression after knockdown was determined by Western blot analysis.
Acknowledgments
This work was supported by NIH Grant GM54068 (LMH), the Cancer Center CORE Grant CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital. We wish to thank Drs. Jeffery Brodsky and Yuichiro Shimizu for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986;102:1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Cabral CM, Liu Y, Sifers RN. Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chillaron J, Haas IG. Dissociation from BiP and retrotranslocation of unassembled immunoglobulin light chains are tightly coupled to proteasome activity. Mol Biol Cell. 2000;11:217–226. doi: 10.1091/mbc.11.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Hellman R, Vanhove M, Lejeune A, Stevens FJ, Hendershot LM. The in vivo association of BiP with newly synthesized proteins is dependent on the rate and stability of folding and not simply on the presence of sequences that can bind to BiP. J Cell Biol. 1999;144:21–30. doi: 10.1083/jcb.144.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Kearney JF, Radbruch A, Liesegang B, Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Knittler MR, Dirks S, Haas IG. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92:1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. Embo J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Kohler G, Howe SC, Milstein C. Fusion between immunoglobulin-secreting and nonsecreting myeloma cell lines. Eur J Immunol. 1976;6:292–295. doi: 10.1002/eji.1830060411. [DOI] [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Kokame K, Agarwala KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J Biol Chem. 2000;275:32846–32853. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Ma J, Kearney JF, Hendershot LM. Association of transport-defective light chains with immunoglobulin heavy chain binding protein. Mol Immunol. 1990;27:623–630. doi: 10.1016/0161-5890(90)90004-j. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. Herp is dually regulated by both the endoplasmic reticulum stress-specific branch of the unfolded protein response and a branch that is shared with other cellular stress pathways. J Biol Chem. 2004;279:13792–13799. doi: 10.1074/jbc.M313724200. [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Nadav E, Shmueli A, Barr H, Gonen H, Ciechanover A, Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem Biophys Res Commun. 2003;303:91–97. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai X, Kokame K, Shiraishi H, Kawamura Y, Miyata T, Yanagisawa K, Komano H. The ubiquitin-like domain of Herp is involved in Herp degradation, but not necessary for its enhancement of amyloid beta-protein generation. FEBS Lett. 2003;553:151–156. doi: 10.1016/s0014-5793(03)01009-3. [DOI] [PubMed] [Google Scholar]
- Sambrook JF, Russell DW. Molecular cloning. 3. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schulze A, Standera S, Buerger E, Kikkert M, van Voorden S, Wiertz E, Koning F, Kloetzel PM, Seeger M. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J Mol Biol. 2005;354:1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Shen Y, Ballar P, Apostolou A, Doong H, Fang S. ER stress differentially regulates the stabilities of ERAD ubiquitin ligases and their substrates. Biochem Biophys Res Commun. 2007;352:919–924. doi: 10.1016/j.bbrc.2006.11.121. [DOI] [PubMed] [Google Scholar]
- Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Skowronek MH, Hendershot LM, Haas IG. The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc Natl Acad Sci U S A. 1998;95:1574–1578. doi: 10.1073/pnas.95.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Laar T, Schouten T, Hoogervorst E, van Eck M, van der Eb AJ, Terleth C. The novel MMS-inducible gene Mif1/KIAA0025 is a target of the unfolded protein response pathway. FEBS Lett. 2000;469:123–131. doi: 10.1016/s0014-5793(00)01253-9. [DOI] [PubMed] [Google Scholar]
- Wilhovsky S, Gardner R, Hampton R. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol Biol Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem (Tokyo) 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]