Abstract
The mechanisms involved in the posttranslational targeting of membrane proteins are not well understood. The light-harvesting chlorophyll proteins (LHCP) of the thylakoid membrane are a large family of hydrophobic proteins that are targeted in this manner. They are synthesized in the cytoplasm, translocated across the chloroplast envelope membranes into the stroma, bound by a stromal factor to form a soluble intermediate, “transit complex”, and then integrated into the thylakoid membrane by a GTP dependent reaction. Signal recognition particle (SRP), a cytoplasmic ribonucleoprotein, is known to mediate the GTP dependent cotranslational targeting of proteins to the endoplasmic reticulum. We show that chloroplasts contain an SRP consisting of, cpSRP54, a homologue of SRP54 and a previously undescribed 43-kDa polypeptide (cpSRP43) instead of an RNA. We demonstrate that both subunits of cpSRP are required for the formation of the transit complex with LHCP. Furthermore, cpSRP54, cpSRP43, and LHCP are sufficient to form a complex that appears to be identical to authentic transit complex. We also show that the complex formed between LHCP and cpSRP, together with an additional soluble factor(s) are required for the proper integration of LHCP into the thylakoid membrane. It appears that the expanded role of cpSRP in posttranslational targeting of LHCP has arisen through the evolution of the 43-kDa protein.
The insertion of proteins into membranes is a fundamental process essential for the vitality of all organisms. The paradigm for this process is the targeting mediated by signal recognition particle (SRP), a cytoplasmic ribonucleoprotein. In prokaryotes, the SRP-RNA binds a single 54-kDa-polypeptide subunit, while in eukaryotes, five additional subunits are bound. One of the distinctive features of this targeting mechanism is cotranslational protein insertion. The synthesis of hydrophobic protein domains at the membrane circumvents potential protein folding problems that might otherwise occur in an aqueous environment. However not all hydrophobic proteins are targeted cotranslationally. For example, the major proteins of the thylakoid membrane, the light harvesting chlorophyll proteins (LHCP), are targeted by a posttranslational mechanism. LHCP form a large family of related proteins that have three to four transmembrane domains. They are synthesized in the cytoplasm, and are targeted to the thylakoid membrane through three aqueous compartments: the cytoplasm, the inter-envelope space, and the stroma (1, 2). The factors that mediate the posttranslational targeting of members of the LHCP family have not been definitively identified.
The LHCP are inserted into the thylakoid membrane in a reaction requiring GTP and stroma (3–5). It was found previously that a stromal factor binds LHCP to form a soluble intermediate, designated transit complex, that maintains the solubility of these proteins as they are transported through the stroma (6, 7). The transit complex is the only soluble form of LHCP that accumulates when stroma is mixed with LHCP in vitro and is recognized as a characteristic band on nondenaturing gels (6). LHCP are not stable in the stroma, and though, they are transported through this compartment, they are not detected in the soluble fraction under normal circumstances. Several lines of evidence indicate that the chloroplast homologue of the 54-kDa subunit of the signal recognition particle (cpSRP54) is a component of transit complex (7). (i) cpSRP54 is the predominant stromal protein that is crosslinked to LHCP. (ii) LHCP in transit complex can be coimmunoprecipitated with cpSRP54. (iii) Stroma immunodepleted of cpSRP54 is not capable of forming transit complex.
The transit complex is thought to be an intermediate in the targeting of LHCP to the thylakoid membranes based on the findings that the transit complex contains a productive form of LHCP capable of integrating into the thylakoid membrane upon addition of stroma and GTP (6) and that LHCP integration does not occur if stroma is immunodepleted of cpSRP54 (7) because no transit complex forms in the absence of cpSRP54. The requirement for the presence of transit complex and additional stroma has fueled the speculation that two stromal factors are involved in LHCP integration: one factor binds LHCP to form the transit complex and the second facilitates membrane insertion (6). cpSRP54 was required but not sufficient to form transit complex suggesting that even this complex has more than one stromal component (7). In the present work, we have identified a 43-kDa protein that binds to cpSRP54 and we show that the complex of both proteins is required for the biogenesis of LHCP.
MATERIALS AND METHODS
Plasmids and Strains.
cpSRP43, beginning with A60, was fused to the C terminus of glutathione S-transferase (GST) as follows: The forward oligonucleotide AAGGATCCATGGCCGCCGTACAAAGAAAC and the reverse oligonucleotide TCGGGATCCAATGATCTTGTTCAC were used to amplify a 121-bp PCR product from the cpSRP43 precursor. This fragment was digested with BamHI, cloned into the BamHI site in pLMC11 (V.I.K., F.P.-C., M. Havaux, P. Contard, D.S., K. Meiherhoff, P. Gourt, J.D.G.J., N.E.H., and L.N., unpublished material), and clones with the proper orientation were selected and designated, pGEX4Tchaos(m). pGEX4Tchaos(m) was transformed into BL21 cells to form strain BL2143. The double expressing strain, BL21cpSRP, was made by transforming BL2143 with pACYC54his. pACYC54his was constructed by ligation of the purified 111-bp XhoI-NcoI and the 1,490-bp NcoI -HindIII fragments from pNH4 (8) into pACYC184 digested with SalI and HindIII. The resulting plasmid encodes mature cpSRP54 fused to a C-terminal hexahistidine tag.
Antibodies and Immunoblot Quantitation.
Antibodies against cpSRP54 were raised in rabbits injected with maltose-binding protein fused to residues 394–564 of cpSRP54 as described (13). Antibodies against cpSRP43 were raised in chickens injected with GST fused to residues 94–376 of cpSRP43 (as described in V.I.K., F.P.-C., M. Havaux, P. Contard, D.S., K. Meiherhoff, P. Gourt, J.D.G.J., N.E.H., and L.N., unpublished material). Both antibodies recognize a single protein in chloroplast stroma. The amount of cpSRP54 translation product was measured by quantitative immunoblotting. Three dilutions of sample were compared with a dilution series of antigen to ensure that the sample was in the linear range of the assay. Antigen concentration was determined by quantitative amino acid analysis performed at the Stanford Protein and Nucleic Acid facility. Blots were quantitated as described (8). Primary antibodies were detected by enhanced chemiluminescence with secondary antibodies conjugated to peroxidase (goat anti-rabbit antibodies for cpSRP54 and rabbit anti-chicken antibodies for cpSRP43). Films were scanned and quantitated using imagequant software from Molecular Dynamics.
Purification of cpSRP43.
Chloroplast stroma was depleted of RUBISCO and ribosomes by centrifugation for 2 h at 60,000 RPM on 5–20% sucrose gradients in a SW-60 rotor. The top 1 ml of the gradients were pooled and adjusted to 0.3 M KCl, 1% Tween-20 by addition of an equal volume of a 2× solution. Extract was incubated overnight at 4°C with anti-cpSRP54 crosslinked to protein A-Sepharose (9). The beads were washed 3 times in batch with 20 mM Hepes-KOH (pH 8.0), 0.3 M KCl, and 1% Tween-20, transferred to a column and washed with an additional 15 ml of the same buffer and 10 ml of buffer lacking detergent. cpSRP subunits were eluted with 8 M urea in 20 mM Hepes-KOH (pH 8.0). Protein was concentrated by trichloroacetic acid precipitation and analyzed by SDS/PAGE on 12% polyacrylamide gels. For pea cpSRP43, molecular mass was 43 kDa; for arabidopsis cpSRP43, molecular mass was 42 kDa.
Expression and Purification of Recombinant cpSRP43 and cpSRP.
To express cpSRP43, BL2143 cells were grown to an A600 of 0.6–1.0 and incubated with 0.3 mM isopropyl β-d-thiogalactoside for 1 h. Cells were harvested, frozen and sonicated in lysis buffer (50 mM Tris⋅HCl, pH 8.0/300 mM NaCl/1 mM DTT/1 mM EDTA/1 mM phenylmethylsulfonyl fluoride/1 μg/ml leupeptin). cpSRP43 was bound to glutathione Sepharose that was then washed successively with lysis buffer and thrombin cleavage buffer (50 mM Tris, pH 8.0/150 mM NaCl/2.5 mM CaCl2). cpSRP43 was eluted by overnight treatment at 4°C with 20 units of thrombin per liter of original cells. The eluate was recovered and further purified by gel filtration chromatography as described in the legend to Fig. 1. To express cpSRP, BL21cpSRP cells were grown as above and incubated with 0.1 mM isopropyl-β-d-thiogalactoside for 3 h. cpSRP was bound to glutathione Sepharose as above, but was eluted with 10 mM glutathione in lysis buffer. The eluate was applied to Ni2+-nitrilotriacetic acid agarose, washed successively with lysis buffer, then 20 mM Hepes-KOH (pH 8.0), and eluted in 20 mM Hepes-KOH (pH 8.0), 1 mM DTT, 200 mM imidazole, and 1 mM of 4-(2-aminoethyl)benzenesulfonyl fluoride.
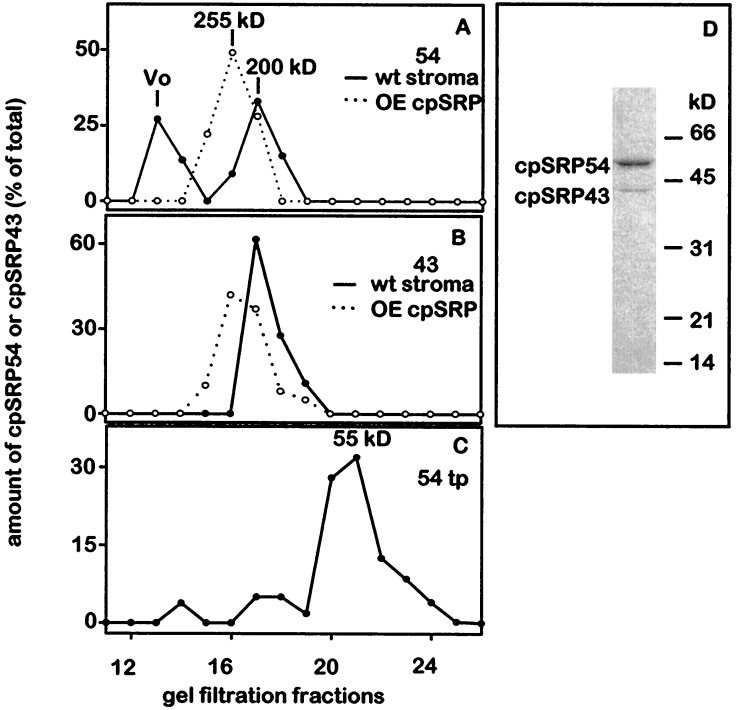
Figure 1.
cpSRP is an oligomer containing 54- and 43-kDa subunits. Pea stroma (A and B, solid lines) (5), wheat-germ translated cpSRP54 (C), or cpSRP produced in E. coli (OEcpSRP; A and B, dotted lines) were fractionated by gel filtration and cpSRP54 (A and C) or cpSRP43 (B) were detected by immunoblot analyses (8). The molecular mass of OEcpSRP is larger than that of cpSRP because the former contains cpSRP43 fused to GST. (D) Coimmunoprecipitation of cpSRP43 with antisera against cpSRP54. Coomassie stain of a gel loaded with 2 μg of protein. Stroma was prepared by lysing chloroplasts in 20 mM Hepes-KOH (pH 8.0), 5 mM MgCl2, 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. Samples were fractionated on a Protein Pak 300 SW column in 20 mM of Hepes-KOH (pH 8.0), 5 mM MgCl2, and 180 mM NaCl at 0.5 ml/min. The column was calibrated using the following proteins as standards: bovine thyroglobulin, 670 kDa; sweet potato β-amylase, 200 kDa; BSA, 66 kDa; ovalbumin, 45 kDa; and cytochrome c, 12 kDa.
Reconstitution of Transit Complex.
Radiolabeled LHCP precursor (8 × 104 cpm), synthesized in wheat germ extracts (10), was mixed with either arabidopsis stroma (equivalent to 36 μg of Chl), recombinant cpSRP43 (50 ng), or cpSRP54 translation product (160 ng) in 55 mM sorbitol, 10 mM Hepes-KOH (pH 8.0), 10 mM MgCl2, and 1 mM ATP in a final volume of 15 μl for 15 min at 25°C. The amounts of cpSRP43 and cpSRP54 added were determined empirically. First we held cpSRP43 constant and varied the amount of cpSRP54. We then chose the optimum amount of cpSRP54 from the first experiment, held it constant, and varied the amount of cpSRP43. All samples contained equal amounts of wheat germ extract. Complex formation using proteins purified from Escherichia coli was carried out as described above with 0.75 μg of the refolded pLHCP (11, 12) and 0.5 μg of recombinant cpSRP in a final volume of 40 μl. Immunoprecipitation was performed in 240 μl of 55 mM of sorbitol, 10 mM Hepes-KOH (pH 8.0), 10 mM MgCl2, 1 mM ATP, 0.3 M KCl, and 1% Tween-20 containing anti-cpSRP54-IgG (0.48 mg) crosslinked to 3 mg of protein A Sepharose beads. After end over end rotation for 2 h at 4°C, the beads were transferred into Wizard minicolumns (Promega) and washed with 5 ml of a solution of 20 mM Hepes-KOH (pH 8.0), 0.3 M KCl, and 1% Tween-20 followed by 2 ml of the same buffer lacking detergent. Bound protein was eluted with 20 μl of 4× SDS/PAGE sample buffer and subjected to immunoblot analysis with enhanced chemiluminescence detection.
Immunodepletion of Stroma.
3 mg protein A Sepharose was swollen, washed in 1× TBS, resuspended in ≈500 μl 1× TBS, and rotated end over end overnight at 4°C with anti-cpSRP54 IgGs (0.48 mg). The beads were washed three times with 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1% Tween-20 and two times with 10 mM Hepes-KOH (pH 8.0). The beads were transferred into Wizard minicolumns and excess fluid was removed by centrifugation in a microfuge. The beads were incubated with 120 μl of stroma (equivalent to 360 μg of chlorophyll) for 15 min. After incubation, stroma was recovered by a short centrifugation and the process was repeated two additional times.
RESULTS
cpSRP54 Forms a Complex with a 43-kDa Polypeptide.
Although we observed that cpSRP54 was the principal stromal factor that interacts with LHCP, cpSRP54 alone does not bind to LHCP (7, Fig. 2). To identify factors associated with cpSRP54 that might also be required for transit complex formation, we fractionated stroma by FPLC and used antibodies to quantitate cpSRP54 in the different fractions. As shown in Fig. 1A, approximately half of the cpSRP54 eluted in the void volume where it was bound to 70 S ribosomes (13). The remainder of the protein eluted as a 200 kDa species. In agreement with previous results (6), the 200 kDa fraction formed the transit complex with LHCP. In contrast recombinant cpSRP54 (data not shown), or cpSRP54 synthesized in wheat germ extracts was eluted from the gel filtration columns as a 55 kDa monomer suggesting that the 200 kDa complex contained additional subunits (Fig. 1C).
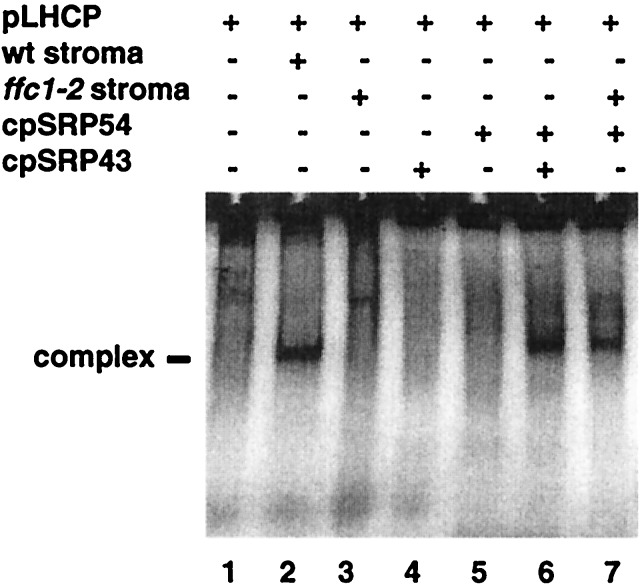
Figure 2.
Evidence that transit complex is comprised of cpSRP43, cpSRP54, and LHCP. The indicated components: pLHCP, 8 × 104 cpm synthesized in wheat germ extracts; arabidopsis stroma (equivalent to 36 μg of chlorophyll); cpSRP54 (160 ng) synthesized in wheat germ extracts; purified recombinant cpSRP43 (50 ng) were mixed as described in materials and methods and assayed for the formation of transit complex on nondenaturing gels (6) by radioimaging .
To analyze the polypeptides of the 200-kDa complex, we purified it by monospecific antibodies raised against cpSRP54. As shown in Fig. 1D, a polypeptide of 43-kDa coimmunoprecipitated with cpSRP54. The sequence of the N terminus and four internal peptide fragments derived from protease cleavage of the 43 kDa polypeptide corresponded to the predicted amino acid sequence of CHAOS, the protein encoded by a plant-specific gene termed, Cao (V.I.K., et al., manuscript in preparation GenBank accession no. AF013115). Furthermore, antibodies against CHAOS recognized the 43-kDa protein and additionally, a mutation in Cao caused a defect in LHCP biogenesis (V.I.K., et al., unpublished data). These data demonstrated that CHAOS is a constituent of cpSRP and is required for LHCP integration into the thylakoid membranes; therefore we designated CHAOS, cpSRP43. cpSRP43 is unrelated to any previously described SRP subunit (V.I.K., et al., unpublished data).
cpSRP43 Does Not Bind to 70S Ribosomes.
To examine the distribution of cpSRP43 in the stroma, FPLC fractionated stroma was also assayed for cpSRP43 by immunoblot analysis. A single peak of cpSRP43 was observed in the 200 kDa fraction coeluting with cpSRP54; no cpSRP43 was observed in the void volume (Fig. 1B). Likewise, stroma fractionated on 5–20% sucrose gradients was assayed for both proteins. As observed previously, cpSRP54 was found in 4S and 70S fractions (13). cpSRP43 cosedimented with cpSRP54 in the 4S fraction but was absent from the 70S fraction (data not shown). These data reveal that cpSRP43 does not associate with 70S ribosomes. Thus while cpSRP43 appears to be largely complexed with cpSRP54, a second ribosomal associated form of cpSRP54 is present that can be distinguished from the stromal form by the absence of cpSRP43.
Evidence That the Transit Complex Is Comprised of cpSRP43, cpSRP54, and LHCP.
When in vitro translated, radiolabeled pLHCP is mixed with arabidopsis stroma containing endogenous cpSRP54 and cpSRP43, transit complex forms (Fig. 2, lane 1 vs. lane 2). Previously, we observed that LHCP did not form transit complex in stroma that was immunodepleted by antisera raised against cpSRP54 (7). We now know that this procedure removes cpSRP43 in addition to cpSRP54 (Fig. 1D). Stroma from the arabidopsis mutant ffc1–2, which lacks cpSRP54 but contains cpSRP43 (P. Amin, D. A. C. Sy, M. L. Pilgrim, and N.E.H., unpublished manuscript), is also unable to form the transit complex (Fig. 2, lane 3). However, when exogenous cpSRP54 is added to this stroma, transit complex now forms clearly demonstrating the requirement for cpSRP54 (Fig. 2, lane 7). From our previous results (7), transit complex did not form when cpSRP54 alone was added to stroma lacking cpSRP. Together, these results suggested that cpSRP43 was also required.
To determine the importance of cpSRP43 in the formation of transit complex, we assayed the activity of purified recombinant protein in combination with cpSRP54 synthesized in vitro. As expected, cpSRP43 or cpSRP54 alone did not bind to LHCP (Fig. 2, lanes 4 and 5). However, when purified cpSRP43 and cpSRP54, synthesized in a wheat germ extract, were both added to LHCP in the absence of stroma, a complex of the same electrophoretic mobility as authentic transit complex forms (Fig. 2, lane 6 vs. lanes 2 and 7). This data suggested that the only stromal factors needed to form the complex were cpSRP54 and cpSRP43. However as wheat germ extract might contain some stromal contaminants, we tested whether a complex would also form by using only the three highly purified proteins (Fig. 3).
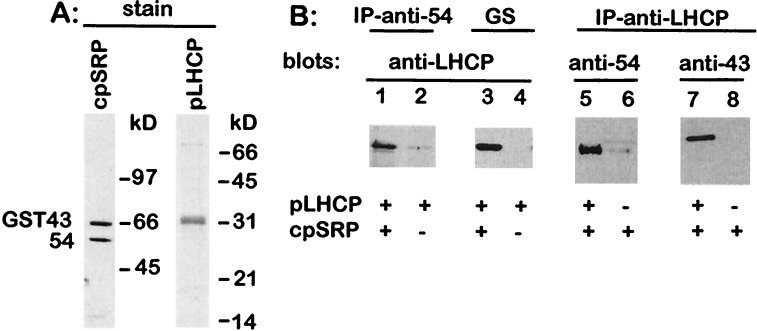
Figure 3.
Purified cpSRP interacts with purified LHCP. (A) Silver-stained gel of recombinant cpSRP protein; 8% polyacrylamide gel, 0.3 μg of protein. Coomassie gel of recombinant pLHCP protein; 12% polyacrylamide gel, 2 μg of protein. (B) Coprecipitation of pLHCP with anti-cpSRP54 antibodies (lanes 1 and 2) or glutathione Sepharose beads (GS; lanes 3 and 4), which bind GST-43, occurs in the presence of cpSRP. Coprecipitation of cpSRP54 and cpSRP43 with anti-LHCP occurs in the presence of pLHCP (lanes 5–8).
A highly purified cpSRP was obtained, under native conditions, when affinity tagged cpSRP54 was coexpressed with GST43 (Fig. 3A). The E. coli expressed cpSRP eluted as a 255 kDa complex, when fractionated by FPLC suggesting it has the same subunit composition as stromal cpSRP (Fig. 1 A and B). The slightly larger molecular mass is expected because cpSRP43 is expressed as a GST fusion. Purified LHCP precursor, obtained from E. coli inclusion bodies (Fig. 3A), was found to associate with this cpSRP as measured by coimmunoprecipitation of LHCP with antibodies raised against cpSRP54, coprecipitation of LHCP with glutathione Sepharose, and coimmunoprecipitation of cpSRP54 and cpSRP43 with antibodies raised against LHCP (Fig. 3B). These data prove that cpSRP43 and cpSRP54 are sufficient to form a complex with LHCP.
Reconstitution of LHCP Integration in Stroma Immunodepleted of cpSRP.
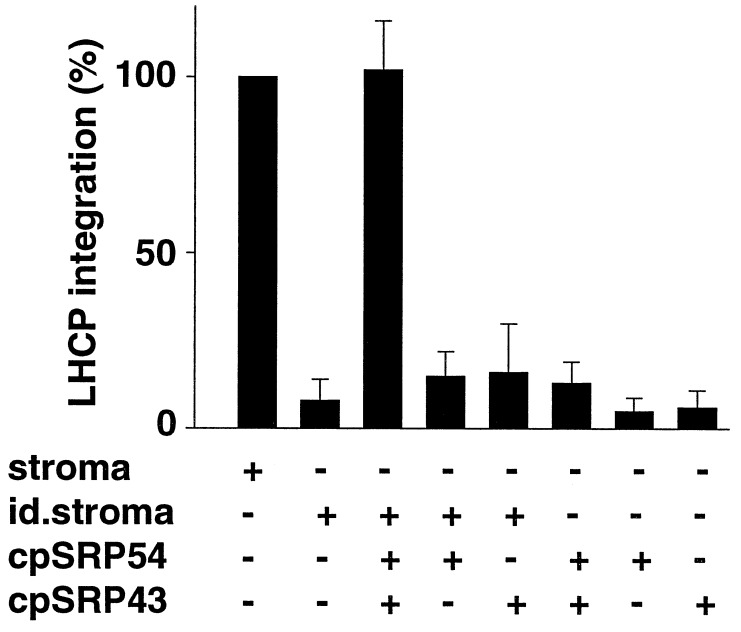
We next sought to test the activity of cpSRP on the integration of LHCP into thylakoid membranes. As seen in Fig. 4 and observed previously (7), LHCP does not integrate into the thylakoids in the presence of stroma immunodepleted of cpSRP. Individual cpSRP subunits had little if any effect, but the simultaneous addition of both cpSRP43 and cpSRP54 to the immunodepleted stroma resulted in the integration of LHCP into the membranes. Although cpSRP could restore integration activity to the immunodepleted stromal extract, the stroma was required to promote the insertion of LHCP into the thylakoid membrane. Hence, stromal factors in addition to cpSRP are required for LHCP biogenesis.
Figure 4.
Reconstitution of LHCP integration in stroma immunodepleted of cpSRP by adding back cpSRP54 and cpSRP43. Integration assays were performed as described (5). Stroma (30 μl), cpSRP54-immunodepleted stroma (30 μl), recombinant cpSRP43 (150 ng), and/or in vitro translated cpSRP54 (500 ng) were added as indicated. After 30 min at 25°C, thylakoids were treated with 0.1 mg/ml trypsin for 30 min at 0°C, followed by washing with 0.1 N NaOH. Each treatment is expressed relative to the positive control. The values indicated represent the average of three experiments. (Bars = SEM.)
DISCUSSION
In the present work we show that cpSRP54 forms a complex with a 43-kDa chloroplast protein. This complex forms spontaneously in vitro and when both proteins are expressed in E. coli. We also demonstrate that the two proteins form a functional unit that we designate cpSRP. By removing both cpSRP54 and cpSRP43 from stroma (Fig. 1D), antibodies raised against cpSRP54 inhibit two activities associated with LHCP biogenesis, i.e., formation of transit complex and LHCP integration activity (7). Only when both proteins are replenished are the two activities restored.
Before integration, LHCP forms a transit complex with stromal proteins (6). When the transit complex is reisolated from stroma, integration into the thylakoid membrane fails to occur unless more stroma is added (6). Thus transit complex appears to be a productive intermediate that by itself is inactive in LHCP integration (6). Our data show that cpSRP43 and cpSRP54 are sufficient to form a complex with LHCP that has the same properties as the transit complex formed between LHCP and stroma. Both complexes comigrate on nondenaturing gels. Likewise, in both cases, additional stroma is required for LHCP integration.
Although we have not actually proven that transit complex consists of only cpSRP43, cpSRP54, and LHCP, we consider it very likely because the interaction between the three proteins is highly specific. Based on crosslinking and immunoprecipitation experiments (7), we know that cpSRP54 is a component of the transit complex. Yet, it alone cannot interact with LHCP. Likewise, when all stromal proteins except for cpSRP54 are present, as seen in the ffc1-2 mutant stroma, the characteristic transit complex also does not form. However, the complex of the appropriate size forms simply by mixing cpSRP43, cpSRP54, and pLHCP. Given that cpSRP43 and cpSRP54 normally form a complex in the stroma, these data imply that transit complex consists of cpSRP and LHCP.
Our observation that cpSRP is not sufficient for LHCP integration proves that a second stromal factor is required for LHCP biogenesis. We surmise that LHCP, which has been translocated across the envelope into the stroma, first binds cpSRP to form the transit complex. It is possible that LHCP interacts with other stromal factors such as cpn 60 and hsp70 before binding to cpSRP (17, 18). However a specific role of either chaperone in LHCP biogenesis has yet to be demonstrated (6, 19). In a second step, an additional stromal factor and GTP are required. These factors may bind the transit complex producing an activated form of LHCP that is integration competent. The additional stromal factor cannot be tightly associated with cpSRP because it is not coimmunoprecipitated with the two subunits and remains in the stroma. This additional factor is not likely to be an RNA, because RNase treatment of stroma had no effect on LHCP biogenesis (20). Work is in progress to identify the remaining components required for reconstituting the soluble phase of LHCP transport. One candidate for such a factor is a chloroplast homologue of FtsY, a soluble protein in E. coli (14) that resembles the α subunit of the SRP receptor and regulates the GTPase activity of SRP54 (15, 16).
The molecular mass of the stromal form of cpSRP is estimated to be 200 kDa by gel filtration (Fig. 1 A and B). The calculated molecular masses of cpSRP43 and cpSRP54 based on the polypeptide predicted from the cDNA sequences are 35 and 54 kDa, respectively. One possibility is that cpSRP is a tetramer comprised of two polypeptides of each subunit. If so, the size of the tetramer is predicted to be 178 kDa for the stromal form and 236 kDa for the recombinant form containing GST-43. Both roughly correspond to the particle masses estimated by gel filtration (200 and 255 kDa, respectively), suggesting they both have the same subunit stoichiometry (Fig. 1 A and B). Furthermore, an active cpSRP consists of just the two subunits as judged by complex formation with LHCP (Figs. 2 and 3) and the specificity of the immunodepletion experiments where activity is removed or restored by the removal or addition of just the two subunits (Fig. 1D, Fig. 4). Consistent with this model, preliminary evidence indicates that cpSRP43 is a dimer (D.S. and N.E.H., unpublished results). Hence a tetramer should form if each subunit of cpSRP43 binds one subunit of cpSRP54.
Whereas the function of cpSRP depends on cpSRP43, the function of cytosolic SRP depends upon an ancillary RNA that is tightly bound to the SRP protein subunits. We were unable to coimmunoprecipitate an RNA with cpSRP54. Furthermore as mentioned above, RNase treatment of stroma had no effect on LHCP biogenesis (20). These results, plus the finding that purified cpSRP54 and cpSRP43 assemble into an active transit complex, establish that cpSRP does not require an RNA to function; this distinguishes it from all cytoplasmic SRPs.
cpSRP also differs from cytoplasmic SRP in its ability to interact with substrates posttranslationally. Previous crosslinking studies demonstrated that cpSRP54 directly binds to LHCP (7). The requirement for cpSRP43 for this interaction suggests that at least one of its roles is to enable cpSRP54 to bind substrate. Possibly the evolution of cpSRP43 has led to an expanded role for cpSRP54 in the posttranslational targeting of membrane proteins. Many chloroplast-encoded proteins appear to be cotranslationally targeted to the thylakoid membrane (21). An interesting question is whether cpSRP has retained the ability to function in cotranslational targeting. It is pertinent to note that of the two cpSRP subunits, only cpSRP54 is associated with the ribosome. One possibility is that the ribosome bound form of cpSRP54 represents a second, cytoplasmic-like cpSRP, and is engaged in cotranslational targeting, while the form bound to cpSRP43 only functions posttranslationally. Recently, putative SRP-RNA homologues have been identified in the plastid genome of the red alga, Porphyra purpurea, and the diatom, Odontella sinensis, though, potential homologues in the green algae and higher plants have not yet been identified (22). Further studies on the ribosomal association of cpSRP54, and its possible role in the biogenesis of chloroplast encoded proteins, may provide clues as to whether an ancillary SRP-RNA and/or a second cpSRP exists in plastids from higher plants.
Acknowledgments
We thank Arthur Grossman for critically reviewing the manuscript and many helpful suggestions. This work was supported by grants from National Science Foundation (N.E.H.), U.S. Department of Agriculture (N.E.H.), and Deutsche Forschungsgemeinschaft (D.S.). The Carnegie Institution of Washington publication number for this work is 1373.
ABBREVIATIONS
- SRP
signal recognition particle
- cpSRP
chloroplast SRP
- GST
glutathione S-transferase
- LHCP
light-harvesting chlorophyll protein
References
- 1.Cline K, Fulsom D R, Viitanen P V. J Biol Chem. 1989;24:14225–14232. [PubMed] [Google Scholar]
- 2.Reed J E, Cline K, Stephens L C, Bacot K O, Viitanen P V. Eur J Biochem. 1990;194:33–42. doi: 10.1111/j.1432-1033.1990.tb19423.x. [DOI] [PubMed] [Google Scholar]
- 3.Cline K. J Biol Chem. 1986;261:14804–14810. [PubMed] [Google Scholar]
- 4.Chitnis P R, Nechushtai R, Thornber J P. Plant Mol Biol. 1987;10:3–11. doi: 10.1007/BF00014181. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman N E, Franklin A E. Plant Physiol. 1994;105:295–304. doi: 10.1104/pp.105.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Payan L A, Cline K. J Cell Biol. 1991;112:603–613. doi: 10.1083/jcb.112.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X X, Henry R, Yuan J G, Cline K, Hoffman N E. Proc Natl Acad Sci USA. 1995;92:3789–3793. doi: 10.1073/pnas.92.9.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilgrim M L, Wijk K-J v, Parry D H, Sy D A C, Hoffman N E. Plant J. 1998;13:177–186. doi: 10.1046/j.1365-313x.1998.00021.x. [DOI] [PubMed] [Google Scholar]
- 9.Harlow E, Lane D. Antibodies. A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 10.Adam Z, Hoffman N E. Plant Physiol. 1993;102:35–43. doi: 10.1104/pp.102.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulsen H, Rumler U, Rudiger W. Planta. 1990;181:204–211. doi: 10.1007/BF02411539. [DOI] [PubMed] [Google Scholar]
- 12.Cline K, Henry R, Li C J, Yuan J G. EMBO J. 1993;12:4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin A E, Hoffman N E. J Biol Chem. 1993;268:22175–22180. [PubMed] [Google Scholar]
- 14.Gill D R, Salmond G P C. Mol Gen Genet. 1987;210:504–508. doi: 10.1007/BF00327204. [DOI] [PubMed] [Google Scholar]
- 15.Miller J D, Bernstein H D, Walter P. Nature (London) 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 16.Powers T, Walter P. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 17.Lubben T H, Donaldson G K, Viitanen P V, Gatenby A A. Plant Cell. 1989;1:1223–1230. doi: 10.1105/tpc.1.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madueno F, Napier J A, Gray J C. Plant Cell. 1993;5:1865–1876. doi: 10.1105/tpc.5.12.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan J G, Henry R, Cline K. Proc Natl Acad Sci USA. 1993;90:8552–8556. doi: 10.1073/pnas.90.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulson D, Cline K. Plant Physiol. 1988;88:1146–1153. doi: 10.1104/pp.88.4.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagendorf A T, Michaels A. Plant Sci. 1990;70:137–145. [Google Scholar]
- 22.Packer J C L, Howe C J. Mol Microbiol. 1998;27:508–510. doi: 10.1046/j.1365-2958.1998.00709.x. [DOI] [PubMed] [Google Scholar]