Abstract
Resistance in tomato to the bacterial pathogen Pseudomonas syringae pathovar tomato requires Pto and Prf. Mutations that eliminate Prf show a loss of both Pto resistance and sensitivity to the organophosphate insecticide fenthion, suggesting that Prf controls both phenotypes. Herein, we report that the overexpression of Prf leads to enhanced resistance to a number of normally virulent bacterial and viral pathogens and leads to increased sensitivity to fenthion. These plants express levels of salicylic acid comparable to plants induced for systemic acquired resistance (SAR) and constitutively express pathogenesis related genes. These results suggest that the overexpression of Prf activates the Pto and Fen pathways in a pathogen-independent manner and leads to the activation of SAR. Transgene-induced SAR has implications for the generation of broad spectrum disease resistance in agricultural crop plants.
Genetic analyses of plant disease resistance have demonstrated that resistance is controlled by single resistance genes in the host that specifically recognize pathogen strains containing complementary avirulence genes (1). Avirulence genes encode pathogen elicitor molecules that are either directly or indirectly recognized by the product of the corresponding plant disease resistance genes and specifically control the induction of plant host defenses to a narrow range of pathogens. The biochemical events responsible for plant disease resistance remain elusive, but recognition at the site of infection induces a signaling cascade resulting in changes in plant gene expression that leads to hypersensitive cell death in host cells and inhibition of pathogen growth (2). Expression of the local hypersensitive response (HR) also activates systemic acquired resistance (SAR), which results in nonspecific plant immunity to a broad range of pathogens in the distal portions of the plant (3).
A number of resistance and avirulence genes have been cloned from a wide variety of plants and pathogens respectively. The predicted protein products of most avirulence genes share very little homology with each other or with known protein sequences in several databases (4). On the other hand, resistance genes show a high degree of similarity, even though they are involved in resistance to a diverse array of pathogens (5, 6). The majority of resistance genes cloned to date contain both a nucleotide-binding site and a carboxy-terminal leucine-rich repeat domain. These genes are often referred to as the “NBS/LRR” superfamily of plant disease resistance genes. Although complementary pairs of resistance and avirulence genes have been cloned for several plant–pathogen interactions, the molecular events that define specificity have yet to be elucidated.
The cellular location of the receptors, hypothesized to be the protein products of resistance genes, should reflect the cellular location of the avirulence component. One class of resistance genes, predominantly responding to fungal pathogens, carry an amino terminal signal motif and a transmembrane domain, which would place the leucine-rich repeat region extracellularly (7–9). The avirulence components associated with these resistance genes do not enter the plant cell but rather are presumed to bind to the receptor extracellularly to induce disease resistance. Recent studies with these resistance genes suggest that the specificity of this resistance response lies within the extracellular leucine-rich repeat region (10).
Plant bacterial pathogens appear to carry a type III protein secretion system (11, 12) that has been shown in animal bacterial pathogens to be involved in the delivery of bacterial proteins directly into the host cell (13). Thus, it is hypothesized that the type III secretion system in phytopathogenic bacteria is involved in the delivery of bacterial proteins into the plant cell that are recognized intracellularly by the product of plant disease resistance genes. Furthermore, the transient introduction of bacterial avirulence proteins into the plant cell are sufficient to induce plant defenses (14, 15), providing further evidence that bacterial avirulence proteins are most likely recognized inside the plant cell.
Resistance in tomato to the bacterial pathogen Pseudomonas syringae pathovar tomato (P. s. tomato) is dependent on the tomato resistance genes Pto and Prf (16, 17). Pto encodes a protein kinase with serine–threonine specificity (18). Prf is a member of a superfamily of plant disease resistance genes that contain leucine-rich repeat and nucleotide-binding site protein motifs (19). The specific interaction of AvrPto, the avirulence protein from P. s. tomato (20), and Pto (15, 21) is presumed to initiate a kinase cascade that ultimately activates plant defenses. Mutations in Prf simultaneously display a loss of avrPto-specified resistance and a loss in sensitivity to the organophosphate insecticide fenthion, which also requires the Fen gene, a Pto-related kinase (22). Current hypotheses suggest Prf acts in concert with Pto and Fen or more likely acts downstream of Pto and Fen to induce signal transduction events leading to plant resistance.
Recognition of an avirulent pathogen by the plant results in the local induction of plant defenses and the systemic induction of SAR. The activation of SAR is associated with increased levels of salicylic acid in distal regions of the plant (23). The introduction of salicylate hydroxylase, a bacterial protein that converts salicylic acid into the inactive form catechol, into plants results in the loss of both SAR (24) and local resistance (25) associated with the recognition of an avirulence protein. The mechanism by which salicylic acid functions is still unclear, but the systemic spread of SAR may be associated with the spread of microscopic HRs throughout the plant after recognition of an avirulent pathogen (26), which could function to activate salicylic acid production. Also associated with SAR is the systemic activation of pathogenesis related genes (27). SAR can be activated by a specific avirulent pathogen; however, it functions against a diverse range of virulent pathogens.
One of the major goals of designing effective strategies for broad spectrum plant disease resistance has been to exploit the SAR pathway by either chemical or genetic means. Constitutive induction of the SAR pathway has been achieved through the application of the chemical inducers 2,6-dichloroisonicotinic acid (28) and benzothiadiazole (29) and the isolation and characterization of plant mutants that constitutively induce the SAR pathway (30–35). The induction of SAR in these mutants is often associated with the activation of the HR that leads to the development of localized cell death throughout the plant (30–33). Such lesion-mimic mutations have been effective in designing resistance to powdery mildew in barley (36, 37); however, the formation of cell death lesions must be tightly regulated to avoid uncontrolled cell death throughout the plant. Dwarfism also is commonly associated with mutations that constitutively induce SAR (34, 35). A challenge for genetic engineering has been to develop plants that can express the SAR pathway without such deleterious side effects.
Considering that Prf is presumed to function downstream of the avrPto recognition event, we hypothesized that overexpression of Prf could lead to the constitutive activation of this defense pathway and thus provide a tool for engineering transgene-induced SAR. Here, we report that low levels of Prf mRNA overexpression are sufficient for the induction of SAR but insufficient for activation of HR. Furthermore, fenthion sensitivity is increased in plants that overexpress Prf mRNA, however avrPto-specified resistance is unaffected.
MATERIALS AND METHODS
Pathogen Strains and Plant Lines.
The P. s. tomato strains used in this study were DC3000, a strain that carries avrPto and T1, a strain that lacks avrPto activity. DC3000 and T1 were kindly provided by D. Cuppels (Agriculture Canada Research Center, London, ON Canada) and G. Bonn (Harrow Research Station, University of California, Davis), respectively. Xanthomonas campestris pv. vesicatoria strain 56 and Ralstonia solanacearum strain 82 were kindly provided by B. Stahl (University of Florida, Gainesville) and L. Sequiera (University of Wisconsin, Madison), respectively. Septoria lycopersici strain 810C was kindly provided by D. Hillard (New York State Agricultural Experiment Station, Cornell University, Ithaca). Tobacco Mosaic Virus (TMV) was acquired from greenhouse grown plants naturally infected with the virus.
The tomato varieties Rio Grande, VF36 and Moneymaker were used in this study. VF36 and Moneymaker both lack the Pto resistance gene, although they are presumed to carry a functional Prf. The Rio Grande 76R used in this study carries the Pto resistance gene and has been referred to as wild-type throughout this work. The Prf mutant prf-3 that shows a 1.1-kb deletion was used for transformation. prf-3 was generated by fast neutron mutagenesis of the wild-type Rio Grande 76R.
Plant Transformation and Organization of T-DNA Inserts.
pSOR2–7, a pCDL04541 T-DNA vector containing plant DNA subcloned into the BamHI site (19) was transformed into tomato varieties Rio Grande prf-3 and VF36 by using Agrobacterium tumefaciens strain LBA4404. pSOR2–7 was shown to carry the Prf gene through complementation of the prf-3 mutant with this cosmid (7). Positive plant transformants were identified through selection on kanamycin (250 μg/ml) and verified by DNA blot analysis. To assess the number of copies of Prf inserted into the plant transformants, the DNA flanking the inserts was analyzed. The pCDL04541 T-DNA vector that was used in the transformation has a unique EcoRI site on the left border side of the inserted plant DNA. To identify each T-DNA insertion, genomic DNA was digested with EcoRI, and analyzed by DNA blot analysis by using a probe from the left border of the T-DNA. Each point of insertion can be identified by its unique bands. For the insert 1/insert 2 (I1/I2) transformant (T0) 100 progeny (T1) from a self-cross of the original transformant were screened for their T-DNA insertions. Two sets of two bands cosegregated, suggesting two-unlinked insertional sites and because each site has multiple bands then each site must contain multiple copies of the T-DNA.
Fenthion and Pathogen Screens.
Growth curves and the fenthion assays were conducted as described (38). P. s. tomato was used at a concentration of 1 × 103 colony-forming units (cfu)/ml for growth curves and 5 × 104 cfu/ml for disease assays. P. s. tomato disease was assayed by pipette infiltration into the mesophyll of individual intact leaves. Disease symptoms were scored 5 days after inoculation. Growth curves using X. c. vesicatoria were initiated by vacuum infiltrating plants with bacteria at a concentration of 1 × 105 cfu/ml. R. solanacearum was inoculated into the vascular tissue of tomatoes at a concentration of 1 × 109 cfu/ml by injecting 25 μl glass pipettes, previously dipped into the bacterial solution, into the stem at the soil surface. Twelve days after inoculation, the stem was cut every 3 cm from the inoculation point and touched onto Pseudomonas agar F (Difco). Plates were grown for 2 days at 28°C. To quantitate the number of bacteria in the stem, 0.1 gm of stem tissue was isolated, ground in 10 mM MgCl2 and dilutions were plated on Pseudomonas agar F with rifampicin 100 μg/ml (a rifampicin resistant strain of R. solanacearum was used in this experiment). Bacterial colonies were counted after 2 days at 28°C. TMV inoculum was obtained by grinding TMV-infected tomato leaves in 10 mM potassium phosphate, pH 7.0, with celite. Lower leaves of 5-wk-old tomatoes were brushed immediately with this inoculum. After 10 days, RNA was isolated from upper leaves. S. lycopersici was maintained on potato dextrose agar in the dark. Spores were obtained by flooding plates with water. Plants were infected with S. lycopersici by brushing leaves with spores at a concentration of 1.5 × 106 spores/ml. The plants were kept under plastic to maintain high humidity levels for 4 days. Disease levels were scored after 7 days.
Introduction of NahG.
Salicylate hydroxylase (NahG) had previously been transformed into the tomato line Moneymaker (39). NahG-containing plants were crossed with I2 and the F1 of this cross was tested for the enhanced resistance. The presence of NahG and I2 in these plants was verified by DNA blot analysis. Because I2 is in tomato variety Rio Grande, the loss of the enhanced resistance could be a function of NahG or the introduction of a Moneymaker background. To control for this effect, I2 was crossed with wild-type Moneymaker and the progeny tested for the enhanced resistance. No difference in growth of virulent P. s. tomato was observed in these plants as compared with I2.
Salicylic Acid Quantitation.
Plant tissue (0.5 g) was collected from axenically grown plants. Wild-type plants were induced for SAR by hand inoculating lower leaves with DC3000 at 1 × 108 cfu/ml and upper leaf tissue collected 48 hr later. Both free and total salicylic acid, after hydrolysis with β-glucosidase, were quantified by using HPLC (40).
RESULTS
Identification of Plants that Overexpress Prf mRNA.
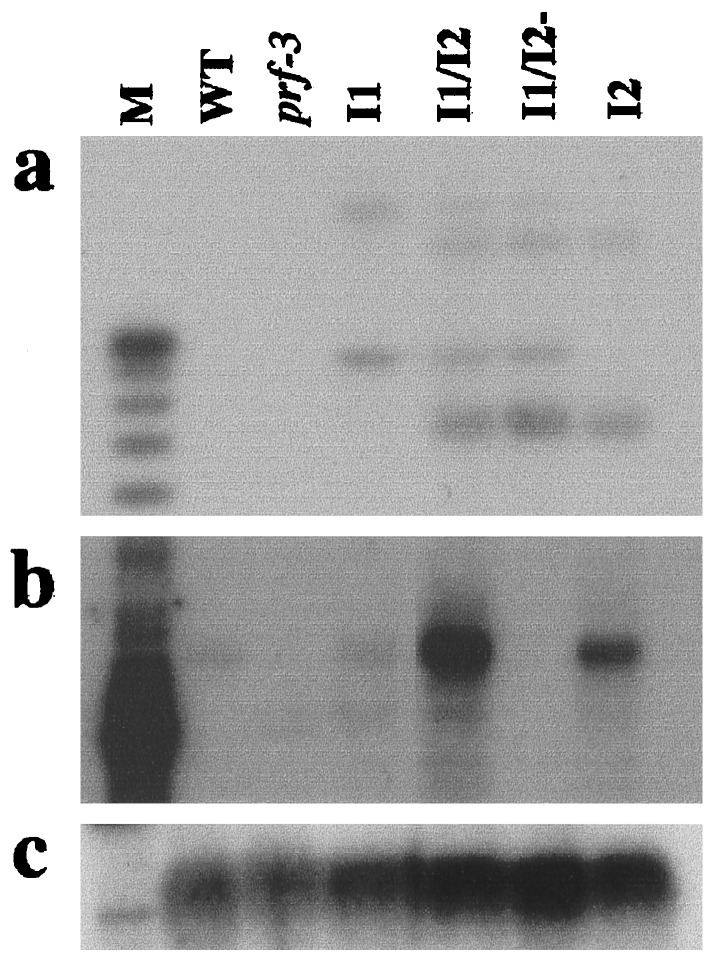
We screened VF36 and prf-3 mutant plants transformed with a cosmid containing Prf for enhanced Prf mRNA expression levels and resistance to T1, a normally virulent strain of P. s. tomato. Seven independent transformants showed both increased Prf mRNA expression and resistance to T1 (data not shown), indicating that Prf overexpression mediates enhanced disease resistance. In four of these plants, Prf mRNA overexpression appears to be a function of multiple insertions of the T-DNA. The transformant showing the highest level of Prf mRNA expression, a prf-3 transformant, contains two unlinked multi-copy T-DNA insertion sites (see Materials and Methods and Fig. 1a). Plants homozygous for either insert were isolated from a self-crossing of the original transformant (Fig. 1a). Those containing T-DNA I1 and I2 exhibited a 1.5- and 6-fold increase, respectively, in Prf mRNA expression over wild-type plants. In contrast, plants carrying both I1 and I2 T-DNA inserts (I1/I2) show a 10-fold increase in Prf mRNA expression (Fig. 1b).
Figure 1.
Multiple insertions of the Prf-containing T-DNA leads to Prf mRNA overexpression. (a) T-DNA copy number was assessed by DNA blot analysis by using the left border of the T-DNA vector as a probe. The multiple bands in I1/I2 indicate multiple copies of the T-DNA. Two sets of bands cosegregate, suggesting two insertion sites I1 and I2. (b) Prf mRNA overexpression as assayed by RNA blot analysis by using a 3′ probe of Prf. (c) RNA blot from b probed with 18S rDNA as a loading control. prf-3, a prf mutant containing a 1.1-kb deletion in the ORF; I1, insert 1; I2, insert 2; I1/I2, insert 1 and insert 2; I1/I2-, insert 1 and insert 2, silenced for Prf.
The Effect of Prf mRNA Overexpression.
Overexpression of Prf mRNA causes increased fenthion sensitivity (Fig. 2a) and enhanced resistance to P. s. tomato T1 (Fig. 2 b and c) but has no effect on avrPto-specified resistance (data not shown). The overexpression of Prf mRNA does not appear to lead to any additional phenotypes. Plant growth and fruit production are equivalent to wild type. Furthermore, no HR-like lesions at the macroscopic or microscopic levels were observed in I2 plants (data not shown). Because SAR leads to immunity to virulent pathogens, the enhanced resistance to the virulent pathogen P. s. tomato T1 suggests that the overexpression of Prf leads to the constitutive activation of SAR.
Figure 2.
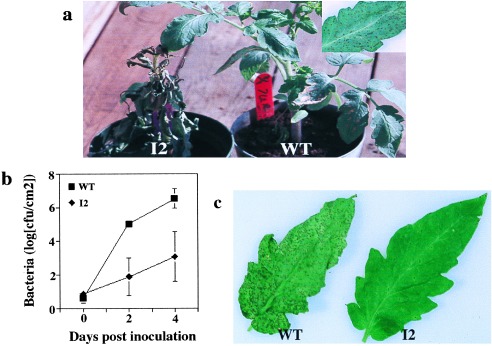
Prf overexpression leads to increased fenthion sensitivity (a) and enhanced resistance to virulent P. s. tomato T1 shown by growth of the bacteria (b) and the level of disease symptoms that develop (c). The Inset in a shows standard fenthion symptoms that are not visible in the wild-type plant at this magnification. Data points in b represent the mean of three replicate experiments ±SD.
Plants with the Highest Level of Prf Expression Show Gene Silencing of Prf.
Within a population of plants homozygous for both inserts (I1/I2), 50% revealed no Prf mRNA, neither the full length mRNA species or the prf-3 deletion mRNA species (I1/I2-, Fig. 1b). This result is presumed to be a function of transgene silencing induced by the overexpression of Prf. Transgene silencing is a common phenomenon in transgenic plants that results in the simultaneous loss of expression of the transgene and the corresponding native gene (41). Transgene silencing is induced by high expression levels of the transgene, which can be generated from high expression promoters, or multiple copies of the T-DNA (42, 43). Of 22 I1/I2 plants silenced for Prf mRNA expression that were analyzed, all were susceptible to P. s. tomato strains carrying avrPto, were insensitive to Fenthion, and did not show the resistance to P. s. tomato T1. These results are phenocopies of Prf loss of function mutants (17) and suggest that the overexpression of Prf is required for the enhanced resistance to virulent P. s. tomato T1.
The Enhanced Resistance to T1 Cosegregates with Prf mRNA Overexpression.
The transgene silencing was used to show cosegregation of Prf overexpression with the enhanced resistance to virulent bacteria. Fourteen I1/I2 plants were analyzed for resistance to P. s. tomato T1, a virulent strain and DC3000, an avirulent strain carrying avrPto. The expression level of Prf mRNA was assayed for each plant. The enhanced resistance phenotype only occurred in transgenic plants that expressed high levels of Prf mRNA (Fig. 3). Plants not expressing Prf mRNA did not display enhanced resistance to T1 or resistance to DC3000. A single transgenic I1/I2 plant (Fig. 3a, lane 7) expressing only wild-type levels of Prf mRNA, did not exhibit resistance to T1 but was resistant to DC3000. Thus wild-type levels of Prf mRNA are sufficient to induce avrPto-specified resistance, but high levels of Prf mRNA are required for enhanced resistance to the virulent strain T1. Transgene silencing has a high degree of specificity, and the cosegregation of Prf mRNA overexpression with resistance to T1 provides strong evidence that Prf overexpression is responsible for the enhanced resistance phenotype.
Figure 3.

Cosegregation of Prf mRNA overexpression with enhanced resistance to P. s. tomato T1. (a) RNA blot analysis showing expression of Prf in a population of plants homozygous for I1 and I2. Plants were screened for their response to virulent P. s. tomato T1 and avirulent P. s. tomato DC3000 after hand infiltration of leaves with bacteria at a concentration of 5 × 104 cfu/ml. (b) The RNA blot from a probed with 18S rDNA as a loading control. Lane 1, wild-type; lane 2, prf-3 mutant; and lanes 3–16, I1/I2 plants. +, disease; −, no disease.
Prf mRNA Overexpression Leads to the Induction of SAR Markers.
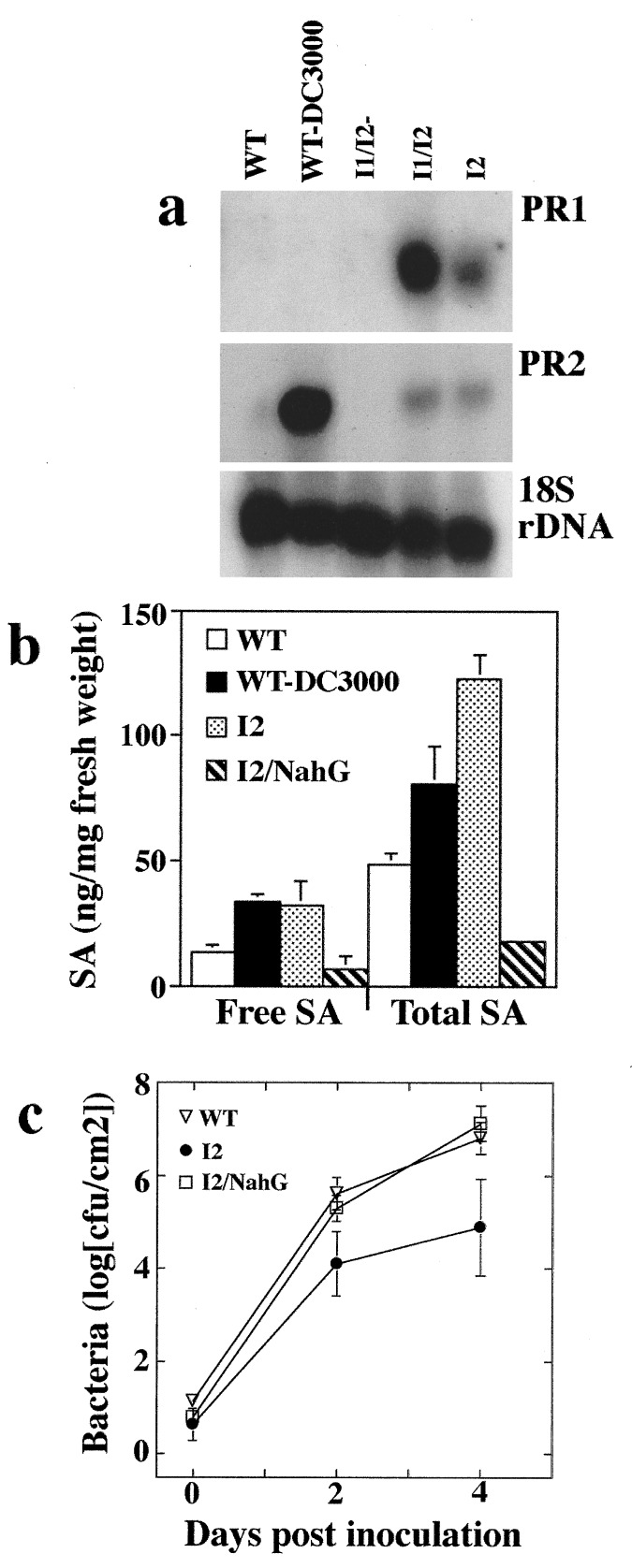
To confirm that the enhanced resistance resulting from Prf overexpression is a function of the activation of SAR, we tested a number of markers of SAR in I2 plants. Increased salicylic acid levels and pathogenesis-related (PR) gene expression correlate with the induction of SAR (23, 27). Furthermore, constitutive expression of PR1 in tobacco leads to increased tolerance to the fungal pathogens Peronospora tabacina and Phytophthora parasitica (44). Both PR1 and PR2 are constitutively expressed in I2 and I1/I2 plants but not in plants silenced for Prf expression (Fig. 4a; induced PR1 mRNA expression in wild-type is observed at longer exposures). Salicylic acid levels were 2- to 3-fold higher in I2 plants compared with wild-type. Similar levels of salicylic acid were found in wild-type plants induced for SAR with DC3000 (Fig. 4b). To test whether increased levels of salicylic acid are required for Prf-enhanced resistance, the salicylic acid-degrading enzyme salicylate hydroxylase (24) (NahG) was introduced into I2 plants (see Materials and Methods). I2/NahG plants were fully susceptible to P. s. tomato T1 (Fig. 4c). Furthermore, the level of salicylic acid in I2/NahG plants was below the level of uninduced 76R plants (Fig. 4b), confirming that increased salicylic acid levels are required for Prf-mediated enhanced resistance.
Figure 4.
Prf overexpression leads to the activation of markers of SAR. (a) Plants that overexpress Prf constitutively express PR1 and PR2. The blot containing total RNA was sequentially probed with PR1, PR2, and an 18S rDNA probe as a loading control. Wild-type plants were induced for defense gene expression by vacuum infiltration with P. s. tomato DC3000 at a concentration of 1 × 107 cfu/ml and tissue collected from inoculated leaves 7 hr later. I1/I2-, insert 1 and insert 2, silenced for Prf; I1/I2, insert 1 and insert 2; and I2, insert 2. (b) Levels of free and total salicylic acid in axenically grown plants. Data points represent the mean of two replicates ±SD. WT-DC3000, wild-type plant induced for SAR. (c) The reduction in salicylic acid levels caused by NahG eliminates the enhanced resistance to P. s. tomato T1 in I2 plants. Data points represent the mean of three replicate experiments ±SD.
Transgene-Induced SAR Leads to Resistance to a Number of Previously Virulent Pathogens of Tomato.
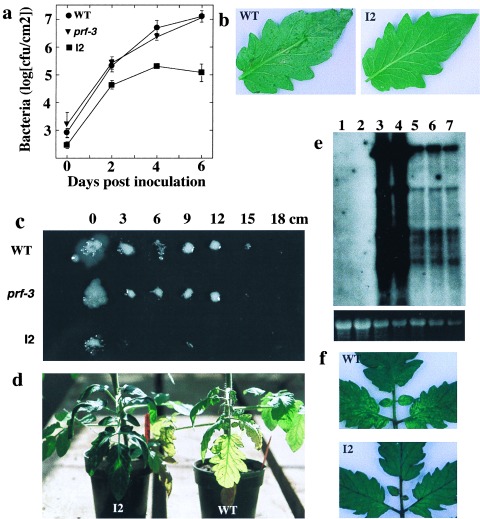
Because SAR appears to be constitutively active in plants that overexpress Prf mRNA, we tested I2 plants for enhanced resistance to additional pathogens. Although SAR provides resistance to a broad range of pathogens, a number of pathogens appear to be unresponsive to SAR resistance (3). Currently the spectrum of pathogens limited by SAR in tomato is not known. I2 plants were resistant to all pathogens tested (Fig. 5), except S. lycopersici, a fungal pathogen of tomato (data not shown). Growth of X. c. vesicatoria, a foliar bacterial pathogen, was reduced ≈100-fold in I2 plants (Fig. 5a) and disease symptoms associated with this pathogen were reduced in I2 plants (Fig. 5b). R. solanacearum is a vascular bacterial pathogen that multiplies in the xylem, and moves systemically, resulting in wilt symptoms that can ultimately kill the plant. I2 plants restricted bacterial spread to the point of vascular inoculation, whereas wild-type plants allowed spread of the bacteria 15 cm up the stem (Fig. 5c). We quantified bacterial titers in the stems and demonstrated that I2 plants reduced bacterial growth 1,000-fold at 9 cm distance from the inoculation point as compared with wild-type. Disease symptoms caused by R. solanacearum include leaf chlorosis and wilting. These symptoms were reduced in I2 plants (Fig. 5d). TMV, a single stranded RNA virus, rapidly spreads throughout the plant from the initial inoculation point. This systemic spread was suppressed in I2 plants, as assayed by levels of viral RNA in upper uninoculated leaves (Fig. 5e). The mosaic symptoms caused by this virus also were reduced in upper leaves of I2 plants (Fig. 5f). In summary, we observed 100- to 1,000-fold reductions in pathogen titers of the various pathogens tested, suggesting that Prf overexpression may prove useful in generating durable field resistance to a broad range of pathogens.
Figure 5.
Prf overexpression leads to enhanced resistance to diverse pathogens. (a) Growth of X. c. vesicatoria. Data points represent the mean of three replicate experiments ±SD. (b) Water-soaked lesions caused by X. c. vesicatoria on leaves of infected plants. (c) R. solanacearum assayed from the stem of infected plants. The numbers indicate the distance up the stem from the point of inoculation. (d) symptoms of R. solanacearum-infected plants. (e) RNA blot analysis of TMV-infected plants. The movement protein gene of TMV was used as a probe. The multiple bands represent the subgenomic processing of the TMV genome. The EtBr stain of rRNA from the RNA blot is shown below as a loading control. Lane 1, wild-type uninfected; lane 2, I2 uninfected; lane 3, prf-3 TMV infected; lane 4, wild-type TMV infected; and lanes 5–7, three I2 plants TMV infected. (f) TMV mosaic symptoms on upper uninoculated leaves infected by TMV through the systemic spread of the virus.
DISCUSSION
We have shown that resistance to a broad spectrum of pathogens can be achieved by the overexpression of Prf mRNA, a component of the Pto resistance pathway. By testing markers of SAR, we have demonstrated that this resistance is most likely a function of constitutively activated SAR. This data suggests that the overexpression of Prf can activate this defense pathway independent of fenthion or avrPto. Furthermore, the overexpression of Prf leads to increased sensitivity to fenthion, but has no effect on avrPto-specified resistance. The response to fenthion suggests that Prf is a limiting factor in this resistance pathway. However, if this is the case, there should be an effect of overexpression of Prf on avrPto-specified resistance. Prf overexpression may increase the response to avrPto; however, it could be that the level of bacterial resistance specified by avrPto recognition is already functioning at its maximum level in the wild-type situation.
Mutations that lead to constitutive SAR often have yield-reducing pleiotropic phenotypes such as the development of lesions on the leaves of the plant or dwarfism (45). The development of cell death lesions in these mutants has been shown to be a function of the activation of the HR (31, 32). Cell death and the induction of SAR are associated with the activation of the Prf pathway through the recognition of the elicitors AvrPto and fenthion. However, the activation of this pathway through the overexpression of Prf mRNA does not lead to either macroscopic or microscopic HR, although it does lead to the activation of SAR. The induction of SAR has been hypothesized to be dependent on the activation of the HR. This data suggests that the activation of SAR is independent of cell death. The signaling leading to HR may be activated in plants that overexpress Prf mRNA, but the threshold of activation could be insufficient for HR induction but sufficient for the activation of SAR. An alternative explanation is that SAR and HR are induced by the action of two separate pathways, both of which are normally activated after the recognition of an avirulence component, but only the SAR pathway is activated by Prf overexpression. This hypothesis is supported by the apparently separate functions of the Pto interactors Pti1 and Pti4/5/6. Pti1 appears to be involved in the activation of the HR (46), after recognition of AvrPto, whereas Pti4/5/6 are transcription factors that are proposed to activate the expression of the PR genes (47).
The induction of SAR without deleterious side effects makes Prf-mediated transgenic SAR a target for the production of broad spectrum-enhanced resistance in agricultural crops. This is especially important in regards to X. c. vesicatoria and R. solanacearum because both of these pathogens cause serious diseases in tomato for which there is no effective genetic resistance. The hypothesis that Prf overexpression can generate transgenic SAR in additional crop plants is being currently tested. Genetic engineering holds the key for future agriculture, and the development of broad spectrum resistance may have major implications for world food production.
Acknowledgments
We thank J. Dangl, W. Gassmann, T. McNellis, M. Mudjett, P. Repetti, and M. Sainz for advice and critical reading of the manuscript. We thank B. Hall and E. Torok for generation of plant transformants. We thank D. Dahlbeck for technical assistance. This work was supported in part by National Science Foundation Cooperative agreement BIR-8920216 to the Center for Engineering Plant Resistance Against Pathogens, a National Science Foundation Science and Technology Center and by the Center for Engineering Plant Resistance Against Pathogens corporate associates Calgene, Inc., Novartis Biotechnology Corporation, Sandoz Seeds, and Zeneca Seeds.
ABBREVIATIONS
- HR
hypersensitive response
- SAR
systemic acquired resistance
- cfu
colony-forming units
- PR
pathogenesis-related
- I1
insert 1
- I2
insert 2
- TMV
Tobacco Mosaic virus
References
- 1.Staskawicz B J, Asubel F M, Baker B J, Ellis J E, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 2.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryals J A, Neuenschwander U H, Willits M J, Molina A, Steiner H-Y, Hunt M. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 5.Dangl J L. Cell. 1995;80:363–366. doi: 10.1016/0092-8674(95)90485-9. [DOI] [PubMed] [Google Scholar]
- 6.Bent A F. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones D A, Thomas C M, Hammond-Kosack K E, Balint-Kurti P J, Jones J D G. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- 8.Song W, Wang G L, Chen L L, Kim H S, Pi L Y, Holsten T, Gardner J, Wang B, Zhai W X, Zhu L H, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 9.Dixon M S, Jones D A, Keddie J S, Thomas C M, Harrison K, Jones J D G. Cell. 1996;84:451–459. doi: 10.1016/s0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- 10.Thomas C M, Jones D A, Parniske M, Harrison K, Balint-Kurti P J, Hatzixanthis K, Jones J D G. Plant Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mudgett M, Staskawicz B J. Curr Opin Microbiol. 1998;1:109–115. doi: 10.1016/s1369-5274(98)80150-1. [DOI] [PubMed] [Google Scholar]
- 13.Lee C A. Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 14.Gopalen S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 16.Martin G B, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 17.Salmeron J M, Barker S J, Carland F M, Mehta A Y, Staskawicz B J. Plant Cell. 1994;6:511–520. doi: 10.1105/tpc.6.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh Y, Martin G B. Plant Physiol. 1995;108:1735–1739. doi: 10.1104/pp.108.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmeron J M, Oldroyd G E D, Rommens C M T, Scofield S R, Kim H, Lavelle D T, Dahlbeck D, Staskawicz B J. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 20.Ronald P C, Salmeron J M, Carland F M, Staskawicz B J. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X, Frederick R D, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 22.Martin G B, Frary A, Wu T, Brommonschenkel S, Chunwongse J, Earle E D, Tanksley S D. Plant Cell. 1994;6:1543–1552. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malamy J, Carr J P, Klessig D F, Raskin I. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- 24.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 25.Delaney T P, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto N, Gaffney T, Manuela G-R, Kessman H, Ward E. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez M E, Pennell R I, Meijer P, Ishikawa A, Dixon R A, Lamb C. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 27.Ward E R, Uknes S J, Williams S C, Dincher S S, Wiederhold D L, Alexander D C, Ahl-Goy P, Métraus J-P, Ryals J A. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawton K A, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- 30.Bowling S A, Guo A, Gordon A S, Klessig D F, Dong X. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietrich R A, Delaney T P, Uknes S J, Ward E R, Ryals J A, Dangl J L. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 32.Greenberg J T, Guo A, Klessig D F, Asubel F M. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 33.Bowling S A, Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, I., Parker, J. & Bent, A. F. (1998) Proc. Natl. Acad. Sci. USA, 95, in press. [DOI] [PMC free article] [PubMed]
- 36.Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. Mol Gen Genet. 1993;239:122–128. doi: 10.1007/BF00281610. [DOI] [PubMed] [Google Scholar]
- 37.Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 38.Carland F M, Staskawicz B J. Mol Gen Genet. 1993;239:17–27. doi: 10.1007/BF00281596. [DOI] [PubMed] [Google Scholar]
- 39.Brading P A. Ph.D. thesis. Norwich, UK: University of East Anglia; 1993. [Google Scholar]
- 40.Uknes S, Winter A M, Delaney T, Vernooij B, Morse A, Friedrich L, Nye G, Potter S, Ward E, Ryals J. Mol Plant Microbe Interact. 1993;6:692–698. [Google Scholar]
- 41.Jorgensen R A, Atkinson R G, Forster R L S, Lucas W J. Science. 1998;279:1486–1487. doi: 10.1126/science.279.5356.1486. [DOI] [PubMed] [Google Scholar]
- 42.Hobbs S L A, Warkentin T D, Delong C M O. Plant Mol Biol. 1993;21:17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- 43.Cluster P D, O’Dell M, Metzlaff M, Flavell R B. Plant Mol Biol. 1996;32:1197–1203. doi: 10.1007/BF00041406. [DOI] [PubMed] [Google Scholar]
- 44.Alexander A, Goodman R M, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E, et al. Proc Natl Acad Sci USA. 1993;90:7327–7331. doi: 10.1073/pnas.90.15.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dangl J L, Dietrich R A, Richberg M H. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Loh Y, Bressan R A, Martin G B. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 47.Zhou J, Tang X, Martin G B. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]