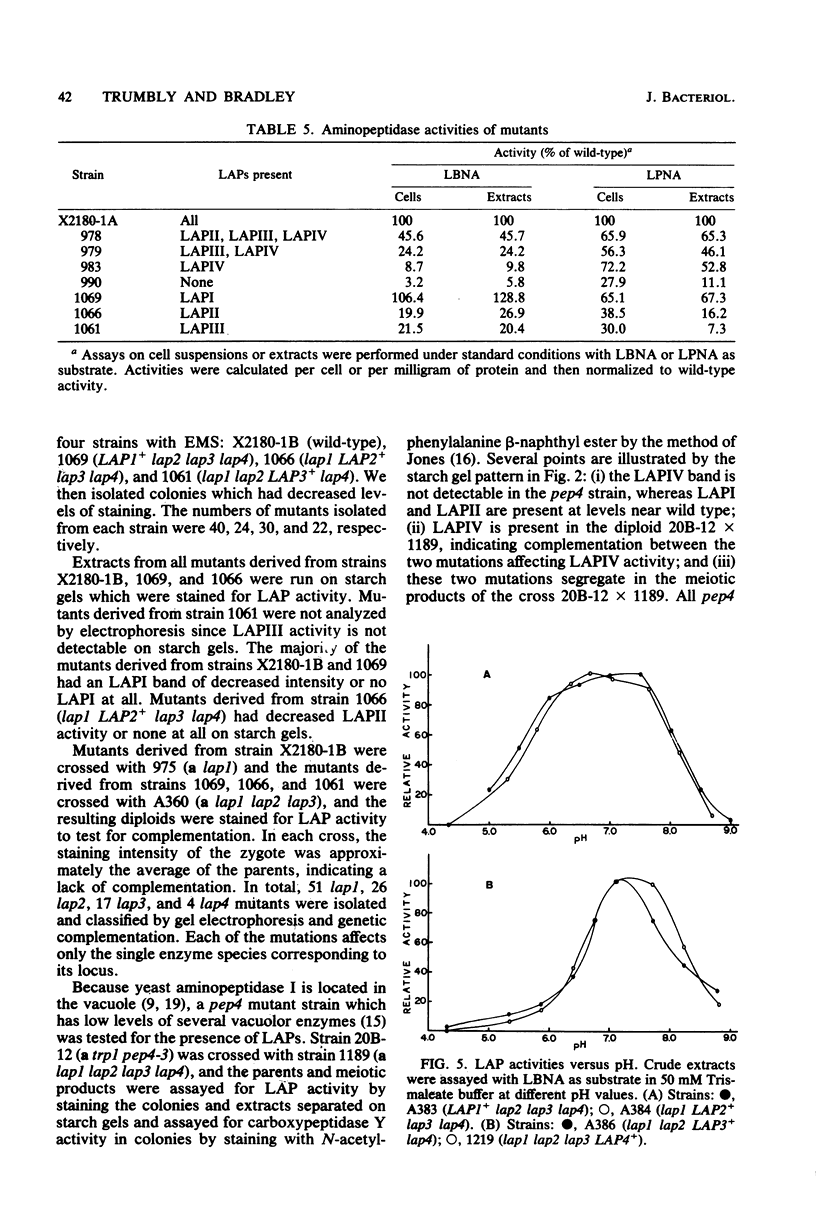
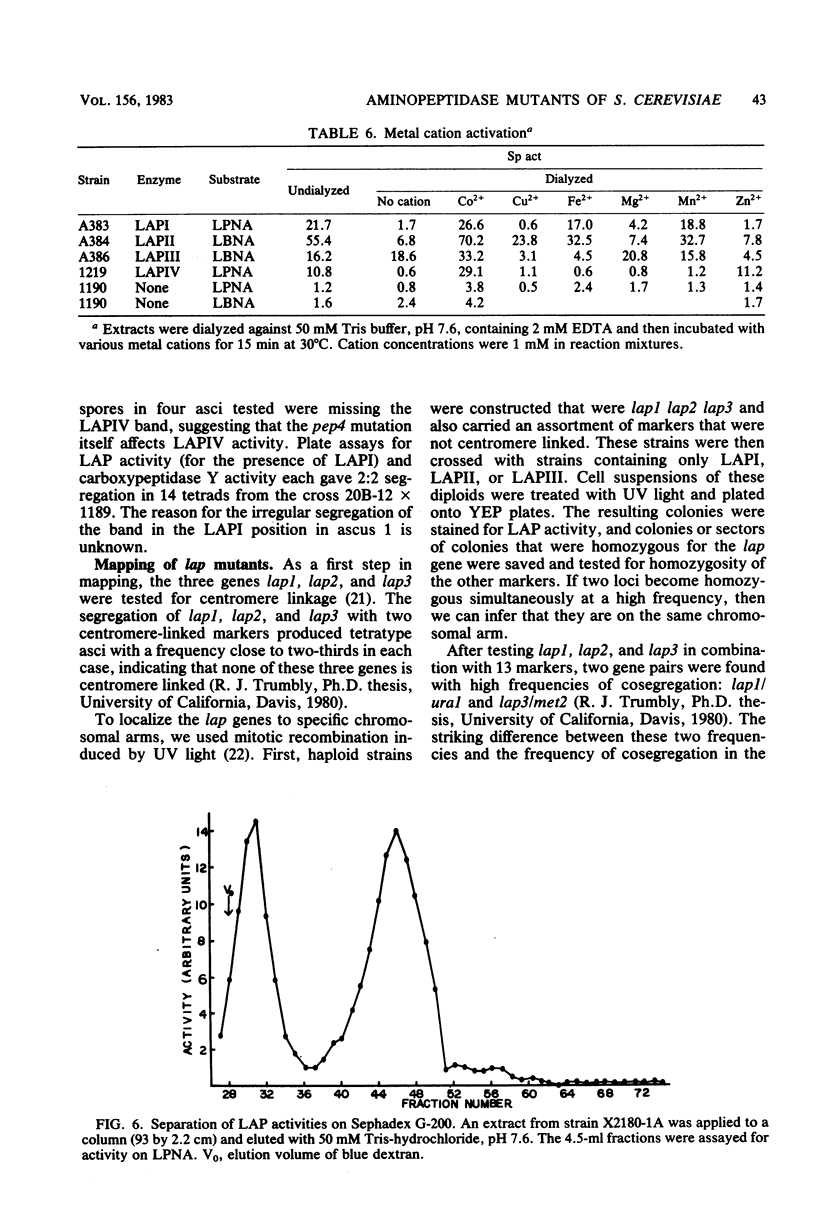
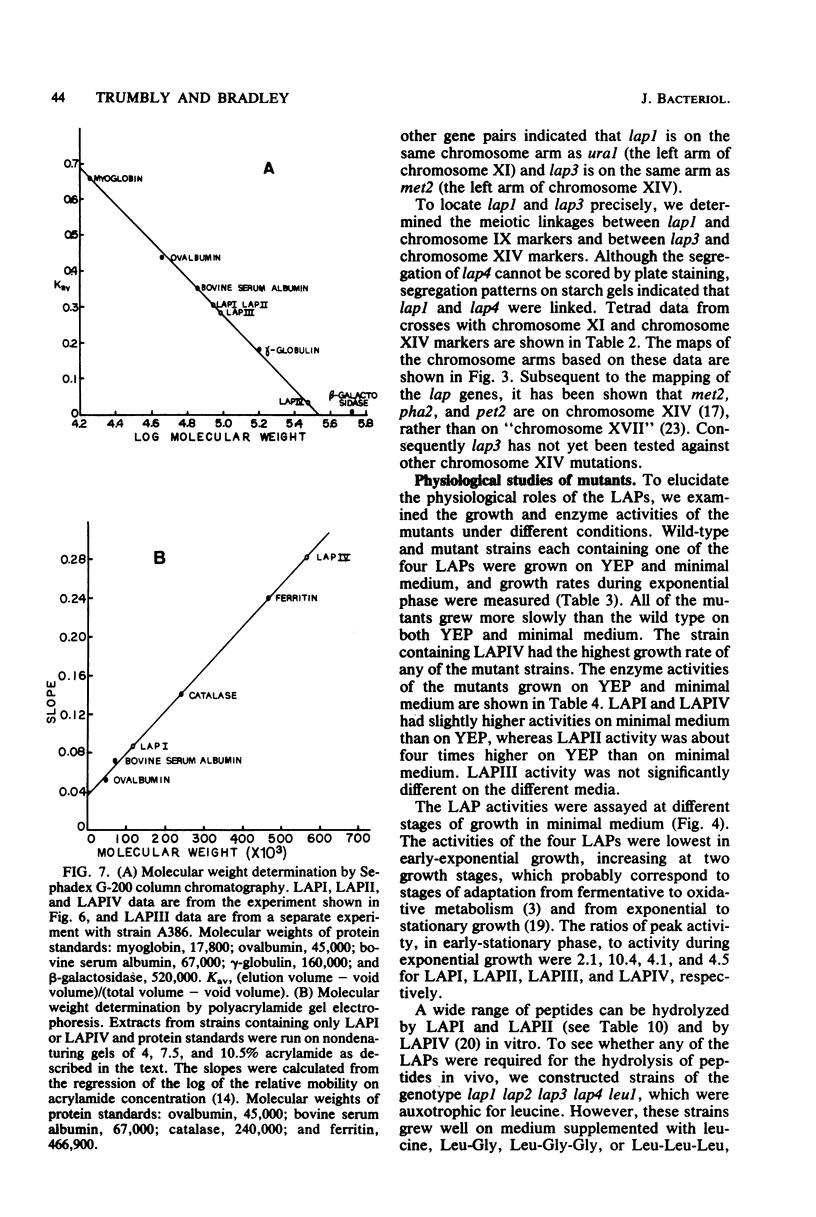
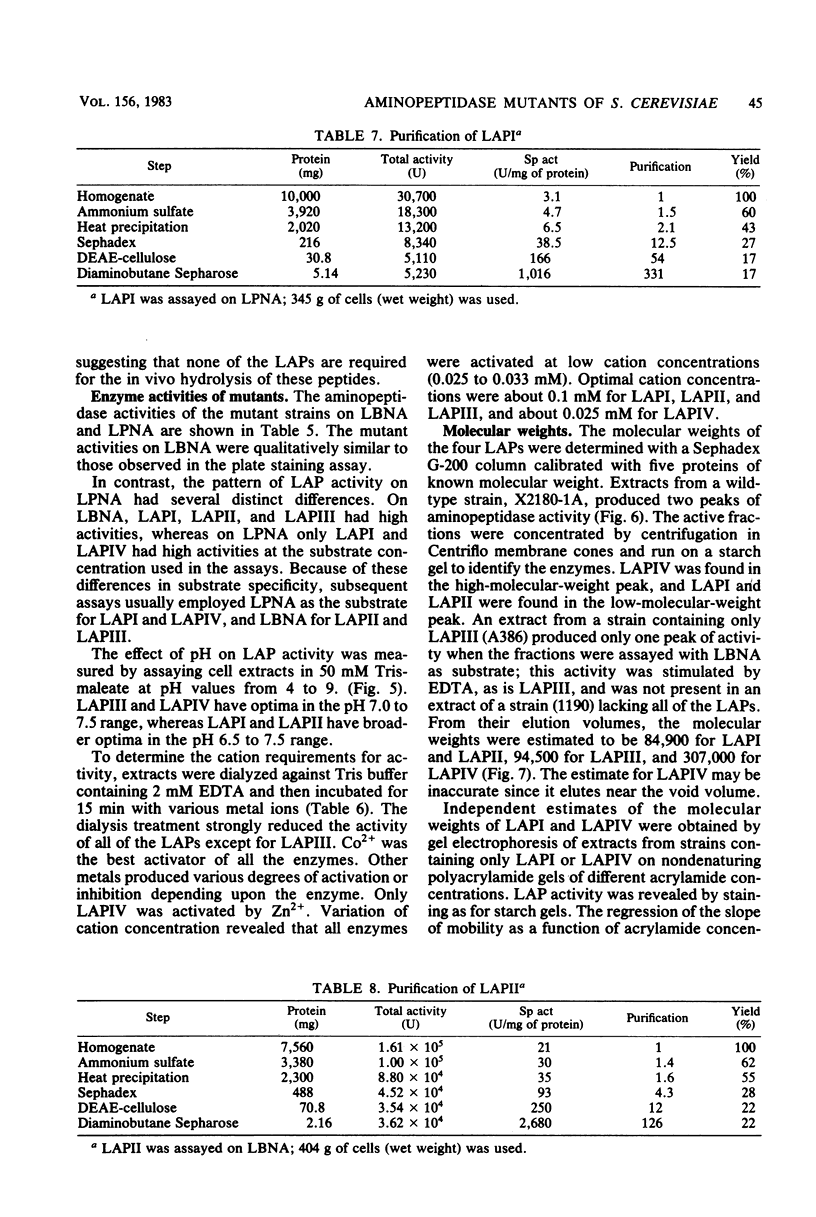
Abstract
Mutants of Saccharomyces cerevisiae were isolated which have decreased ability to hydrolyze leucine beta-naphthylamide, a chromogenic substrate for amino-peptidases. The mutations were shown by starch gel electrophoresis to affect one of four different aminopeptidases. Mutations affecting a given enzyme belong to a single complementation group. The four genes were symbolized lap1, lap2, lap3, and lap4, and the corresponding enzymes LAPI, LAPII, LAPIII, and LAPIV. Both lap1 and lap4 were mapped to the left arm of chromosome XI, and lap3 was mapped to the left arm of chromosome XIV. Strains which possessed only one of the four leucine aminopeptidases (LAPs) were constructed. Crude extracts from these strains were used to study the properties of the individual enzymes. Dialysis against EDTA greatly reduced the activity of all the LAPs except for LAPIII. Of the cations tested, Co2+ was the most effective in restoring activity. LAPIV was the only LAP reactivated by Zn2+. LAPI was purified 331-fold and LAPII was purified 126-fold from cell homogenates. Both of the purified enzymes had strong activity on dipeptides and tripeptides. The activity levels of the LAPs are strongly dependent on growth stage in batch culture, with the highest levels in early-stationary phase. Strains lacking all four LAPs have slightly lower growth rates than wild-type strains. The ability of leucine auxotrophs to grow on dipeptides and tripeptides containing leucine is not impaired in strains lacking all four LAPs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J., Hansche P. E. Population studies in microorganisms. I. Evolution of diploidy in Saccharomyces cerevisiae. Genetics. 1974 Feb;76(2):327–338. doi: 10.1093/genetics/76.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BINKLEY F., TORRES C. Spectrophotometric assay of peptidase activity. Arch Biochem Biophys. 1960 Feb;86:201–203. doi: 10.1016/0003-9861(60)90404-5. [DOI] [PubMed] [Google Scholar]
- Beck C., von Meyenburg H. K. Enzyme pattern and aerobic growth of Saccharomyces cerevisiae under various degrees of glucose limitation. J Bacteriol. 1968 Aug;96(2):479–486. doi: 10.1128/jb.96.2.479-486.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Ciejek E., Thorner J. Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell. 1979 Nov;18(3):623–635. doi: 10.1016/0092-8674(79)90117-x. [DOI] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Durán A., Bowers B., Cabib E. Chitin synthetase zymogen is attached to the yeast plasma membrane. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3952–3955. doi: 10.1073/pnas.72.10.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Röhm K. H. External and internal forms of yeast aminopeptidase II. Eur J Biochem. 1979 Jun;97(1):169–173. doi: 10.1111/j.1432-1033.1979.tb13099.x. [DOI] [PubMed] [Google Scholar]
- Frey J., Röhm K. H. Subcellular localization and levels of aminopeptidases and dipeptidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1978 Nov 10;527(1):31–41. doi: 10.1016/0005-2744(78)90253-x. [DOI] [PubMed] [Google Scholar]
- Gancedo C. Inactivation of fructose-1,6-diphosphatase by glucose in yeast. J Bacteriol. 1971 Aug;107(2):401–405. doi: 10.1128/jb.107.2.401-405.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALVORSON H. Intracellular protein and nucleic acid turnover in resting yeast cells. Biochim Biophys Acta. 1958 Feb;27(2):255–266. doi: 10.1016/0006-3002(58)90332-9. [DOI] [PubMed] [Google Scholar]
- Hansen R. J., Switzer R. L., Hinze H., Holzer H. Effects of glucose and nitrogen source on the levels of proteinases, peptidases, and proteinase inhibitors in yeast. Biochim Biophys Acta. 1977 Jan 24;496(1):103–114. doi: 10.1016/0304-4165(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Hedrick J. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968 Jul;126(1):155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. Chromosomes XIV and XVII of Saccharomyces cerevisiae constitute a single linkage group. Mol Cell Biol. 1982 Nov;2(11):1399–1409. doi: 10.1128/mcb.2.11.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder R., Becker J. M., Naider F. Peptide transport in yeast: utilization of leucine- and lysine-containing peptides by Saccharomyces cerevisiae. J Bacteriol. 1977 Sep;131(3):906–916. doi: 10.1128/jb.131.3.906-916.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz G., Röhm K. H. Yeast aminopeptidase I. Chemical composition and catalytic properties. Biochim Biophys Acta. 1976 May 13;429(3):933–949. doi: 10.1016/0005-2744(76)90338-7. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic Mapping in Saccharomyces IV. Mapping of Temperature-Sensitive Genes and Use of Disomic Strains in Localizing Genes. Genetics. 1973 May;74(1):33–54. doi: 10.1093/genetics/74.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae. Microbiol Rev. 1980 Dec;44(4):519–571. doi: 10.1128/mr.44.4.519-571.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrydziak D. M., Demain A. L., Tannenbaum S. R. Regulation of extracellular protease production in Candida lipolytica. Biochim Biophys Acta. 1977 Apr 27;497(2):525–538. doi: 10.1016/0304-4165(77)90209-4. [DOI] [PubMed] [Google Scholar]
- Rose B., Becker J. M., Naider F. Peptidase activities in Saccharomyces cerevisiae. J Bacteriol. 1979 Jul;139(1):220–224. doi: 10.1128/jb.139.1.220-224.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren J., Pählman S., Glad M., Hjertén S. Hydrophobic interaction chromatography on non-charged Sepharose derivatives. Binding of a model protein, related to ionic strength, hydrophobicity of the substituent, and degree of substitution (determined by NMR). Biochim Biophys Acta. 1975 Nov 18;412(1):51–61. [PubMed] [Google Scholar]
- Röhm K. H. Properties of a highly purified dipeptidase (EC 3.4.13.?) from brewer's yeast. Hoppe Seylers Z Physiol Chem. 1974 Jun;355(6):675–686. doi: 10.1515/bchm2.1974.355.1.675. [DOI] [PubMed] [Google Scholar]
- Snow R. Maximum likelihood estimation of linkage and interference from tetrad data. Genetics. 1979 May;92(1):231–245. doi: 10.1093/genetics/92.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Leibowitz M. J. Mak mutants of yeast: mapping and characterization. J Bacteriol. 1979 Oct;140(1):154–160. doi: 10.1128/jb.140.1.154-160.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]