Abstract
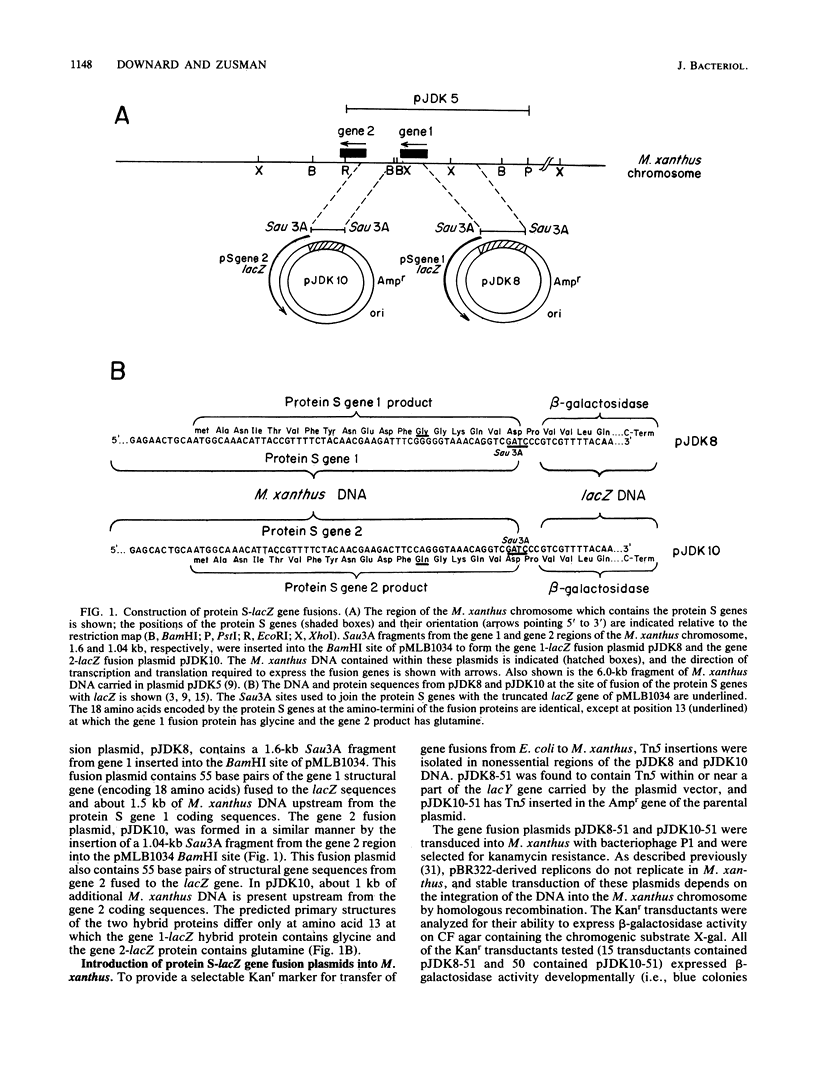
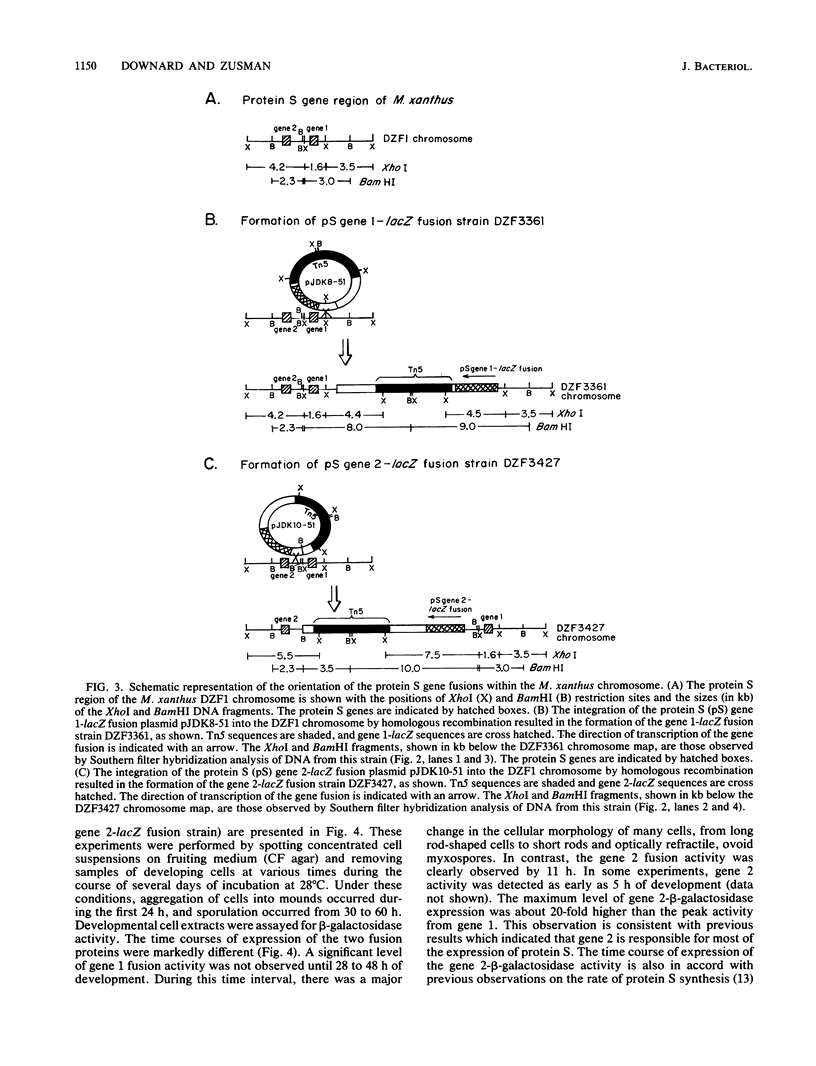
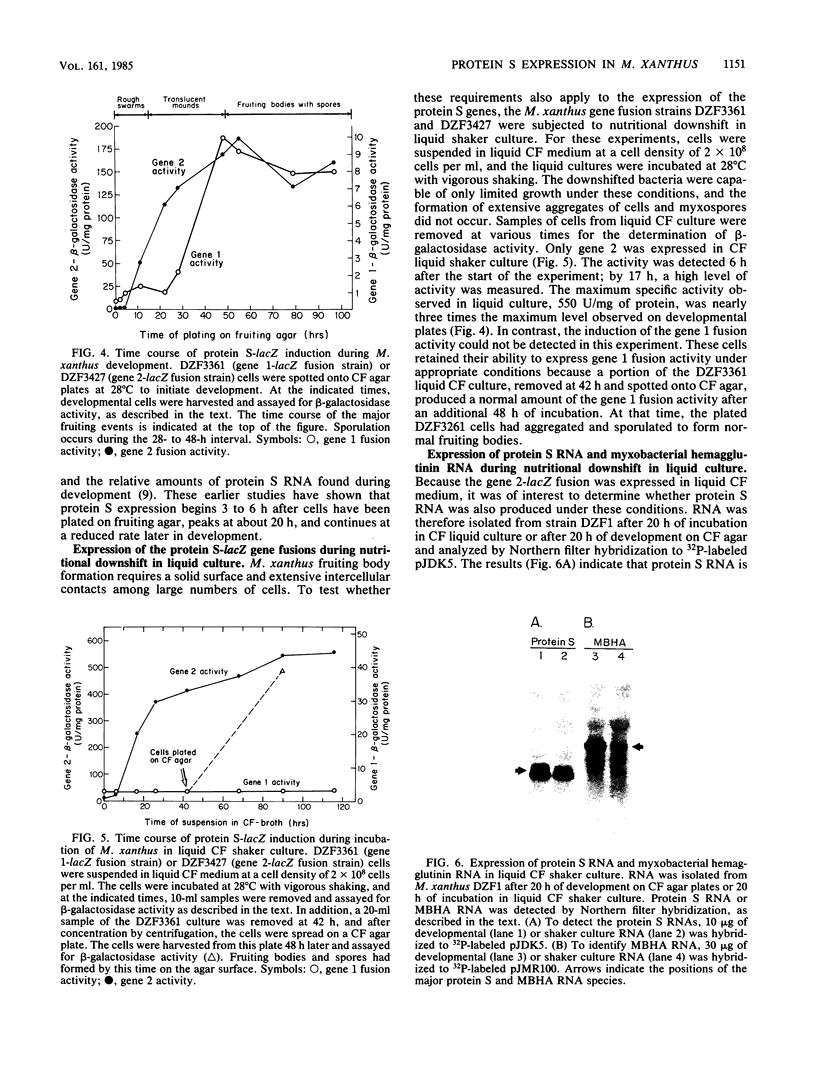
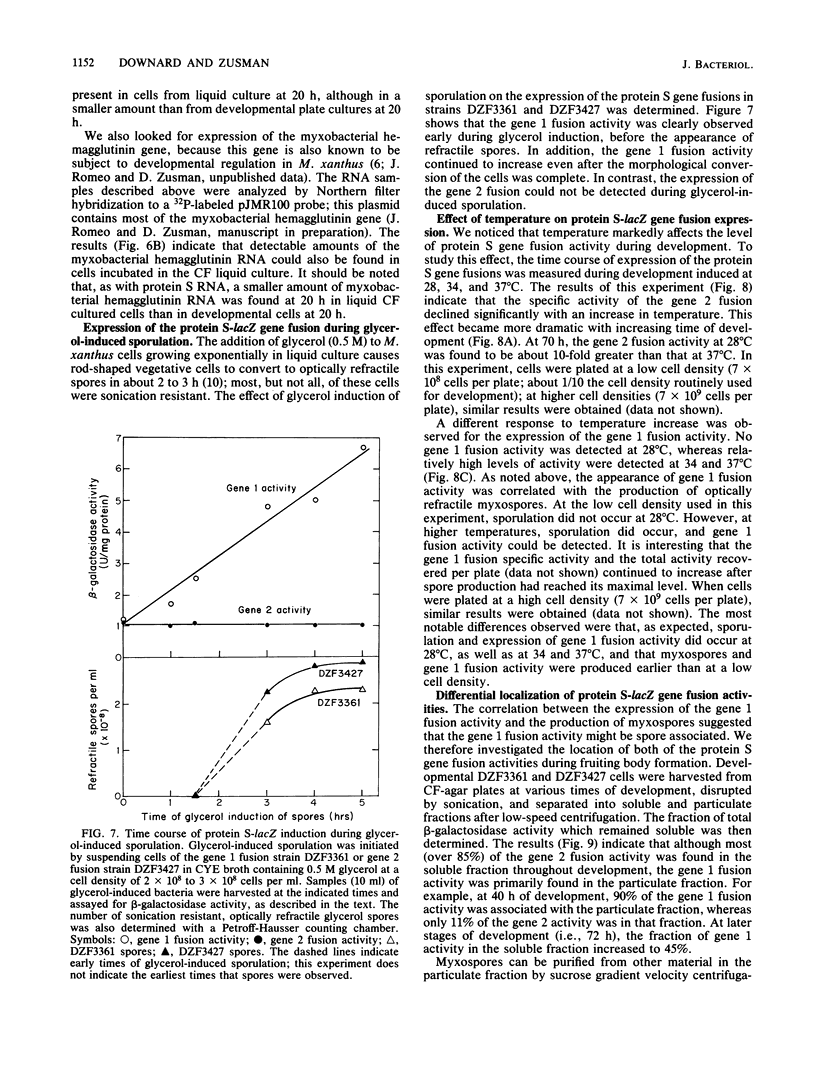
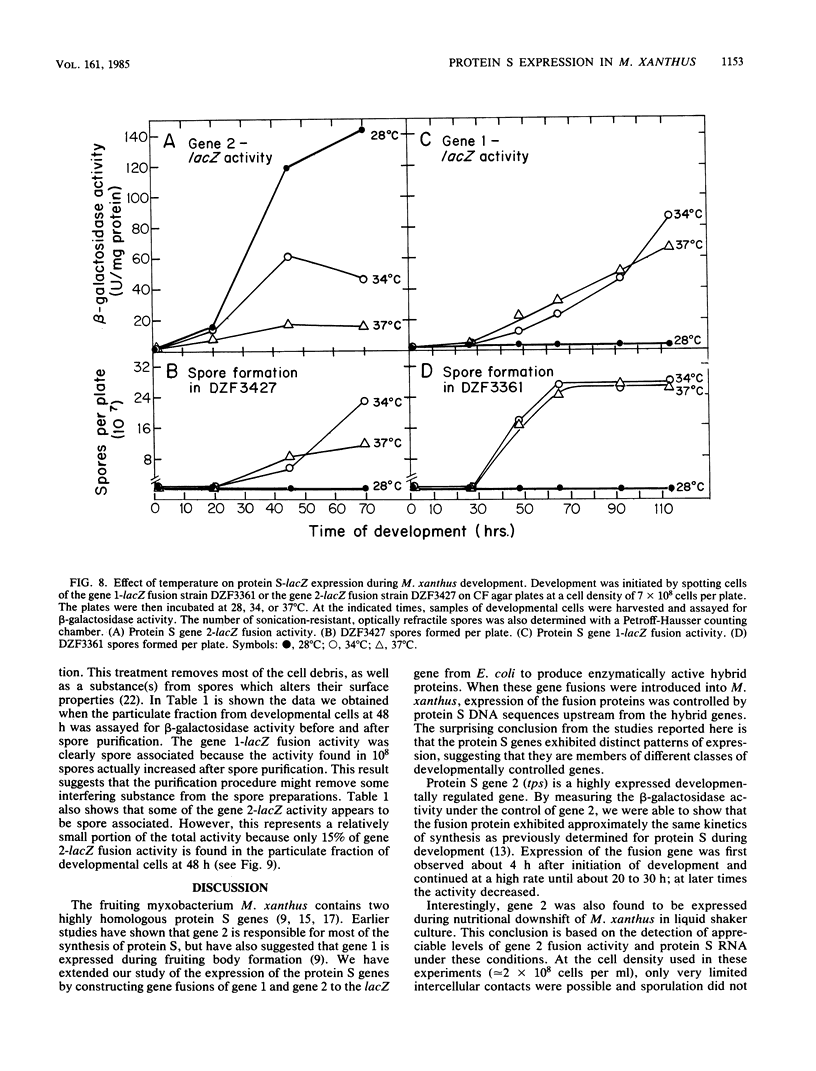
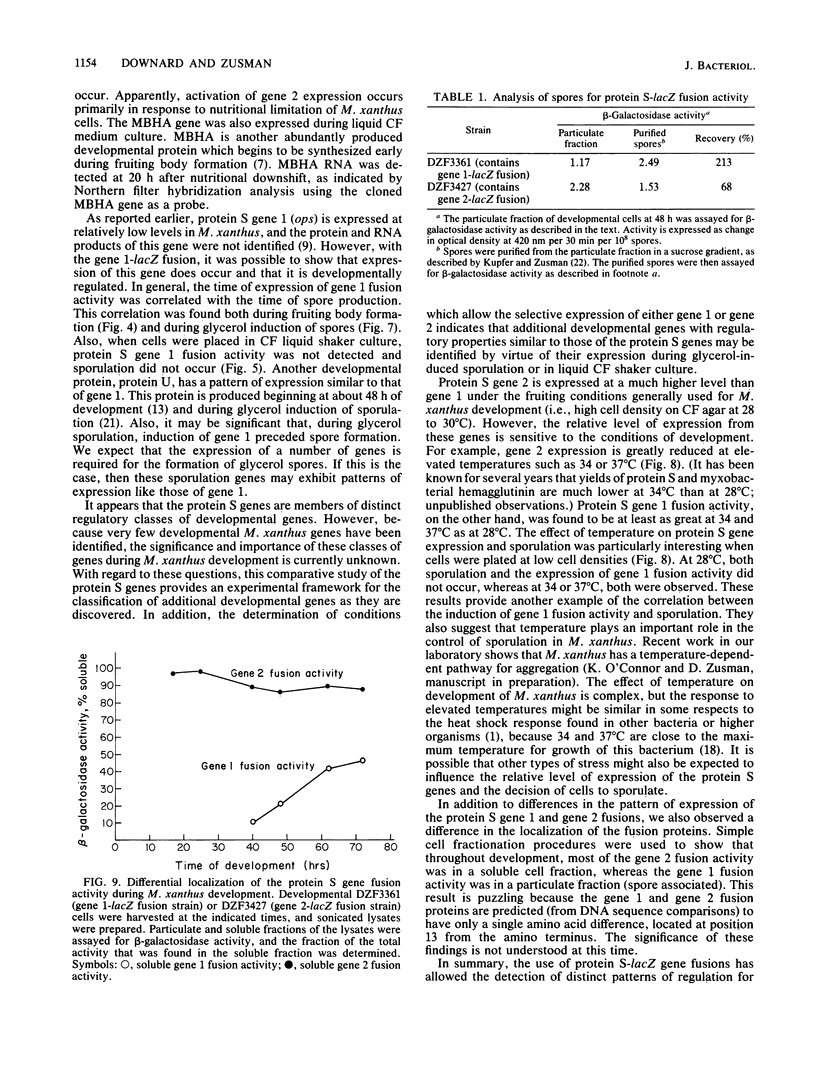
Protein S, the most abundant protein synthesized during development of the fruiting bacterium Myxococcus xanthus, is coded by two highly homologous genes called protein S gene 1 (ops) and protein S gene 2 (tps). The expression of these genes was studied with fusions of the protein S genes to the lacZ gene of Escherichia coli. The gene fusions were constructed so that expression of beta-galactosidase activity was dependent on protein S gene regulatory sequences. Both the gene 1-lacZ fusion and the gene 2-lacZ fusion were expressed exclusively during fruiting body formation (development) in M. xanthus. However, distinct patterns of induction of fusion protein activity were observed for the two genes. Gene 2 fusion activity was detected early during development on an agar surface and could also be observed during nutritional downshift in dispersed liquid culture. Gene 1 fusion activity was not detected until much later in development and was not observed after downshift in liquid culture. The time of induction of gene 1 fusion activity was correlated with the onset of sporulation, and most of the activity was spore associated. This gene fusion was expressed during glycerol-induced sporulation when gene 2 fusion activity could not be detected. The protein S genes appear to be members of distinct regulatory classes of developmental genes in M. xanthus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avery L., Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191(1):99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Cumsky M. G., Zusman D. R. Binding properties of myxobacterial hemagglutinin. J Biol Chem. 1981 Dec 10;256(23):12596–12599. [PubMed] [Google Scholar]
- Cumsky M. G., Zusman D. R. Purification and characterization of myxobacterial hemagglutinin, a development-specific lectin of Myxococcus xanthus. J Biol Chem. 1981 Dec 10;256(23):12581–12588. [PubMed] [Google Scholar]
- Cumsky M., Zusman D. R. Myxobacterial hemagglutinin: a development-specific lectin of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5505–5509. doi: 10.1073/pnas.76.11.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DWORKIN M., GIBSON S. M. A SYSTEM FOR STUDYING MICROBIAL MORPHOGENESIS: RAPID FORMATION OF MICROCYSTS IN MYXOCOCCUS XANTHUS. Science. 1964 Oct 9;146(3641):243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
- Downard J. S., Kupfer D., Zusman D. R. Gene expression during development of Myxococcus xanthus. Analysis of the genes for protein S. J Mol Biol. 1984 Jun 5;175(4):469–492. doi: 10.1016/0022-2836(84)90180-3. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hagen F., Young E. T. Regulation of synthesis of bacteriophage T7 lysozyme mRNA. Virology. 1973 Sep;55(1):231–241. doi: 10.1016/s0042-6822(73)81026-8. [DOI] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Gene expression during development of Myxococcus xanthus: pattern of protein synthesis. Dev Biol. 1979 Feb;68(2):579–591. doi: 10.1016/0012-1606(79)90228-8. [DOI] [PubMed] [Google Scholar]
- Inouye S., Franceschini T., Inouye M. Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6829–6833. doi: 10.1073/pnas.80.22.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Harada W., Zusman D., Inouye M. Development-specific protein S of Myxococcus xanthus: purification and characterization. J Bacteriol. 1981 Nov;148(2):678–683. doi: 10.1128/jb.148.2.678-683.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Ike Y., Inouye M. Tandem repeat of the genes for protein S, a development-specific protein of Myxococcus xanthus. J Biol Chem. 1983 Jan 10;258(1):38–40. [PubMed] [Google Scholar]
- Janssen G. R., Wireman J. W., Dworkin M. Effect of temperature on the growth of Myxococcus xanthus. J Bacteriol. 1977 Apr;130(1):561–562. doi: 10.1128/jb.130.1.561-562.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D., Manoil C., Dworkin M. Myxobacteria: cell interactions, genetics, and development. Annu Rev Microbiol. 1979;33:595–639. doi: 10.1146/annurev.mi.33.100179.003115. [DOI] [PubMed] [Google Scholar]
- Komano T., Furuichi T., Teintze M., Inouye M., Inouye S. Effects of deletion of the gene for the development-specific protein S on differentiation in Myxococcus xanthus. J Bacteriol. 1984 Jun;158(3):1195–1197. doi: 10.1128/jb.158.3.1195-1197.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Inouye S., Inouye M. Patterns of protein production in Myxococcus xanthus during spore formation induced by glycerol, dimethyl sulfoxide, and phenethyl alcohol. J Bacteriol. 1980 Dec;144(3):1076–1082. doi: 10.1128/jb.144.3.1076-1082.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer D., Zusman D. R. Changes in cell surface hydrophobicity of Myxococcus xanthus are correlated with sporulation-related events in the developmental program. J Bacteriol. 1984 Aug;159(2):776–779. doi: 10.1128/jb.159.2.776-779.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morrison C. E., Zusman D. R. Myxococcus xanthus mutants with temperature-sensitive, stage-specific defects: evidence for independent pathways in development. J Bacteriol. 1979 Dec;140(3):1036–1042. doi: 10.1128/jb.140.3.1036-1042.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Cumsky M. G., Zusman D. R. Localization of myxobacterial hemagglutinin in the periplasmic space and on the cell surface of Myxococcus xanthus during developmental aggregation. J Biol Chem. 1981 Dec 10;256(23):12589–12595. [PubMed] [Google Scholar]
- Nelson D. R., Zusman D. R. Evidence for long-lived mRNA during fruiting body formation in myxococcus xanthus. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1467–1471. doi: 10.1073/pnas.80.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Zusman D. R. Transport and localization of protein S, a spore coat protein, during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1983 May;154(2):547–553. doi: 10.1128/jb.154.2.547-553.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983 Jul;155(1):317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]