Abstract
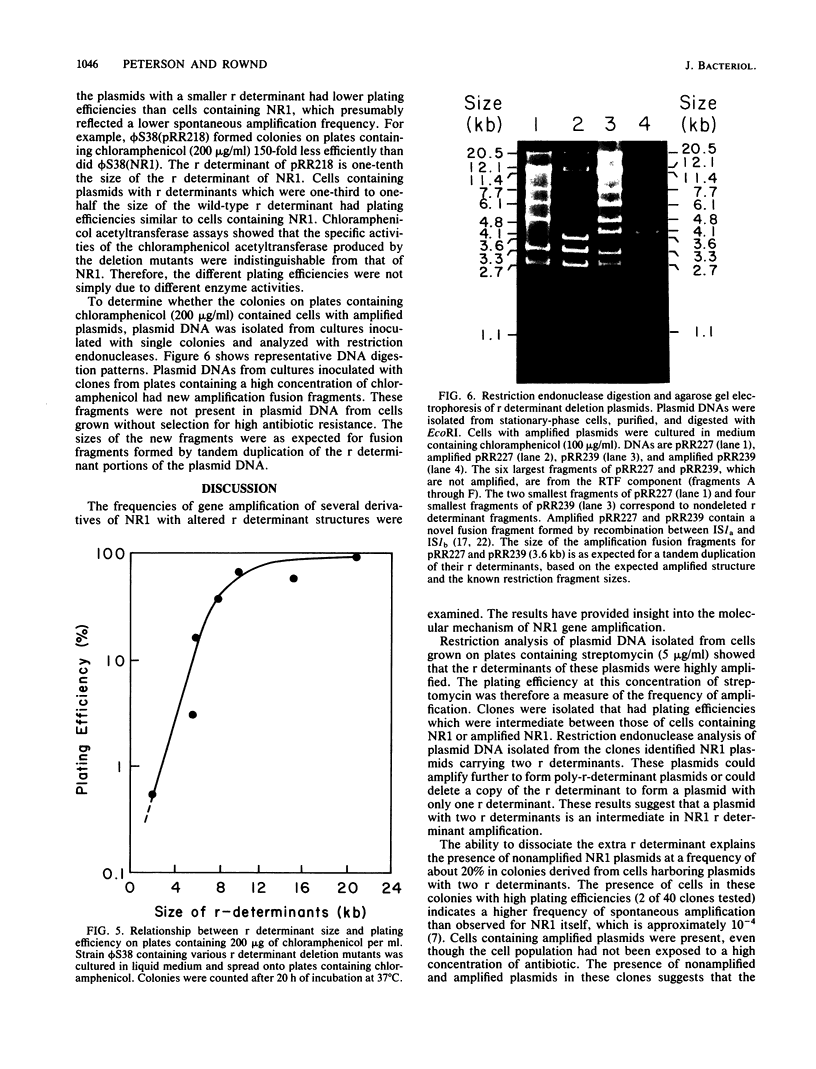
Drug resistance gene amplification of derivatives of plasmid NR1 having various amounts of resistance (r) determinant DNA was examined with two types of NR1 derivatives. The first was an NR1 derivative that carried two tandem copies of the r determinant component which was isolated as an intermediate in the amplification process. The plating efficiency of host cells and restriction endonuclease analysis of the plasmid DNA indicate that plasmids with two tandem copies of the r determinant undergo spontaneous amplification to a more highly amplified state at a frequency 150-fold higher than that of wild-type NR1. The second class of derivatives consisted of plasmids in which different regions of the r determinant component had been deleted. The relationship between spontaneous amplification frequency and r determinant size was examined with these plasmids. Plating efficiency of host cells indicated that plasmids with a smaller r determinant undergo spontaneous amplification at a lower frequency than do plasmids with a larger r determinant. These results suggest that there is an ordered sequence of events in the amplification of the r determinant of NR1.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P., Roth J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci U S A. 1981 May;78(5):3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chandler M., de la Tour E. B., Willems D., Caro L. Some properties of the chloramphenicol resistance transposon Tn9. Mol Gen Genet. 1979 Oct 3;176(2):221–231. doi: 10.1007/BF00273216. [DOI] [PubMed] [Google Scholar]
- Fishman S. E., Hershberger C. L. Amplified DNA in Streptomyces fradiae. J Bacteriol. 1983 Aug;155(2):459–466. doi: 10.1128/jb.155.2.459-466.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Shaw W. V. Chloramphenicol acetyltransferases specified by fi minus R factors. Antimicrob Agents Chemother. 1973 Jan;3(1):99–104. doi: 10.1128/aac.3.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes R., Burkardt H. J., Schmitt R. Repetition of tetracycline resistance determinant genes on R plasmid pRSD1 in Escherichia coli. Mol Gen Genet. 1979 Jan 10;168(2):173–184. doi: 10.1007/BF00431443. [DOI] [PubMed] [Google Scholar]
- Meyer J., Iida S. Amplification of chloramphenicol resistance transposons carried by phage P1Cm in Escherichia coli. Mol Gen Genet. 1979 Oct 3;176(2):209–219. doi: 10.1007/BF00273215. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C. F., Hashimoto H., Mickel S., Rownd R. Round of replication mutant of a drug resistance factor. J Bacteriol. 1974 Jun;118(3):855–866. doi: 10.1128/jb.118.3.855-866.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya R., Rownd R. Transduction of R factors by a Proteus mirabilis bacteriophage. J Bacteriol. 1971 Jun;106(3):773–783. doi: 10.1128/jb.106.3.773-783.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Brodsky R. F. Characterization of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol. 1968 Jan;95(1):28–36. doi: 10.1128/jb.95.1.28-36.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies T., Laufs R., Riess F. C. Amplification of resistance genes in Haemophilus influenzae plasmids. J Bacteriol. 1983 Aug;155(2):839–846. doi: 10.1128/jb.155.2.839-846.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K., Schraml S., Shales S. W., Schmitt R. Tetracycline resistance transposon Tn1721: recA-dependent gene amplification and expression of tetracycline resistance. J Bacteriol. 1981 Sep;147(3):851–859. doi: 10.1128/jb.147.3.851-859.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Warren R. L., Rownd R. H. Partially supercoiled replication intermediates of R plasmid DNA are resistant to relaxation. Nature. 1976 Dec 16;264(5587):676–678. doi: 10.1038/264676a0. [DOI] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Plasmid-determined tetracycline resistance in Streptococcus faecalis: tandemly repeated resistance determinants in amplified forms of pAMalpha1 DNA. J Mol Biol. 1976 Apr 15;102(3):583–600. doi: 10.1016/0022-2836(76)90336-3. [DOI] [PubMed] [Google Scholar]