Abstract
In C. elegans, assembly of hypodermal hemidesmosome-like structures called fibrous organelles is temporally and spatially coordinated with the assembly of the muscle contractile apparatus, suggesting that signals are exchanged between these cell types to position fibrous organelles correctly. Myotactin, a protein recognized by monoclonal antibody MH46, is a candidate for such a signaling molecule. The antigen, although expressed by hypodermis, first reflects the pattern of muscle elements and only later reflects the pattern of fibrous organelles. Confocal microscopy shows that in adult worms myotactin and fibrous organelles show coincident localization. Further, cell ablation studies show the bodywall muscle cells are necessary for normal myotactin distribution. To investigate myotactin's role in muscle-hypodermal signaling, we characterized the myotactin locus molecularly and genetically. Myotactin is a novel transmembrane protein of ∼500 kd. The extracellular domain contains at least 32 fibronectin type III repeats and the cytoplasmic domain contains unique sequence. In mutants lacking myotactin, muscle cells detach when embryonic muscle contraction begins. Later in development, fibrous organelles become delocalized and are not restricted to regions of the hypodermis previously contacted by muscle. These results suggest myotactin helps maintain the association between the muscle contractile apparatus and hypodermal fibrous organelles.
Keywords: Caenorhabditis elegans, cell adhesion, cell ablations, muscle, hemidesmosome-like structures
Throughout the bodies of complex multicellular organisms, different cell types must interact to form the organs and tissues necessary for diverse biological functions. One of the best-characterized examples where different cell types coordinate their behaviors to achieve a specific function occurs at the neuromuscular junction where a nerve and muscle cell function together during synaptic transmission, an event requiring the correct spatial relationship between pre- and post-synaptic protein complexes. This relationship is initiated by contact between the muscle and nerve, and is established through a signaling pathway involving the basement membrane protein agrin (reviewed in 10, 23) and a muscle-specific kinase 14.
Similarly, in Caenorhabditis elegans, bodywall muscles and the hypodermis function coordinately during locomotion. The contractile apparatus of the bodywall muscle consists of thick and thin filaments that slide past each other to create force. During locomotion, the force generated by the contractile apparatus must be transmitted through the hypodermis to the cuticular exoskeleton. This is achieved through a specialized basement membrane situated between the muscle cells and hypodermis, and through hypodermal cell adhesion structures called fibrous organelles 22. Together with muscle and muscle membrane proteins, the basement membrane and the fibrous organelles form a molecular link between the contractile apparatus and the cuticle 21 22.
Each hypodermal fibrous organelle is made up of two substructures, both of which are morphologically similar to vertebrate hemidesmosomes 22. Like vertebrate hemidesmosomes, the substructures have the classical membrane plaque and associated intermediate filaments 22 48. One hemidesmosome-like plaque of each fibrous organelle is found on the hypodermal membrane adjacent to the muscle basement membrane, and the other is found on the hypodermal membrane adjacent to the cuticle 22. The associated intermediate filaments extending between the plaques on either surface probably span the cells.
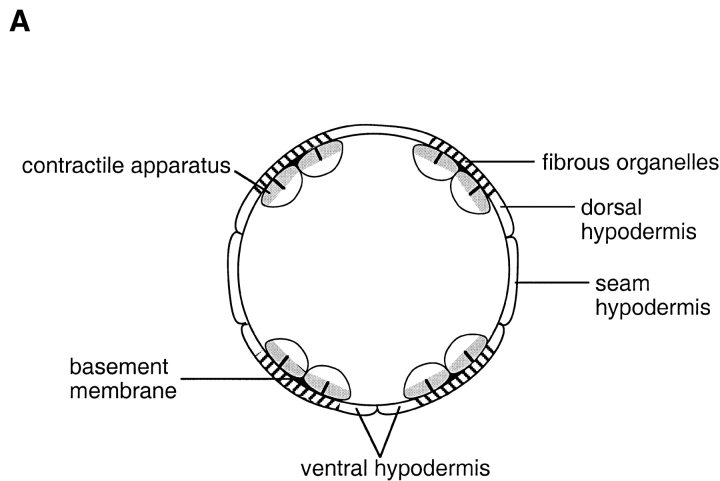
The bodywall muscle contractile apparatus and the hypodermal fibrous organelles are closely associated (see Fig. 1 A). The contractile apparatus is positioned at the muscle cell membrane that faces the hypodermis (reviewed in 55) and fibrous organelles are localized to regions of the hypodermis adjacent to muscle 22. These types of spatial and functional relationships between structures in different cells suggest signals are passed between the cells to establish the association, and that molecules exist to maintain the correct alignment of the structures during the life of the organism.
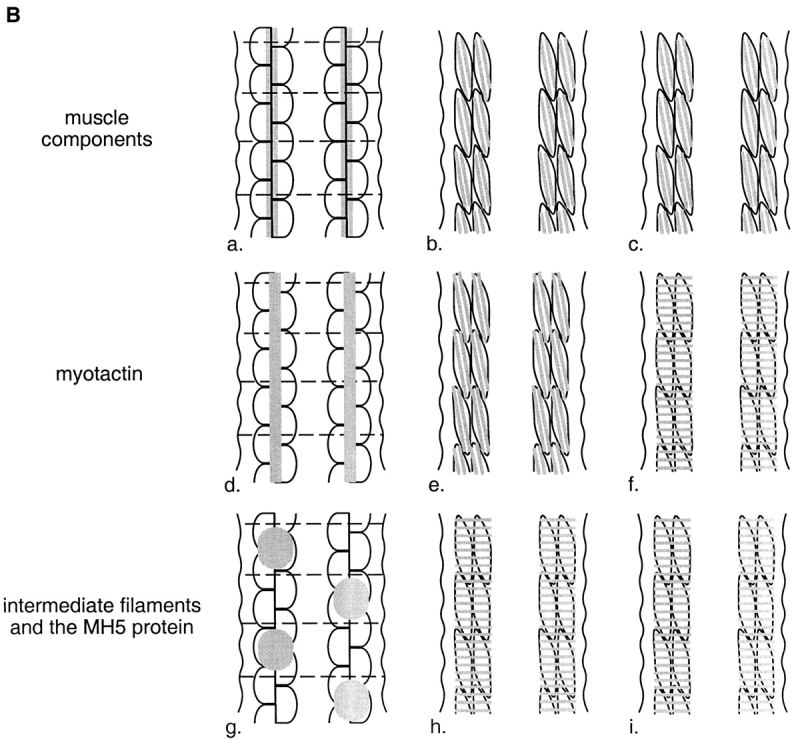
Figure 1.
(A) Schematic representation of a cross-section through a threefold stage embryo depicting the spatial relationships between the contractile apparatus of the bodywall muscle (oval-like shapes), the muscle basement membrane (black) and hypodermal fibrous organelles (hatched areas). Dorsal is toward the top of the page. The four muscle quadrants are shown, each consisting of two rows of muscle cells, with the long axis of the cells perpendicular to the page. The contractile apparatus is restricted to the portion of the muscle cell closest to the muscle cell membrane facing the hypodermis. Hypodermal fibrous organelles are restricted to the region of the hypodermis adjacent to bodywall muscle cells. A specialized basement membrane, positioned between muscle cells and the hypodermis, helps to anchor muscle cells. Myotactin is thought to be an integral membrane protein expressed on the basal hypodermal membrane with its large extracellular domain extending into the basement membrane toward the adjacent muscle cells. (B) Schematic representations of the localization of muscle, fibrous organelle–associated intermediate filaments, the MH5 protein and myotactin in dorsal views of progressively older embryos. Intermediate filament proteins and the MH5 protein, an immunologically defined protein that localizes near fibrous organelles, show the same distribution at all developmental stages. Schematics a, d, and g represent an early stage of development (just before the start of elongation); b, e, and h represent an intermediate stage (twofold); and c, f, and i represent a later stage (threefold). In each schematic (a–i) a portion of the two dorsal muscle quadrants is shown. Each quadrant consists of two rows of mononucleated muscle cells with the long axis of the cells oriented along the anterior-posterior axis of the worm. Anterior is toward the top of the page. Wavy, black, vertical lines mark the lateral edges of the dorsal hypodermis. Horizontal, dashed black lines in a, d, and g represent the boundaries between adjacent dorsal hypodermal cells. These lines do not appear in b, c, e, f, h, and i because at these later developmental stages the dorsal hypodermal cells have fused to form a syncytium. Gray areas represent the localization of components of the muscle cell contractile apparatus (a–c), myotactin (d–f), and the fibrous organelle–associated intermediate filaments and MH5 protein (g–i). Schematics a–c are focused on the plane of the muscle cells, while d–i are focused on the dorsal hypodermal cells. Although we discuss only the dorsal surface of the embryo, similar events occur on the ventral surface. At stages earlier than those shown, muscle cells are adjacent to lateral (seam) hypodermis, and muscle proteins are diffusely distributed in muscle cells. Similarly, the hypodermal proteins are diffusely distributed in dorsal hypodermal cells. As muscle cells migrate to contact dorsal hypodermal cells, muscle components accumulate at the membrane where muscle cells contact each other and the hypodermis (a). At about the same time, myotactin, intermediate filament proteins and the MH5 protein are recruited to specific regions of the hypodermis adjacent to some muscle cells. Myotactin accumulates adjacent to where the contractile apparatus is forming (d) while intermediate filaments and the MH5 protein accumulate in a single broad patch in each hypodermal cell adjacent to one muscle quadrant (g). By the twofold stage, the sarcomeric organization of the obliquely striated muscle is observed (b). At this stage myotactin is still adjacent to the forming contractile apparatus and in fact is organized into rows of dots running oblique to the long axis of the worm and following the oblique striations of the muscle (e). In contrast, intermediate filaments and the MH5 protein are organized in circumferentially oriented bands that are restricted to the small regions of the hypodermis adjacent to muscle (h). During the threefold stage, myotactin colocalizes with intermediate filaments and the MH5 protein (f and i).
The molecular link between the contractile apparatus and the fibrous organelles is set up during embryogenesis and, like the neuromuscular junction, appears to be initiated by cell–cell contact (30; summarized in Fig. 1 B). During embryogenesis, the post-mitotic developing muscle cells migrate from their initial position adjacent to the lateral hypodermal cells onto the neighboring dorsal and ventral hypodermal cells. Components of the muscle contractile apparatus, which are until this time diffusely distributed, coalesce at the membrane near the point of contact with the dorsal/ventral hypodermal cells. Similarly, and within minutes, the fibrous organelle components, which are initially diffusely distributed in the hypodermis, coalesce in the hypodermis adjacent to muscle. Throughout further development, the components of the fibrous organelles remain restricted to the small portion of the dorsal and ventral syncytial hypodermis adjacent to muscle cells 30. The single exception is that in larval stages, when mechanosensory neurons develop, fibrous organelles also appear overlying these neuronal processes 22. These neuron-associated fibrous organelles play an adhesion role similar to that of the muscle-associated fibrous organelles, affixing the neuron to the cuticle.
Both the myofilament lattice and the fibrous organelles assemble in specific and different patterns as development continues. The muscle proteins show an oblique longitudinal striation (reviewed in 55) and the fibrous organelles become organized in circumferentially oriented, regularly spaced bands, limited to regions of the hypodermis adjacent to muscle cells 22. Each hypodermal band consists of multiple smaller complexes, each corresponding to a fibrous organelle. The spacing of the fibrous organelle–containing bands correlates with the spacing of cuticular ridges called annuli.
Three proteins that reflect the fibrous organelle pattern of organization have been identified immunologically 22. All three localize to small dots which together form a banded pattern in regions of the hypodermis which are known to contain fibrous organelles by electron microscopic analysis (22; see also Fig. 8 A). One is an intermediate filament subunit recognized by monoclonal antibody MH4 and by the antibody IFA raised against mammalian intermediate filaments 22. The other two proteins, recognized by MH5 and MH46, respectively, are large and are closely associated with the hypodermal membrane. The MH5 protein is found at both the cuticular and the muscle basement membrane surfaces of the hypodermis while the MH46 protein is specific for the muscle basement membrane surface 22. As will be discussed, the MH46 protein plays a role in muscle–cell adhesion and therefore we have named the protein myotactin. Intermediate filaments, myotactin, and the MH5 protein are also found in the pharynx, an organ at the anterior of the worm used in feeding 1 6 22.
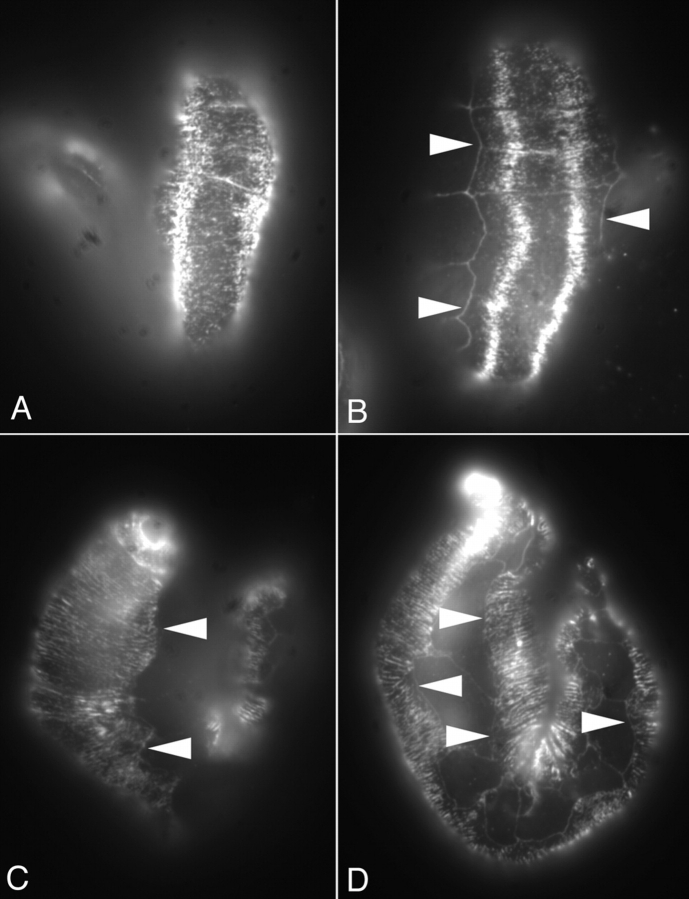
Figure 8.
Wild-type and myotactin mutant embryos stained for intermediate filaments. Mixed stage embryos were fixed for immunofluorescence and labeled with MH4 (intermediate filament protein) and MH27. MH27 marks the boundaries between hypodermal cells and appears as a grid pattern on the embryos in B and D. (A and B) Dorsal view of a wild-type (A) and a st456 homozygous (B) embryo. In both embryos, the MH4-dependent signal is concentrated in the regions of the hypodermis adjacent to muscle. (C) Dorsal view of a st456 homozygote older than the one shown in B. MH4-dependent fluorescence is seen all through the dorsal hypodermis. (D) Lateral view of an embryo at a similar stage to the one shown in C. The MH4-dependent staining extends to the boundaries between the dorsal or ventral hypodermis and the seam hypodermis. The fluorescence pattern seen in C and D is one of circumferentially oriented bands. Arrowheads mark the boundaries between dorsal or ventral and seam hypodermis.
The change in the myotactin staining pattern as development progresses suggests it might be involved in establishing or maintaining the correct spatial relationship between the contractile apparatus and hypodermal fibrous organelles. Unlike the MH4-containing intermediate filaments and the MH5 protein, myotactin appears to colocalize with muscle proteins early in embryonic development, and only later organizes into the circumferential bands of the fibrous organelles (30; see Fig. 1 B). In this study, confocal microscopy was used to show that myotactin localizes at or near fibrous organelles in adult C. elegans. We also show that the early muscle-like distribution of myotactin depends on signals from the bodywall muscle cells. To investigate the role myotactin plays in muscle-hypodermal signaling, we have begun a molecular and genetic characterization of the myotactin gene. We have cloned and sequenced cDNAs that encode myotactin, and characterized mutants in the corresponding gene. The sequence suggests myotactin is a transmembrane protein containing at least 32 fibronectin type III (FNIII)1 repeats extracellularly and unique sequences intracellularly. The phenotype of the mutants suggests myotactin is involved in muscle cell attachment and possibly in maintaining the position of the fibrous organelles within regions of the hypodermis adjacent to muscle.
Materials and Methods
Strains and General Maintenance of Stocks
Worms were grown on bacterial strain OP50 spread on NGM (nematode growth media) agar plates according to Brenner 11. The Bristol variety of the N2 strain of C. elegans is used as the wild-type strain, and the genotypes of the mutant strains used are as follows: RW3625, let-805(st456)/qC1; BC4534, unc-45(rh450)let-805(s2764)/sC1; and let-805(st456)/dpy-1(e1)daf-2(e1370).
Cloning
A clone encoding an MH46 epitope was isolated from a genomic library made from randomly sheared C. elegans DNA cloned into the vector λgt11 4. The screen was performed according to Maniatis 36 with a few exceptions. Filters were washed in TTBS (20 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20) and blocked in TTBS containing 1% normal goat serum (GIBCO BRL) and 5% fish gelatin (Sigma). The MH46 primary antibody was detected using alkaline phosphatase-conjugated goat anti–mouse IgG (Tago) diluted 1:2,500 in block buffer and the secondary antibody was localized using 0.137 mg/ml 5-bromo-4-chloro-3-indolyl phosphate (Sigma), 0.33 mg/ml nitro blue tetrazolium (Sigma) and 0.013 mg/ml phenazine methosulfate (Sigma) according to Ey and Ashman 19. One clone (λG-cc9) was obtained from a screen of 106 PFU.
The genomic fragment from λG-cc9 was labeled using the Prime-It-II kit (Stratagene) and α-P32-labeled ATP (Amersham) according to Stratagene. The labeled DNA was used to screen a randomly primed cDNA library made from RNA of mixed stage worms and cloned into the pACT vector. The screen was performed according to Maniatis 36 except hybridizations were done in 6× SSC, 1% SDS, and 100 mg/ml salmon sperm DNA and washes were done in 10 mM Tris, pH 7.5, containing 1% SDS. The largest clone (pC-cc1A) was sequenced and shown to be identical to the sequence of λG-cc9 in the region of overlap. Using the sequence of pC-cc1A, probes were designed to screen the library for clones containing sequences 5′ and 3′ to pC-cc1A. Similar screens were performed until we had evidence we had cloned the 5′ and 3′ ends of the message.
Sequencing
The overlapping cDNA clones were sequenced by fluorescent sequencing techniques using dye-labeled primers or dye-labeled dideoxynucleotides (ABI). The reactions were performed according to the manufacturer and reactions were run on an ABI 373A gel analyzer.
Nested deletions of the clone pC-cc1A were made using Erase-a-Base (Promega) according to the manufacturer, and the deletion clones were sequenced.
The templates used to obtain the sequence of the ends of each cDNA isolated subsequent to pC-cc1A were amplified by PCR. The sequence of the primers used was as follows: primer 1, 5′-GATGATGAAGATACCCCACC-3′; primer 2, 5′-AGTTGAAGTGAACTTGCGGG-3′; primer 3, 5′-GCGTGTAAAACGACGGCCAGTGATGATGAAGATACCCCACC-3′; primer 4, 5′-GCGTGTAAAACGACGGCCAGTAGTTGAAGTGAACTTGCGGG-3′. The underlined bases in primers 3 and 4 contain a universal priming site. Primers 1 and 3 are complementary to the same sequence 5′ of the pACT cloning site, and 2 and 4 are complementary to the same sequence 3′ of the pACT cloning site. Primer pairs 1 and 4, or 2 and 3, were used to amplify the cDNA insert from each clone and to add the universal priming site to the 5′ or the 3′ end of the amplification products. The products were treated with 0.2 units exonuclease I (United States Biochemicals) and 0.2 units shrimp alkaline phosphatase (United States Biochemicals) for 45 min at 37°C to degrade the unused primer 57 and the enzymes were inactivated for 20 min at 80°C. The products were then sequenced directly. This technique was also used to obtain the sequence of exons 6–9 and 13. These exons were not encoded by any of the cloned cDNAs but were thought to be expressed due to the high degree of homology between the C. elegans and C. briggsae genomic sequences in these regions. In these cases, templates (RT-PCR 1, 2, and 3; see Fig. 3) were amplified from the random-primed library using gene-specific primers complementary to the sequence of the exons flanking the region of interest. As above, one primer used in each reaction was fused to the universal priming site.
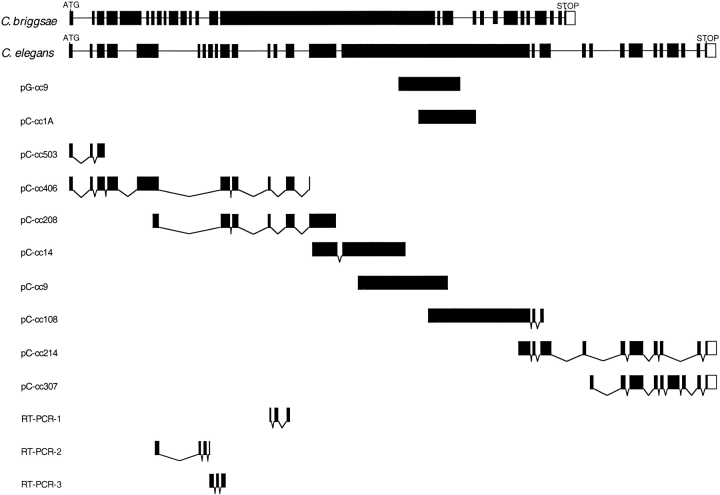
Figure 3.
Schematic diagram of the partial myotactin cDNAs and genomic clones. The top two lines represent C. briggsae and C. elegans genomic clones, respectively. Exons are depicted as filled boxes and introns as lines. pG-cc9 is the genomic clone isolated using the MH46 antibody as a probe (see Materials and Methods) and pC-ccXX(X) are the overlapping cDNA clones. RT-PCR-1, 2 and 3 (see Materials and Methods) were sequenced to confirm the intron–exon boundaries between exons 5 and 6, 6 and 7, 7 and 8, 8 and 9, 9 and 10, 12 and 13, and 13 and 14. The open boxes at the 3′ ends of pC-cc214, pC-cc307, and the C. elegans and C. briggsae genes represent noncoding sequence. The positions of the predicted translational start (ATG) and the stop codon (TAA) are indicated. pC-cc503 contains eight bases at the 5′ end identical to the eight 3′ bases of the SL1 spliced leader sequence. Note that exons 6–9, 13, 19A, 19B, 24 and 25 are all differentially spliced. The combinations with which these exons are used has not been determined. The complete sequence of the myotactin cDNAs is attainable through Genbank. Two cDNA sequences were submitted: Form A includes exon 19A but excludes exons 19B, 24, and 25 (accession number AF148954) and form B includes exons 19B, 24, and 25 but excluded 19A (accession number AF148953). The two forms reflect the differences observed between cDNA pC-cc214 and pC-cc307.
Finally, libraries from each cDNA clone were constructed in the m13 vector to obtain templates to completely sequence each cDNA clone. Each insert was amplified using PCR and primers 1 and 2 (see above). A pool of amplification products was fragmented using a nebulizer and 30 psi nitrogen gas 9, treated with mung bean nuclease to generate blunt ends, and then cloned into the SmaI site of m13. DNA was isolated from random m13 clones from each library and sequenced.
The sequence obtained was edited for quality using the trace editor TED 26 and assembled into contigs using XBAP (a sequence assembly program for X windows; reference 13). The identity of each base was confirmed by obtaining the sequence of both strands, by obtaining the base using two different chemistries (dye-labeled primers or dye-labeled dideoxy-terminators) or by comparison to the genomic sequence generated by the C. elegans genome project.
Mapping
The genomic clone was placed on the physical map by hybridization of a P32-labeled DNA probe (Prime-It II kit; Stratagene) to YAC clones representing ∼95% of the C. elegans genome 12. The YAC clones are spotted on a nitrocellulose sheet in a grid pattern. Each YAC represented on the grid has been ordered into contigs by fingerprint analysis and the contigs placed into linkage groups representing each C. elegans chromosome.
The st456 mutation (see Results) was mapped by standard three factor mapping techniques. Recombinants were cloned from st456/dpy-1(e1)daf-2(e1370) strain, and their progeny were scored to determine where the recombination event took place. Recombination took place between dpy-1 and st456 6 times and between st456 and daf-2 twice, placing the mutation between dpy-1 and daf-2. The st456 mutation failed to complement a mutation, let-805(s2764), isolated in an unrelated lethal screen (Baillie, D., personal communication) performed as described by Stewart et al. 49. This result suggests st456 is an allele of the let-805 gene.
Protein Analysis
Protein similarities were identified using the program BLAST (version 2.0; reference 2) to search the nonredundant protein database and the est database (dbest) through the NCBI server (search date 8 April 1999). Searches were also done of an ftp site containing data from the two C. elegans Genome Sequencing Centers (http://genome.wustl.edu).
FNIII repeats were defined using hidden Markov models (HMM) profile 18 32. The model used is the fn3 HMM from the Pfam protein domain database (release 1.0; reference 47). This HMM is based on a multiple alignment of 456 FNIII domains from Swissprot 33. The search was performed using the programs hmmls and hmnfs from the HMMER hidden Markov model software package 17. Alignment scores over 15 bits (a log odds score, base two) were considered significant.
The fn3 model identifies only the first six of the seven β-strands of the FNIII repeat. Therefore, a new model was generated to identify the amino acids that form the seventh strand in each repeat, and to produce the alignment shown in Fig. 5. The new model was generated using the program hmmb 17, and starting with a seed alignment of the four FNIII repeats of fibronectin 33. The seed alignment was expanded to include repeats identified in the myotactin sequence that had significant scores (in this case >20 bits). The expanded alignment was used to generate a new HMM model. These steps were repeated until no new repeats could be identified in the myotactin sequence. The 32 repeats identified in this way were then reanalyzed using the fn3 model and 30 of the 32 repeats had significant scores of >15. The exceptions are repeats 8 and 27 which had scores of 13.19 and 13.94, respectively.
Figure 5.

Alignment of the 32 putative FNIII repeats. With the exception of repeats 8 and 27, the repeats have scores over 15 when analyzed by the fn3 HMM program (see Materials and Methods).
Secondary structure predictions were made using the program PHD 43 44.
In Situ Hybridization
In situ hybridizations to mixed stage C. elegans embryos were done according to the protocol of Seydoux and Fire 45 with a few exceptions. Embryos were obtained by alkaline hypochlorite treatment of gravid adults 52 and then treated in batch in microfuge tubes rather than on microscope slides. After each incubation the embryos were collected by gentle centrifugation. Embryos were fixed in 3% formaldehyde and 0.25% glutaraldehyde in 85 mM K2HPO4, pH 7.2 31. The probes used were made by PCR amplification according to Seydoux and Fire 45.
Immunological Techniques
Embryos were prepared for antibody staining and staged as described in Hresko et al. 30. Fluorescence was viewed on a Bmax-60F microscope (Olympus) equipped with Nomarski and fluorescence optics. Images were taken with a Pentamax camera containing a Kodak KAF-1400 chip (1,317 × 1,035) with 6.8-mm pixels (Princeton Instruments). The camera was controlled by an Optiplex GXPro200 computer (DELL Computer Corp.) running WinView software (v. 1.6.2.1, Princeton Instruments). After acquisition the raw 12-bit WinView images were transferred to a Power Macintosh computer, linearly scaled to 8-bits ([max pixel value − min pixel value]/256), inverted to conform to the Macintosh intensity scale, reduced to 256 shades of gray, and saved as TIFF files using a batch file conversion program (Spe2Tiff; written by and freely available on request from waddle@hamon.swmed.edu). Images were assembled and annotated using Adobe Photoshop 3.0.5 (Adobe Systems, Inc.) and printed on a Tektronix Phaser 440 printer (Tektronix).
Adult worm fragments were prepared for antibody staining using a French press as described by Francis and Waterston 21. After fragmenting the worms, the pieces were extracted three times for 30 min in 0.5% NP-40 in low salt buffer (7.5 mM Na2PO4, pH 7.0, 40 mM NaCl, 1 mM EDTA, and 1 mM PMSF), for 15 min in 0.5 M KSCN in low-salt buffer and for 15 min in low-salt buffer 22. The extracted fragments were then fixed for 20 min in −20°C MeOH. The samples were viewed using a Zeiss Axioplan microscope equipped for confocal imaging with a Bio-Rad MRC 1000 laser.
Ablations
Bodywall muscle precursor cells were ablated at the 28-cell stage using a nitrogen pulse laser (Laser Sciences, Inc.) as described 3. The beam was directed through a Zeiss Axioplan microscope with the 100× objective and laser intensity was adjusted by the use of neutral density filters. In one set of experiments MS.ap and MS.pp were ablated and in another set C.ap was ablated. MS.ap and MS.pp collectively give rise to six anterior bodywall muscle cells from each dorsal quadrant and three from each ventral quadrant 51. C.ap gives rise to nine posterior bodywall muscle cells from the left dorsal quadrant and seven from the left ventral quadrant 51. After ablations were performed, the embryos were allowed to develop to a specific development stage, fixed by freeze fracture 50 and stained with appropriate antibodies. Confirmation that the correct cell(s) was ablated was obtained by staining the embryos with an antibody against bodywall muscle myosin.
Mutant Screen
The myotactin genomic fragment (from pG-cc9) maps in the physical interval between the par-2 and ben-1 genes, corresponding to the region of the genetic map containing the dpy-1 gene. We reasoned mutations in the myotactin gene might be lethal and screened for lethal mutations linked to the dpy-1 gene. Homozygous dpy-1(e1) hermaphrodites were mutagenized with 50 mM EMS (ethylmethanesulfonate) 52. Mutagenized hermaphrodites were mated to wild-type males (N2 strain) and individual L4 hermaphrodites were cloned from the F1 progeny. The progeny of each cloned hermaphrodite were examined for the loss of dpy animals, suggesting a mutagenesis event causing a lethal mutation linked to the dpy-1 gene had occurred in that strain. Embryos from strains segregating few or no dpy animals were stained with MH46 to identify mutants negative for the antibody. One mutant was identified. The mutant strain was outcrossed six times to eliminate other mutations that might be in the background.
Germline Transformation
Germline transformation was carried out according to the protocol of Mello et al. 39. The DNA fragment used is ∼30 kb and contains the C. briggsae myotactin gene (95% identical to the C. elegans protein), including 8 kb upstream of the translational start site and ∼2 kb downstream of the stop codon. This plasmid, along with a plasmid containing the rol-6 gene carrying a dominant mutation, was injected into dpy-1(e1)let-805(st456)/qC1 hermaphrodites. The qC1 chromosome contains a rearrangement that suppresses recombination between dpy-1 and let-805. Rolling L4 hermaphrodites were picked 3 d later and their progeny were examined for the presence of rescued animals by looking for animals with the dpy phenotype.
RNA-mediated Interference
Two templates were made from each clone (pC-cc307 and 406) using the enzyme PWO (Boerhinger Mannhiem Biologicals) and primers homologous to the vector. The T7 polymerase binding site was added to the 5′ end of the coding (template 1) or the noncoding (template 2) strand of the templates during the amplification 41. Sense and antisense RNA's were synthesized from templates 1 and 2, respectively, using mCAP (Stratagene) according to the manufacturer. Equimolar amounts of sense and antisense RNA were annealed to yield double-stranded RNA. The double-stranded RNA was injected into hermaphrodites. Hermaphrodites were allowed to lay eggs for 36 h, then the hermaphrodites were transferred to new plates. The plates were examined for non–wild-type animals to determine the phenotype caused by injection of the RNA.
To determine if the phenotype caused by RNA-mediated interference was similar to that of the let-805(st456) homozygotes, embryos were collected 10–15 h after transfer of injected hermaphrodites to new plates and the arrested embryos were fixed by freeze fracture 50 and stained with antibodies.
Results
Myotactin Colocalizes with Hypodermal Intermediate Filaments
Myotactin and fibrous organelle–associated intermediate filaments colocalize in adult C. elegans worms. Adult worm fragments were fixed and double labeled with MH46 to localize myotactin and a rabbit polyclonal antibody against intermediate filaments 22, and viewed by confocal microscopy. The distribution of myotactin and intermediate filaments in adult worms is similar (Fig. 2). Both proteins are organized in thin bands running circumferentially around the worm, and both are restricted to regions of the hypodermis adjacent to muscle and some mechanosensory neurons. The myotactin- and intermediate filament-dependent staining within each band is not uniform, but rather consists of a large number of discrete punctate structures. Merging of the rhodamine and fluorescein images shows the punctate staining patterns seen with the two antibodies are coincident as evidenced by the yellow fluorescence (Fig. 2 C). The correlation between the distribution of myotactin and intermediate filaments suggests myotactin localizes close to fibrous organelles.
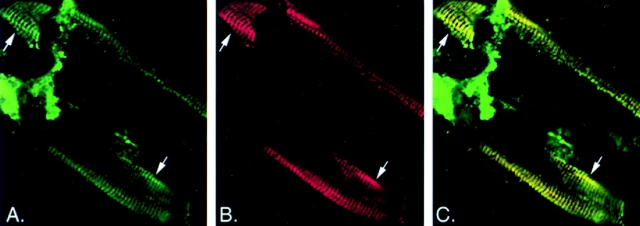
Figure 2.
Confocal images of a wild-type adult worm fragment stained for intermediate filaments (green) and myotactin (red). (A) Intermediate filaments are concentrated in thin bands running circumferentially around the worm (arrows). The intermediate filament-dependent staining within each band is not uniform but is associated with discrete punctate structures within the band. (B) Like intermediate filament-dependent staining, myotactin-dependent staining is associated with punctate structures that are organized into bands (arrows). (C) Merging of the images shown in A and B shows the punctate patterns seen with the two antibodies are coincident and thus produce the yellow fluorescence.
Cloning and Sequencing
As a first step in understanding the role myotactin plays in signaling between muscle and the hypodermis, we cloned a genomic fragment (pG-cc9) encoding an MH46 epitope from an expression library 4. We used the cloned genomic fragment to start cloning a set of overlapping cDNAs (Fig. 3; see Materials and Methods) extending from the likely 5′-end to the stop codon. The 5′ most sequence obtained encodes eight bases identical to the eight 3′ bases of the SL1 spliced leader, which is trans-spliced to the 5′ end of many C. elegans messages, indicating this sequence represents the 5′ end of the myotactin message. Typically, the first ATG after SL1 encodes the initiator methionine, and in >90% of the messages examined to date, the initiator methionine is <30 nucleotides from the 3′ end of SL1 8. In the sequence presented here, there are 12 nucleotides separating SL1 and the first ATG which we have designated as the initiator. The 3′ most clone, pC-cc307, was isolated from an oligo dT primed cDNA library, and contains sequence identical to that of a 3′ expressed sequence tag (CEMSD21; reference 37), suggesting the sequence encoded by pC-cc307 is close to the 3′ end of the message. Furthermore, the sequence encoded by pC-cc307 and CEMSD21 contains a stop codon at the end of a long open reading frame beginning with the initiating ATG. This stop codon is followed by sequence consistent with 3′ noncoding region with multiple stop codons in all three reading frames.
While the cDNAs were being sequenced, the genomic sequence of the Caenorhabditis elegans gene (Chissoe, S., personal communication) and a Caenorhabditis briggsae homologue (Marra, M., personal communication) became available (the C. elegans genome sequencing project) which allowed us to define the intron–exon boundaries over most of the gene (Fig. 3). The exon boundaries are conserved between the C. elegans and the C. briggsae genes with one exception: exons 15 and 16 of the C. elegans gene are encoded by only one exon in the C. briggsae gene. However, the intron sizes are very different, with the C. elegans introns generally being larger than the corresponding C. briggsae introns.
Myotactin is predicted to be a large transmembrane protein. A Northern blot of total RNA from mixed stage worms probed with the insert from pG-cc9 shows three RNAs hybridize to the probe, the smallest being ∼15 kb (Fig. 4). Furthermore, the longest open reading frame encoded by the DNA fragments depicted in Fig. 3 predicts a protein of 4,450 amino acids with a calculated molecular mass of ∼498 kD.
Figure 4.

Multiple messages are transcribed from the myotactin gene, the smallest being ∼15 kb. 20 μg of total RNA from mixed stage worms was run on a 0.7% agarose gel in the presence of formaldehyde and then transferred to nitrocellulose. A 32P-labeled DNA fragment corresponding to the sequence of pC-cc1A was used to probe the Northern blot. Molecular weight markers are indicated at the left, and were determined by either hybridization of the same blot with a twitchin probe (22 kb; 7) or a dynein heavy chain probe (14 kb; 35) or by the position of the 28S (3.5 kb) and 18S (1.75 kb) rRNAs of C. elegans.
Analysis of the hydrophobicity identifies three regions of the myotactin sequence sufficiently hydrophobic to insert into the membrane. One at the amino terminus (amino acids 1–20) probably represents a signal sequence suggesting the protein is secreted or is a transmembrane protein. The other two regions, encoded by exons 19A and B, respectively, are long enough to span the membrane, and therefore are likely to be transmembrane sequences. These exons are used alternatively (see Fig. 3) suggesting at least two protein isoforms are expressed, each with a different transmembrane domain.
The putative extracellular domain of myotactin contains at least 32 repeats of ∼100 amino acids in length with homology to fibronectin type III repeats (FNIII; Fig. 5) as identified by a HMM for FNIII domains (47; see Materials and Methods). Additional sequences (amino acids 331–425, 559–662, 670–770, 3658–3755, and 3777–3891) contained in the putative extracellular domain may represent another 5 FNIII repeats. These sequences are not identified as FNIII repeats by the fn3 model, but are each ∼100 amino acids in length and are predicted to contain 5–7 β-strands by the secondary structure program PHD 43 44. This is similar to predictions concerning the secondary structure of the 32 predicted FNIII repeats and is consistent with the crystal structure of the FNIII repeat 33.
The putative cytoplasmic domain of myotactin has two serine-rich regions suggesting this sequence might be modified in response to condition or stimuli. The amino acids encoded by exons 22–23 and those encoded by exons 26 and 27 are each 14% serine (10/69 and 8/55, respectively), and those encoded by differentially expressed exons 24 and 25 (see Fig. 3) collectively, are 18% serine (32/173), while the average protein is only 7% serine 15.
C. elegans myotactin and the presumed orthologue from C. briggsae appear to be novel transmembrane proteins. To date, a protein consisting of multiple FNIII repeats and lacking other known repeat motifs in its extracellular domain has not been reported. Furthermore, the cytoplasmic domain of the protein is unique. The only proteins in the databases (search date 8 April 1999) with significant similarities to the cytoplasmic domains of the C. elegans and C. briggsae proteins, are proteins rich in serine and an est entry from Onchocerca volvulus, another nematode.
Myotactin Is the MH46 Antigen
Two lines of evidence suggest myotactin is the MH46 antigen. First, the expression of myotactin message is tightly correlated with the presence of the MH46 epitope. Myotactin message is first detected by in situ hybridization in embryos before elongation (Fig. 6A and Fig. B). Signal is detected in two groups of cells: one group on the dorsal surface of the embryo flanking the dorsal midline, in the position of the dorsal hypodermal cells (Fig. 6 A); and a second group on the lateral edges of the embryo about midway between the dorsal and ventral surfaces, in the position of the ventral hypodermal cells (Fig. 6 B). These two groups of cells are also positive for the MH46 antibody 30. Later in development, at about the 1.5–1.75-fold stage, myotactin message is also detected in the pharynx (data not shown), a tissue also positive for the MH46 protein 22.
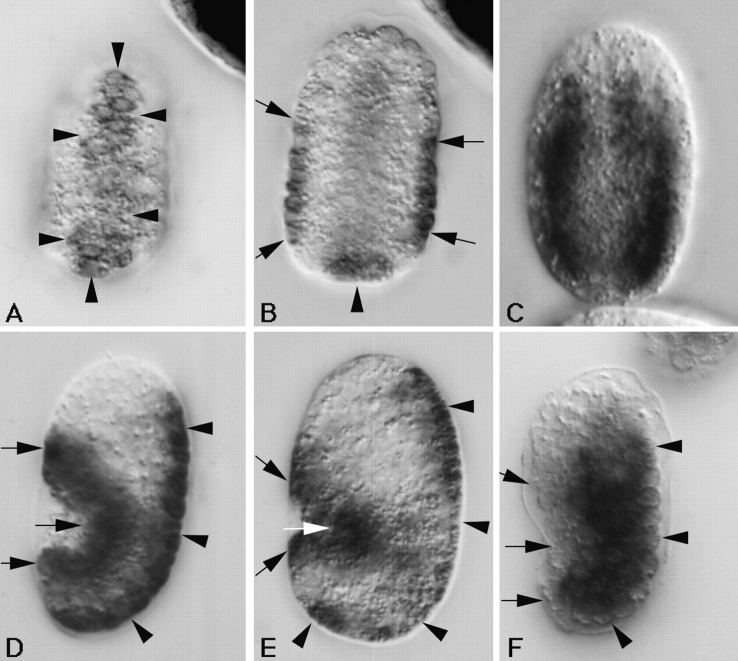
Figure 6.
Embryonic expression of the myotactin gene assayed by in situ hybridization. Mixed stage embryos were fixed and incubated with anti-sense myotactin (A, B, D, and E) or myosin (C and F) single stranded DNA probes labeled with digoxigenin. Probes were visualized using an alkaline-phosphatase-conjugated anti-digoxigenin antibody. Anterior is toward the top and in D–F dorsal is to the right. Arrowheads designate dorsal hypodermal cells and arrows designate ventral hypodermal cells. (A) Dorsal and (B) mid-focal plane images of the same <290-min embryo. The anti-sense myotactin probe is detected on the dorsal surface of the embryo (A) and in cells at the lateral edges (B) of the embryo. These cells are the dorsal and ventral hypodermal cells, respectively. (C) Mid-focal plane of a 300-min embryo. The myosin probe is detected in the bodywall muscle cells giving a different pattern from that of the myotactin probe (compare to B). (D) Lateral view of a 320-min embryo and (E) lateral view of a comma stage embryo (390 min). Anti-sense myotactin probe is detected at the dorsal (dorsal hypodermal cells) and ventral (ventral hypodermal cells) edges of the embryo. (F) Lateral view of a 320-min embryo. The myosin probe is detected in bodywall muscle cells. The positive cells are away from the dorsal and ventral edges of the embryo. Embryos hybridized with either myosin or myotactin sense probes showed no staining.
Similarly, the MH46 protein is not detected in bodywall muscle (although it appears to colocalize with muscle specific antigens during some stages of development) and the myotactin message could not be detected in muscle cells. In situ hybridizations were done using a myosin heavy chain probe to mark the position of the muscle cells and to show that the muscle cells are accessible to DNA probes under the conditions used. When myosin message is first detected, the positive cells are positioned midway between the dorsal and ventral surfaces, away from the lateral edges of the embryo (Fig. 6 C), indicating these cells are not those detected by the myotactin probe at a comparable developmental stage (compare Fig. 6B and Fig. C). Later in development, the myotactin probe is detected in hypodermal cells at the dorsal and ventral edges of the embryo (Fig. 6D and Fig. E; arrows). At this stage, myosin is detected in cells positioned more interiorly (Fig. 6 F). We found no evidence that myotactin is expressed in bodywall muscle cells at any developmental stage.
The second line of evidence that myotactin encodes the MH46 protein is that both the MH46 epitope and an MH46-negative mutant (st456; see below) map to the same 30 kb of the genome. In wild-type embryos, MH46 staining is seen along the length of the embryo adjacent to the muscle quadrants (Fig. 7A and Fig. B). In contrast, homozygous st456 mutants do not show MH46 staining, although the embryos are positive for myosin and for MH27 (Fig. 7C and Fig. D), an antibody that recognizes the boundaries between hypodermal cells 22. To confirm the st456 mutation maps to the region of the genome encoding the myotactin gene, we used a 30-kb fragment of the C. briggsae genome which encodes a C. briggsae homologue (95% identical to C. elegans myotactin at the amino acid level) to rescue the st456 phenotype. All aspects of the phenotype described below are rescued by this fragment of DNA. A second allele of the gene, s2764, was isolated (Baillie, D., personal communication) as described by Stewart et al. 49, which also fails to stain with MH46 and fails to complement the allele isolated in this study. The gene has been designated let-805 (Baillie, D., personal communication).
Figure 7.
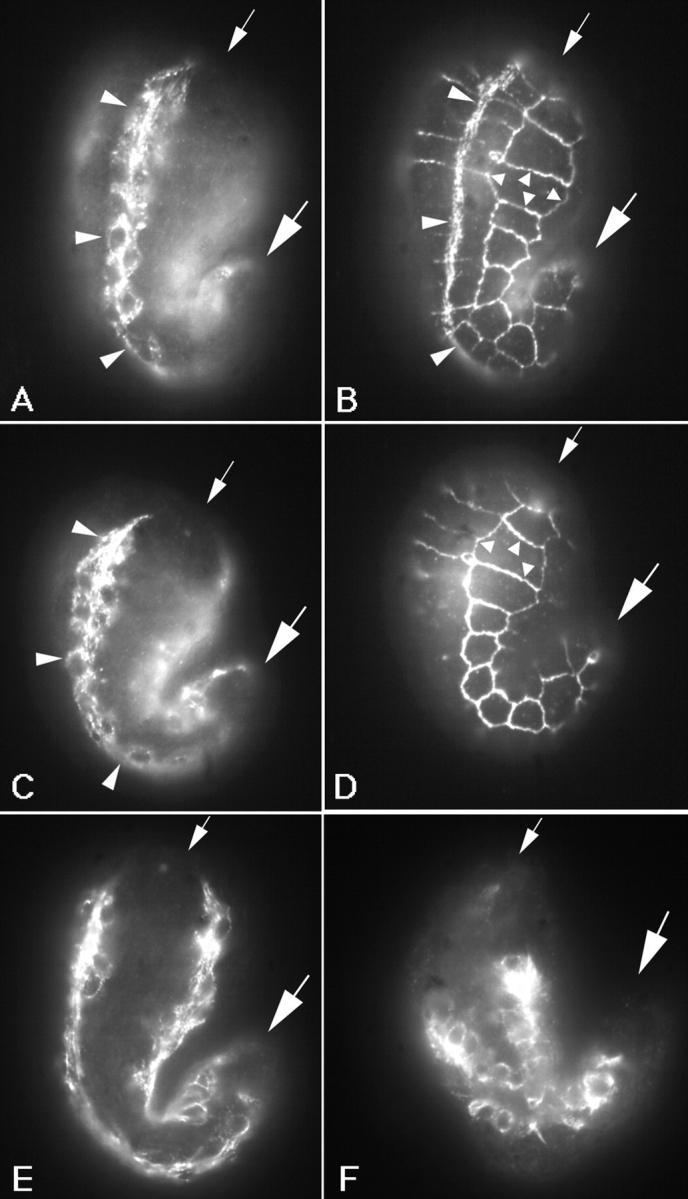
Muscle cells detach in let-805(st456) mutant embryos. Immunofluorescence micrographs of embryos fixed and labeled with monoclonal antibodies MH46 and MH27 (B and D), or a polyclonal antibody against myosin (A, C, E, and F). MH27 recognizes the boundaries between hypodermal cells and is used to visualize the outline of the embryos. The staining pattern appears as a grid on the surface of the embryo (B and D, small arrowheads). Dorsal is to the left in each case, and large and small arrows mark the posterior and anterior ends of the embryos, respectively. (A–D) Lateral views of a wild-type (A and B) or a let-805(st456) homozygous (C and D) embryo at the 1.5-fold stage. Myosin positive cells (A and C) of one dorsal quadrant (large arrowheads) are seen. In both cases the muscle quadrant extends from the anterior to the posterior end of the embryo (the posterior of the quadrant in A is out of the focal plane). Myotactin is localized in the hypodermis adjacent to muscle cells in the wild-type embryo (B, large arrowheads). No myotactin staining is detected in the mutant embryo (D). (E and F) Lateral view of let-805(st456) homozygotes stained for myosin. Note the muscle quadrants do not extend to the anterior or posterior of either embryo. C, E, and F are images of increasingly older animals showing an increase in severity of the muscle cell detachment over time.
Phenotypic Analysis of Myotactin Mutants
The phenotype of let-805(st456) and let-805(s2764) homozygotes (described below) is likely to be the loss-of-function phenotype of the let-805 gene. Both mutations are fully recessive, but more importantly, embryos with a similar phenotype can be produced by a technique referred to as RNA-mediated interference 20 29 41. RNA-mediated interference involves injection of wild-type hermaphrodites with double-stranded RNA homologous to a gene of interest, and results in progeny with the loss-of-function phenotype for that gene (see 41 and references therein). Wild-type hermaphrodites injected with double-stranded RNA homologous to pC-cc406 or pC-cc307, neither of which encodes the MH46 epitope, produced some progeny that were negative for MH46 suggesting myotactin was not made. Furthermore, these MH46-negative embryos were indistinguishable from st456 homozygotes with respect to all the characteristics described below. These data suggest st456 is a loss-of-function allele.
Time-lapse video microscopy shows myotactin mutant embryos have movement and elongation defects. Wild-type embryos begin to elongate at ∼300 min after the first cleavage and elongate three- to fourfold by the time of hatching at 800 min. At the 1.75-fold stage bodywall muscle contractions begin and are first seen as twitches but soon the embryo begins to roll vigorously within the eggshell 51. Like wild-type embryos, myotactin mutants also begin to twitch at about the 1.75-fold stage, but unlike wild-type embryos, the mutants never roll within the eggshell. The mutants fail to elongate beyond the twofold stage, and die before hatching. Since many, if not all mutations that result in failure of embryos to roll also result in arrest of elongation at the twofold stage 5 28 42 54 56 59, the elongation defect associated with myotactin mutations is probably a secondary consequence of the motility defect.
The abnormal movement associated with the mutant myotactin phenotype apparently results from defects in muscle–cell adhesion. In the homozygous mutants, before contractions begin, bodywall muscle quadrants extend from the anterior to the posterior of the embryo (Fig. 7 C), and the sarcomeric structure of the muscle appears wild-type. However, at about the time when muscle contractions begin (1.75-fold stage) the muscle quadrants in the mutants no longer extend the length of the embryo, indicating that the muscle cells have detached (Fig. 7 E). As embryogenesis continues, the detachment worsens until muscle cells are seen only in the mid-section of the embryo and the normal structure of the contractile apparatus is lost (Fig. 7 F). Without connection of the muscle to the exoskeleton, movement is severely impaired.
To ascertain more precisely where the attachment between the muscle cells and the hypodermis is disrupted in the mutants, we examined the localization of other components of muscle attachment structures. At the time in development when the muscle cells detach, fibrous organelle–associated intermediate filaments (Fig. 8 B; see below) and the MH5 protein (an immunologically defined protein that localizes close to fibrous organelles [22 and data not shown]), appear to localize as in wild-type (compare Fig. 8A and Fig. B). By contrast, perlecan, a basement membrane heparin sulfate proteoglycan synthesized by bodywall muscle 42, remains associated with the muscle cells upon their detachment in myotactin mutants (data not shown). These data suggest the detachment is associated with a break at the interface between hypodermal cells and the basement membrane.
Since myotactin in later stages of embryogenesis localizes near fibrous organelles, we examined the effect of the myotactin mutations on the localization of intermediate filaments (Fig. 8) as well as on the localization of the MH5 protein (data not shown). During early developmental stages, the localization of both components appears wild-type in the mutants (compare Fig. 8A and Fig. B). Both are concentrated in regions of the hypodermis adjacent to muscle, and both appear to be organizing into the banded distribution observed for these proteins in older embryos. However, in mutant embryos some time after elongation arrest and muscle cell detachment, both intermediate filaments and the MH5 protein become dispersed throughout the dorsal and ventral hypodermis (Fig. 8C and Fig. D). The staining seen with either MH4 (Fig. 8 D) or MH5 (data not shown) extends all the way to the boundaries between dorsal or ventral hypodermis, and seam cells. The precise correlation of this developmental stage with a wild-type stage is made difficult by the elongation arrest (elongation is a ready marker of developmental events in wild-type embryos). Nonetheless, given the timing of events after elongation arrest, this is likely equivalent to the wild-type threefold stage where MH4- and MH5-dependent staining is restricted to regions of the hypodermis adjacent to muscle (see Fig. 8 A). It is also around this time that the MH46 staining pattern reflects that of the fibrous organelle intermediate filaments.
Although after muscle cell detachment intermediate filaments and the MH5 protein do not remain restricted to the regions of the hypodermis previously contacted by muscle, they do appear to maintain their association with the cuticular annuli since the staining pattern seen is regularly spaced, circumferentially oriented bands (Fig. 8C and Fig. D). As discussed earlier, each fibrous organelle contains two membrane plaques, one associated with the hypodermal membrane adjacent to the muscle basement membrane and one associated with the membrane adjacent to the cuticle. The resolution of the light microscope is not sufficient to distinguish between the two sets, and therefore it is not clear if both sets, or only the set adjacent to the cuticle, is present and/or delocalized in the mutants.
Role of Muscle Cells in Fibrous Organelle Assembly
To understand more about how the muscle acts to establish or maintain the correct spatial relationship between the contractile apparatus and the fibrous organelles, we looked at the localization of fibrous organelle intermediate filaments and myotactin in embryos lacking groups of muscle cells. We previously suggested that the initial recruitment of these proteins to regions of the hypodermis adjacent to muscle was initiated by contact between muscle and dorsal or ventral hypodermis 30. To test this idea, we ablated muscle cell precursors in 28-cell embryos, allowed the embryos to develop for some time, and then fixed and stained the embryos with MH4 (to localize intermediate filaments) or MH46 (to localize myotactin). We ablated either the MS.ap and MS.pp cells, or the C.ap cell, resulting in the loss of 18 anterior or 16 posterior bodywall muscle cells, respectively. To assess the success of the ablations, the embryos were stained for myosin (Fig. 9D and Fig. F). Myotactin protein was absent in regions of the hypodermis that normally contact the missing muscle cells, but was present in regions of the hypodermis adjacent to the remaining muscle cells (Fig. 9C and Fig. E). Where present, myotactin appears organized in the obliquely striated wild-type pattern (Fig. 9 E). The same phenomenon is seen when ablated embryos are stained for intermediate filament subunits (data not shown). That is, intermediate filaments in the ablated embryos are only in regions of the hypodermis contacting muscle, and appear to be organized as in wild-type embryos. These results are consistent with the hypothesis that intermediate filament proteins, myotactin, and possibly other hypodermal proteins organize in response to a signal produced by bodywall muscle cells.
Figure 9.
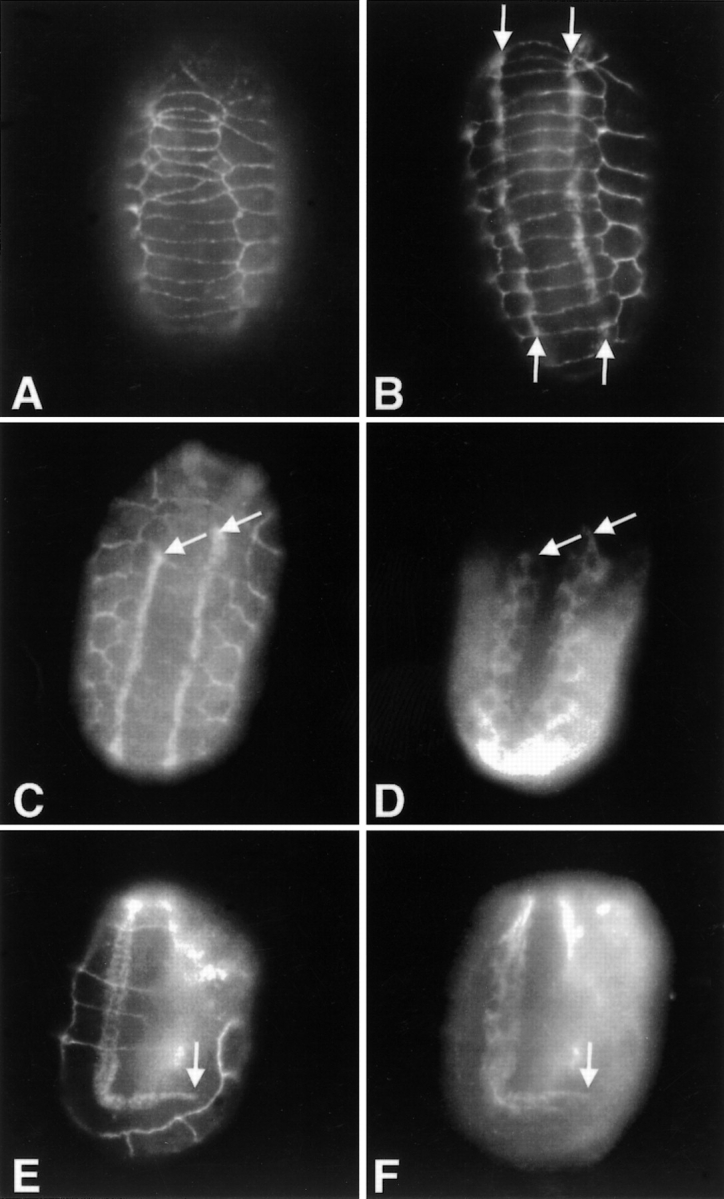
Myotactin organizes in response to muscle cells. (A) 310-min wild-type embryo stained with MH27 to illustrate the MH27 staining pattern. (B) An ∼1.25-fold (∼400 min) wild-type embryo stained with MH27 and MH46. At this stage the MH46 staining extends all the way to the anterior (top) and posterior (not seen in this focal plane) of the embryo. (C and D) Embryo ablated for MS.ap and MS.pp at the 28 cell stage, allowed to develop to the 1.25-fold stage, and then fixed and stained for myosin (D) and with MH46 and MH27 (C). (E and F) Embryo ablated for C.ap and allowed to develop to about the twofold stage and then fixed and stained for myosin (F) and with MH46 and MH27 (E). (B) Arrows indicate the portion of the staining due to MH46. (C–F) Arrows mark the position where the truncated muscle quadrants end, and show the corresponding gap in the organization of myotactin where hypodermal cells have failed to contact muscle.
We do not believe the lack of fibrous organelle staining in ablated regions of the embryo is due to a lack of expression of intermediate filament proteins and myotactin. In wild-type embryos, both proteins are expressed before contact is made between muscle and dorsal or ventral hypodermis 30, making it unlikely that muscle cells are required for proper expression of these proteins. Instead, we think the results of the ablation experiments suggest muscle cells are required for recruitment to and/or stabilization of these proteins in this region of the hypodermis.
Discussion
Myotactin is a novel transmembrane protein. The sequences of the DNA fragments diagrammed in Fig. 3 potentially encode two alternative transmembrane domains (encoded by exons 19A and B). The putative extracellular domain consists mainly of at least 32 FNIII repeats and can be expressed in multiple forms due to differential splicing. Two alternate cytoplasmic domains are also encoded by the gene. The larger predicted cytoplasmic domain (encoded by exons 19B–27) is 519 amino acids in length, and includes an additional 173 amino acids (encoded by exons 24 and 25) not found in the shorter form, 18% of which are serines. A search of the nonredundant protein database using BLASTX (2; search date 8 April 1999) identifies many FNIII repeat–containing proteins, but all of these contain other known extracellular repeat motifs. Furthermore, the only database entries that show any significant homology to the putative cytoplasmic domain are proteins rich in serine, the presumed orthologue from C. briggsae (see Results) and an est entry from Onchocerca volvulus, another nematode.
Analysis of myotactin mutants shows myotactin is required for muscle–cell adhesion (Fig. 7). Myotactin may help mediate the attachment by forming part of a link between specific muscle substructures, such as dense bodies or M-lines, and the hypodermal membrane. During some embryonic stages the distribution of myotactin, a hypodermal transmembrane protein, mirrors that of the forming contractile apparatus 30. This raises the possibility that a specific muscle substructure may be linked, in part through myotactin, to specific sites in the hypodermal membrane. Myotactin, which is anchored in the hypodermal membrane, has the potential to span the basement membrane and interact with a muscle membrane protein. Based on crystallographic data, the length of a FNIII repeat is ∼35Å 33. The 32 predicted FNIII repeats of the extracellular domain of myotactin could extend over 110 nm, or long enough to span the basement membrane, which has been estimated from electron micrographs to be only ∼20 nm wide 58.
If myotactin does indeed interact with the muscle membrane, it is of interest to identify the muscle-associated protein to which it binds. Although the distribution of integrin and perlecan make them likely candidates to interact with myotactin, we do not believe this is the case. Integrin (encoded by the pat-3 gene; reference 25, 59) is a transmembrane receptor positioned at the base of each dense body and M-line 21, and perlecan is a basement membrane proteoglycan concentrated at these same sites 21. However, in integrin and perlecan mutants, myotactin is associated with muscle cells, suggesting myotactin–muscle interactions are not dependent solely on either integrin or perlecan 30. Although myotactin is not organized in oblique striations in integrin and perlecan mutants 30, we think this reflects the loss of muscle structure in the two mutants rather than the loss of a myotactin-binding protein. Thus, early myotactin organization depends on muscle organization.
The initial association between muscle and fibrous organelles is evident when muscle cells migrate onto dorsal or ventral hypodermis and hypodermal proteins organize in response to the contact. During wild-type development, upon contact between muscle and dorsal or ventral hypodermis, intermediate filaments and myotactin accumulate in the region of the hypodermis adjacent to muscle (see Fig. 1 B). Embryos missing certain groups of muscle cells, due to ablation of one or more muscle precursor cells, fail to accumulate these proteins adjacent to the region the missing muscle cells should occupy. This suggests a signal from the muscle cells is required to recruit certain proteins, including some fibrous organelle–associated proteins, to regions of the hypodermis adjacent to muscle.
Myotactin is not involved in receiving the signal from muscle that establishes the association between muscle and fibrous organelles. In myotactin mutants, fibrous organelle–associated intermediate filament proteins and the MH5 protein, both accumulate in regions of the hypodermis adjacent to muscle upon contact between muscle, and dorsal or ventral hypodermis. Only later in development, after the time contraction normally begins, do the hypodermal proteins in the mutants become delocalized and no longer restricted to the regions overlying muscle. The fibrous organelles do however remain associated with the cuticular annuli.
Myotactin May Be Required to Maintain the Association between Muscle and Fibrous Organelles
Fibrous organelles become delocalized in myotactin mutants. In embryos homozygous for the st456 mutation, the localization of intermediate filaments and the MH5 protein appears wild-type until late in embryogenesis when both are seen all through the dorsal and ventral hypodermis. We believe the distribution of intermediate filaments and the MH5 protein in myotactin mutants reflects the distribution of fibrous organelles for two reasons. First, although unlike in wild-type embryos the two proteins are not restricted adjacent to muscle, they do codistribute with one another, and both remain associated with cuticular annuli (see Fig. 8) as in wild-type embryos. Second, the distribution of the two proteins appears wild-type through the embryonic stages when the fibrous organelles form. We suggest that in the myotactin mutants, the fibrous organelles at least partly assemble as in wild-type, but in the absence of myotactin may be allowed to move within the membrane. Movement of another membrane-associated, intermediate filament anchoring structure, the vertebrate hemidesmosome, has been shown by treating 804G cells with cytochalasin D to disrupt the actin cytoskeleton 40. Hemidesmosomes were located subjacent to the nucleus before cytochalasin treatment, but in as little as 30 min after addition of the drug, hemidesmosomes were found at the cell periphery in cell projections. These peripheral hemidesmosomes were associated with cytoplasmic intermediate filaments and with the transmembrane hemidesmosome-associated protein BP180. The speed of the effect suggests relocation of hemidesmosomes is due to movement of a complex of proteins rather than to disassembly and reassembly of the structures 40.
One possibility is that muscle cell detachment is responsible for the delocalization of fibrous organelles in myotactin mutants. However, observations of fibrous organelle–associated proteins in contractile mutants argue this is probably not the case. Examination of myosin A and tropomyosin mutants, where muscle cells cannot contract and appear to remain attached to the hypodermis at the anterior and posterior of the embryo, revealed that fibrous organelles also become delocalized (Hresko, M., and R.H. Waterston, unpublished results). Delocalization of fibrous organelles is clear even in the regions of the hypodermis where the muscle cells appear to remain attached. Interestingly, in both myotactin and contractile mutants, delocalization of fibrous organelles occurs during what might be equivalent to the wild-type threefold stage. This is the stage when the distribution of myotactin first reflects that of fibrous organelles. Perhaps the localization of myotactin to the hypodermal membrane at or near fibrous organelles is important for restricting fibrous organelles at this stage. The delocalization observed in the myotactin mutants would therefore occur because myotactin is absent.
An alternative possibility is that, in the myotactin mutants, the delocalization of fibrous organelles occurs because only the membrane plaques adjacent to the cuticle are present. As discussed earlier, each fibrous organelle consists of two membrane plaques, one at the membrane adjacent to muscle and one at the membrane adjacent to the cuticle. Possibly, in the absence of myotactin, the plaques adjacent to the muscle either do not assemble, or assemble but then disassemble. Due to the absence of the plaques adjacent to muscle, the ones adjacent to the cuticle are not restricted to regions of the hypodermis adjacent to muscle and become delocalized. Since in the myotactin mutants fibrous organelle proteins initially concentrate in regions of the hypodermis adjacent to muscle, and in fact fibrous organelles appear to assemble in these regions of the hypodermis, we think it is unlikely that the plaques adjacent to muscle do not form. Therefore, it seems more likely that myotactin helps in maintaining the correct spatial relationship between muscle and fibrous organelles either by maintaining the integrity of the fibrous organelle structures and preventing their disassembly, or by restricting the movement of fibrous organelles within the hypodermal membrane.
Therefore, we favor a model in which myotactin is essential to maintain the correct spatial relationship between the contractile apparatus of the bodywall muscle and the fibrous organelles of the hypodermis. In this model, during wild-type development, myotactin is synthesized and secreted by the dorsal and ventral hypodermal cells, but remains attached to the hypodermis through a transmembrane domain. As muscle cells migrate and contact the dorsal and ventral hypodermis, the extracellular domain of myotactin extends and interacts with a muscle-associated protein. The interaction between muscle cells and the hypodermal membrane, made through myotactin, is required to prevent detachment of muscle cells when contractions first begin. At some time during development, the distribution of myotactin becomes correlated with the position of fibrous organelles. Myotactin, due to its association with muscle and close proximity to fibrous organelles, would help maintain the correct spatial relationship between the muscle contractile apparatus and the fibrous organelles. This might serve to strengthen the connection between the hypodermis and the muscle.
The resolving power of the light microscope does not allow us to determine if myotactin is a fibrous organelle component. However, if myotactin does interact with fibrous organelles, it may do so only later in development (during the threefold stage), suggesting the association is regulated in some way. Based on the sequence presented in this paper, there are at least two ways in which this regulation might occur. First, regulation might occur through the use of different isoforms. The gene encodes eight differentially used exons which allow for myotactin isoforms that differ in the extracellular, the transmembrane, and the cytoplasmic domains. The sequence encoded by these exons might be important for positioning myotactin near fibrous organelles. The second mechanism by which an association between myotactin and fibrous organelles might be regulated is through phosphorylation. The carboxyl end of the putative cytoplasmic domain is enriched in serine residues. Phosphorylation of these regions might alter the function of the myotactin molecule.
Interestingly, a vertebrate transmembrane hemidesmosome-associated protein, BP180 27 34, that shares no sequence similarity to myotactin, may serve a role similar to that of myotactin. A human patient with GABEB (generalized atrophic benign epidermolysis bullosa) has been described who has a different mutation in each allele of his/her BP180 gene 38. In this patient, hemidesmosomes form as do the anchoring filaments that secure the hemidesmosomes to the connective tissue below. However, the lamina lucida, the junction between the epithelium and the dermis, is widened as if the strength of the connection between the hemidesmosomes and the anchoring filaments was not sufficient to keep the epithelium attached 38. This result suggests BP180, like myotactin, may be responsible for strengthening the connection between the membrane at or near a cell adhesion structure, and the basement membrane. While it is known that β4α6 integrin forms a link between hemidesmosomes and the basement membrane 16 24 46 53, the existence of mutations in transmembrane proteins (BP180 or myotactin) other than integrin which result in detachment at epidermal-basement membrane junctions suggests there are multiple links between cell adhesion structures and the basement membrane or dermis. Furthermore, in mice lacking β4 16 53 or α6-integrin 24, there are some regions of the skin where the basal epithelial cells have not detached from the dermis, again suggesting β4α6 integrin–independent links exist between the epithelium and the basement membrane. Whether BP180 and myotactin perform analogous functions of anchoring cell adhesion complexes in specific regions of the membrane awaits further study as does elucidation of mechanisms by which these types of proteins might function.
Acknowledgments
The authors are grateful to Ross Francis for isolating and characterizing the antibodies, including MH46, used in this study; Drs. Marco Marra, Stephanie Chissoe, and the C. elegans Genome Sequencing Center in St. Louis for the sequence of the C. elegans let-805 gene and the C. briggsae homologue; Dr. David Baillie for sharing the let-805(s2764) mutant; Dr. Sean Eddy for help in identifying and aligning the fibronectin III repeats; Dr. Guy Benian for supplying the Northern blot. We thank Drs. Susan Craig, Jim Skeath, and Pam Hoppe for critical evaluation of this manuscript.
This research was supported by the National Institutes of Health and M.C. Hresko was supported in part by a fellowship from the Muscular Dystrophy Foundation.
Footnotes
1.used in this paper: FNIII, fibronectin type III; HMM, hidden Markov models
References
- Albertson D.G., Thomson J.N. The pharynx of Caenorhabditis elegans . Phil. Trans. R. Soc. Lond. Ser. B. Biol. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLASTa new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Horvitz H.R. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell. 1987;51:1071–1078. doi: 10.1016/0092-8674(87)90593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead R.J., Waterston R.H. The basal component of the nematode dense body is vinculin. J. Biol. Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Barstead R.J., Waterston R.H. Vinculin is essential for muscle function in the nematode. J. Cell Biol. 1991;114:715–724. doi: 10.1083/jcb.114.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnik E., Osborn M., Weber K. Intermediate filaments in muscle and epithelial cells of nematodes. J. Cell Biol. 1986;102:2033–2041. doi: 10.1083/jcb.102.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian G.M., L'Hernault S.W., Morris M.E. Additional sequence complexity in the muscle gene, unc-22, and its encoded protein twitchin, of Caenorhabditis elegans . Genetics. 1993;134:1097–1104. doi: 10.1093/genetics/134.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., Steward K. RNA processing and gene structure. In: Riddle D.L., Blumenthal T., Meyer B.J., Priess J.R., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 117–145. [PubMed] [Google Scholar]
- Bodenteich A., Chissoe S., Wang Y.F., Roe B.A. Shotgun cloning as the strategy of choice to generate templates for high-throughput dideoxy sequencing. In: Venter J.C., editor. Automated DNA Sequencing and Analysis Techniques. Academic Press; London: 1993. pp. 42–50. [Google Scholar]
- Bowe M.A., Fallon J.R. The role of agrin in synapse formation. Annu. Rev. Neurosci. 1995;18:443–462. doi: 10.1146/annurev.ne.18.030195.002303. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson A., Sulston J., Brenner S., Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans . Proc. Natl. Acad. Sci. USA. 1986;83:7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear S., Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T.M., Bowen D.C., Valenzuela D.M., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- Doolittle R.F. Of URFs and ORFsA Primer on How to Analyze Derived Amino Acid Sequences. University Science Books; Mill Valley, CA: 1986. [Google Scholar]
- Dowling J., Yu Q., Fuchs E. β4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R., Mitchison F., Durbin R. Mosimum discrimination hidden Markov models of sequence consensus. J. Comput. Biol. 1995;2:9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Hidden Markov models. Cold Spring Harbor Symp. Quant. Biol. 1996;6:361–365. [Google Scholar]
- Ey P.L., Ashman L.K. The use of alkaline phosphatase-conjugated anti-immunoglobulin with immunoblots for determining the specificity of monoclonal antibodies to protein mixtures. Methods Enzymol. 1986;121:497–509. doi: 10.1016/0076-6879(86)21050-2. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francis G.R., Waterston R.H. Muscle organization in Caenorhabditis eleganslocalization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis G.R., Waterston R.H. Muscle cell attachment in Caenorhabditis elegans . J. Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam M., Noakes P.G., Moscoso L., Rupp F., Scheller R.H., Merles J.P., Sanes J.R. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- Georges-Labouesse E., Messaddeq N., Yehia G., Dadalbert L., Dierich A., LeMeur M. Absence of integrin α6 leads to epidermolysis bullosa and neonatal death in mice. Nat. Genet. 1996;13:370–373. doi: 10.1038/ng0796-370. [DOI] [PubMed] [Google Scholar]
- Gettner S.N., Kenyon C., Reichardt L.F. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans . J. Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson T., Hillier L. A trace display and editing program for data from fluorescence based sequencing machines. Nucleic Acid Res. 1991;19:6481–6483. doi: 10.1093/nar/19.23.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidice G.J., Emery D.J., Diaz L.A. Cloning and primary structural analysis of the bullous pemphigoid autoantigen BP180. J. Invest. Dermatol. 1992;99:243–250. doi: 10.1111/1523-1747.ep12616580. [DOI] [PubMed] [Google Scholar]
- Guo X., Johnson J.J., Kramer J.M. Embryonic lethality caused by mutations in basement membrane collagen of C. elegans . Nature. 1991;349:707–709. doi: 10.1038/349707a0. [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K.J. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative ser-thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hresko M.C., Williams B.D., Waterston R.H. Assembly of bodywall muscle and muscle cell attachment structures in Caenorhabditis elegans . J. Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., McCarter J., Francis R., Schedl T. emo-1, a Caenorhabditis elegans Sec61p g homologue, is required for oocyte development and ovulation. J. Cell Biol. 1996;134:699–714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Brown M., Main I.S., Sjolander K., Haussler D. Hidden Markov models in computational biologyapplications to protein modeling. J. Mol. Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- Leahy D.J., Aukhil I., Erickson H.P. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Li K., Tamai K., Tan E.M.L., Uitto J. Cloning of type XVII collagen. J. Biol. Chem. 1993;268:8825–8834. [PubMed] [Google Scholar]
- Lye J.R., Wilson R.K., Waterston R.H. Genomic structure of a cytoplasmic dynein heavy chain gene from the nematode Caenorhabditis elegans . Cell. Motil. Cytoskel. 1995;32:26–36. doi: 10.1002/cm.970320104. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J. Molecular CloningA Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1982. [Google Scholar]
- McCombie W.R., Adams M.D., Kelly J.M., Fitzgerald M.G., Utterback T.R., Kahn M., Dubnick M., Kerlavage A.R., Venter J.C., Fields C. Caenorhabditis elegans expressed sequence tags identify gene families and potential disease gene homologues. Nat. Genet. 1992;1:124–131. doi: 10.1038/ng0592-124. [DOI] [PubMed] [Google Scholar]
- McGrath J.A., Gatalica B., Christiano A.M., Li K., Owaribe K., McMillan J.R., Eady R.A.J., Uitto J. Mutations in the 180 kd bullous pemphigoid antigen (BPAG2), a benign epidermolysis bullosa. Nat. Genet. 1995;11:83–86. doi: 10.1038/ng0995-83. [DOI] [PubMed] [Google Scholar]
- Mello C., Krammer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegansextrachromosomal maintenance and integration of transforming sequences. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddelle K.S., Hopkinson S.B., Jones J.C.R. Hemidesmosomes in the epithelial cell line 804Gtheir fate during wound closure, mitosis and drug induced reorganization of the cytoskeleton. J. Cell Sci. 1992;103:475–490. doi: 10.1242/jcs.103.2.475. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Downs W.D., Lin R., Wittman C., Bei Y., Cha Y., Ali M., Priess J.R., Mello C. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rogalski T.M., Williams B.D., Mullen G.P., Moerman D.G. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparin sulfate proteoglycan. Genes Dev. 1993;7:1471–1484. doi: 10.1101/gad.7.8.1471. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–77. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Fire A. Whole-mount in situ hybridization for the detection of RNA in Caenorhabditis elegans embryos. Methods Cell Biol. 1995;48:323–337. doi: 10.1016/s0091-679x(08)61394-1. [DOI] [PubMed] [Google Scholar]
- Sonnenberg A., Calafat J., Janssen H., Daams H., van der Raaij-Helmer L.M.H., Falcioni R., Kennel S.J., Aplin J.D., Baker J., Loizidou M., Garrod D. Integrin α6/β4 complex is located in hemidesmosomes, suggesting a major role in epidermal cell-basement membrane adhesion. J. Cell Biol. 1991;113:907–917. doi: 10.1083/jcb.113.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., Eddy S.R., Durbin R. Pfama comprehensive database of protein families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. Structure and function of intercellular junctions. Int. Rev. Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- Stewart H.I., O'Neil N.J., Janke D.L., Franz N.W., Chamberlin H., Howell A.M., Gilchrist E.J., Ha T.T., Kuervers L.M., Vatcher G. Lethal mutations defining 112 complementation groups in a 4.5 sequenced region of Caenorhabditis elegans chromosome III. Mol. Gen. Genetics. 1998;260:280–288. doi: 10.1007/pl00013816. [DOI] [PubMed] [Google Scholar]
- Strome S. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans . J. Cell Biol. 1986;103:2241–2252. doi: 10.1083/jcb.103.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans . Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J. Methods. In: Wood W.B., editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1988. pp. 587–606. [Google Scholar]
- van der Neut R., Krimpenfort P., Calafat J., Niessen C.M., Sonnenberg A. Epithelial detachment due to absence of hemidesmosomes in integrin β4 null mice. Nat. Genet. 1996;13:366–369. doi: 10.1038/ng0796-366. [DOI] [PubMed] [Google Scholar]
- Venolia L., Waterston R.H. The unc-45 gene of Caenorhabditis elegans is an essential gene with maternal expression. Genetics. 1990;126:345–353. doi: 10.1093/genetics/126.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H. Muscle. In: Wood W.B., editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1988. pp. 281–335. [Google Scholar]
- Waterston R.H. The minor myosin heavy chain, mhc A, of Caenorhabditis elegans is necessary for the initiation of thick filament assembly. EMBO (Eur. Mol. Biol. Organ.) J. 1989;8:3429–3436. doi: 10.1002/j.1460-2075.1989.tb08507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werle E., Schneider C., Renner M., Volker M., Fiehn W. Convenient, single-step, one-tube purification of PCR products for direct sequencing. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. The anatomy. In: Wood W.B., editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1988. pp. 81–122. [Google Scholar]
- Williams B.D., Waterston R.H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 1993;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]