Abstract
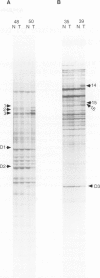
Genomic alterations have been analysed in 65 non-small-cell lung cancer (NSCLC) tissue samples by using the arbitrarily primed polymerase chain reaction (AP-PCR), which is a PCR-based genomic fingerprinting. We have shown that AP-PCR may be applied as a useful and feasible practical method for detection of the genomic alterations that accompany malignancy in NSCLC. Genomic changes detected by us consisted of: allelic losses or gains in anonymous DNA sequences, homozygously deleted DNA sequences and polymorphic DNA sequences. According to these genomic changes, lung tumours evaluated in the present study have been scored into three groups: low, moderate and high genomic damage tumours. The aim of this study was to investigate the effect of genomic damage on patient survival. Survival analysis was carried out in 51 NSCLC patients. Our results revealed that high genomic damage patients showed a poorer prognosis than those with low or moderate genomic damage (P = 0.038). Multivariate Cox regression analysis showed that patients with higher genomic alterations displayed an adjusted-by-stage risk ratio 4.26 times higher than the remaining patients (95% CI = 1.03-17.54). We can conclude that genomic damage has an independent prognostic value of poor clinical evolution in NSCLC.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achille A., Biasi M. O., Zamboni G., Bogina G., Magalini A. R., Pederzoli P., Perucho M., Scarpa A. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996 Aug 15;56(16):3808–3813. [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend S. H., Bernards R., Rogelj S., Weinberg R. A., Rapaport J. M., Albert D. M., Dryja T. P. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986 Oct 16;323(6089):643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- Gomez-Lus P., Fields B. S., Benson R. F., Martin W. T., O'Connor S. P., Black C. M. Comparison of arbitrarily primed polymerase chain reaction, ribotyping, and monoclonal antibody analysis for subtyping Legionella pneumophila serogroup 1. J Clin Microbiol. 1993 Jul;31(7):1940–1942. doi: 10.1128/jcm.31.7.1940-1942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio Y., Takahashi T., Kuroishi T., Hibi K., Suyama M., Niimi T., Shimokata K., Yamakawa K., Nakamura Y., Ueda R. Prognostic significance of p53 mutations and 3p deletions in primary resected non-small cell lung cancer. Cancer Res. 1993 Jan 1;53(1):1–4. [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O. P., Sudar D., Rutovitz D., Gray J. W., Waldman F., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992 Oct 30;258(5083):818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kohno T., Morishita K., Takano H., Shapiro D. N., Yokota J. Homozygous deletion at chromosome 2q33 in human small-cell lung carcinoma identified by arbitrarily primed PCR genomic fingerprinting. Oncogene. 1994 Jan;9(1):103–108. [PubMed] [Google Scholar]
- Martínez-Murcia A. J., Rodríguez-Valera F. The use of arbitrarily primed PCR (AP-PCR) to develop taxa specific DNA probes of known sequence. FEMS Microbiol Lett. 1994 Dec 15;124(3):265–269. doi: 10.1111/j.1574-6968.1994.tb07295.x. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T., Oyama T., Kusano T., Osaki T., Nakanishi R., Shirakusa T. Mutations of the p53 gene as a predictor of poor prognosis in patients with non-small-cell lung cancer. J Natl Cancer Inst. 1993 Dec 15;85(24):2018–2023. doi: 10.1093/jnci/85.24.2018. [DOI] [PubMed] [Google Scholar]
- Okazaki T., Takita J., Kohno T., Handa H., Yokota J. Detection of amplified genomic sequences in human small-cell lung carcinoma cells by arbitrarily primed-PCR genomic fingerprinting. Hum Genet. 1996 Sep;98(3):253–258. doi: 10.1007/s004390050203. [DOI] [PubMed] [Google Scholar]
- Peinado M. A., Malkhosyan S., Velazquez A., Perucho M. Isolation and characterization of allelic losses and gains in colorectal tumors by arbitrarily primed polymerase chain reaction. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10065–10069. doi: 10.1073/pnas.89.21.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perolat P., Merien F., Ellis W. A., Baranton G. Characterization of Leptospira isolates from serovar hardjo by ribotyping, arbitrarily primed PCR, and mapped restriction site polymorphisms. J Clin Microbiol. 1994 Aug;32(8):1949–1957. doi: 10.1128/jcm.32.8.1949-1957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried T., Petersen I., Holtgreve-Grez H., Speicher M. R., Schröck E., du Manoir S., Cremer T. Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res. 1994 Apr 1;54(7):1801–1806. [PubMed] [Google Scholar]
- Slebos R. J., Kibbelaar R. E., Dalesio O., Kooistra A., Stam J., Meijer C. J., Wagenaar S. S., Vanderschueren R. G., van Zandwijk N., Mooi W. J. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990 Aug 30;323(9):561–565. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- Sugio K., Kishimoto Y., Virmani A. K., Hung J. Y., Gazdar A. F. K-ras mutations are a relatively late event in the pathogenesis of lung carcinomas. Cancer Res. 1994 Nov 15;54(22):5811–5815. [PubMed] [Google Scholar]
- Vega F. J., Iniesta P., Caldés T., Sanchez A., López J. A., de Juan C., Diaz-Rubio E., Torres A., Balibrea J. L., Benito M. p53 exon 5 mutations as a prognostic indicator of shortened survival in non-small-cell lung cancer. Br J Cancer. 1997;76(1):44–51. doi: 10.1038/bjc.1997.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T., Stolz W., Landthaler M., Rüschoff J., Schlegel J. Nonradioactive arbitrarily primed polymerase chain reaction: a novel technique for detecting genetic defects in skin tumors. J Invest Dermatol. 1996 Jan;106(1):194–197. doi: 10.1111/1523-1747.ep12329949. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A. Tumor suppressor genes. Science. 1991 Nov 22;254(5035):1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucleic Acids Res. 1991 Oct 11;19(19):5275–5279. doi: 10.1093/nar/19.19.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Q. M., Deng W. G., Xia Z. P., Pang H. H. Polymorphism and genetic relatedness among wild and cultivated rice species determined by AP-PCR analysis. Hereditas. 1995;122(2):135–141. doi: 10.1111/j.1601-5223.1995.00135.x. [DOI] [PubMed] [Google Scholar]