Abstract
Transcription sites are detected by labeling nascent transcripts with BrUTP in permeabilized 3T3 mouse fibroblasts followed by laser scanning confocal microscopy. Inhibition and enzyme digestion studies confirm that the labeled sites are from RNA transcripts and that RNA polymerase I (RP I) and II (RP II) are responsible for nucleolar and extranucleolar transcription, respectively. An average of 2,000 sites are detected per nucleus with over 90% in the extranucleolar compartment where they are arranged in clusters and three-dimensional networklike arrays. The number of transcription sites, their three-dimensional organization and arrangement into functional zones (Wei et al. 1998) is strikingly maintained after extraction for nuclear matrix. Significant levels of total RP II mediated transcription sites (45%) were associated with splicing factor–rich nuclear speckles even though the speckles occupied <10% of the total extranucleolar space. Moreover, the vast majority of nuclear speckles (>90%) had moderate to high levels of associated transcription activity. Transcription sites were found along the periphery as well as inside the speckles themselves. These spatial relations were confirmed in optical sections through individual speckles and after in vivo labeling of nascent transcripts. Our results demonstrate that nuclear speckles and their surrounding regions are major sites of RP II-mediated transcription in the cell nucleus, and support the view that both speckle- and nonspeckle-associated regions of the nucleus contain sites for the coordination of transcription and splicing processes.
Keywords: transcription sites, nuclear matrix, nuclear speckles, splicing factors, laser scanning confocal microscopy
There is an emerging view that the genome and its associated functional domains are dynamically linked in an overall nuclear architecture (Berezney et al. 1995; Jackson and Cook 1995; Nickerson et al. 1995; van Driel et al. 1995). For example, DNA is replicated precisely once in each cell cycle (Laskey et al. 1989) in discrete domains of the cell nucleus (Nakayasu and Berezney 1989; Berezney 1991; O'Keefe et al. 1992; Hassan et al., 1993; Ma et al. 1998) and splicing factors are organized in 20–50 speckled structures with additional diffuse distribution in the regions outside of the nuclear speckles (Spector 1993). Both replication sites (Nakayasu and Berezney 1989) and splicing factor–rich nuclear speckles (Spector et al. 1983; Blencowe et al. 1994) are strikingly maintained after extraction for nuclear matrix.
Identifying sites of transcription in the cell nucleus is an important step for elucidating the relationships of nuclear architecture and genomic function. While RNA polymerase I (RP I)1–mediated transcription of rRNA genes has been definitively localized to the nucleolus (for review see Smetana and Bush, 1974), defining the sites of RNA polymerase II (RP II)–mediated transcription of pre-mRNA has been a daunting task. Using tritiated uridine for in vivo labeling, electron microscopic autoradiography demonstrated that newly synthesized extranucleolar RNA is preferentially located in the perichromatin and euchromatin regions of the mammalian nucleus (Bouteille et al. 1974; Fakan and Puvion 1980; Fakan 1994). Equating the localization of newly transcribed RNA with the actual sites of transcription, however, can be misleading. The rapid rate of synthesis of most pre-mRNA (≤5 min) and coupling with RNA splicing makes it difficult to distinguish in vivo sites of transcription from downstream sites of RNA processing and intranuclear transport.
Recently, a permeabilized cell model has been developed in coordination with incorporation of BrUTP and a fluorescence detection system to label sites of transcription that avoid some of the problems associated with in vivo labeling approaches. Since this is a transcription readout system, the rate of transcription is relatively low and no transport of newly labeled RNA either within or from nucleus to cytoplasm has been detected (Jackson et al. 1993; Wansink et al. 1993). Instead, discrete punctate sites of labeled RNA are observed under the fluorescence microscope in the nucleolar and extranucleolar regions that become more intense with increasing pulse time. Using this nascent RNA labeling method, hundreds of transcription sites were visualized (Jackson et al. 1993; Wansink et al. 1993).
Both ultrastructural (Beyer and Osheim 1988) and biochemical (LeMaire and Thummel 1990; Bauren and Wieslander 1994; Wuarin and Schibler 1994) evidence support the view that RNA splicing of pre-mRNA in eukaryotes occurs cotranscriptionally. Recent findings that hyperphosphorylated RNA polymerase II (RP IIo) is associated with in vitro assembled spliceosomes and interacts via its carboxy-terminal domain (CTD) with serine arginine (SR)–rich splicing factors and several 3′ end cleavage factors, provide further indications that transcription and splicing are coordinated processes (Mortillaro et al. 1996; Vincent et al. 1996; Yuryev et al. 1996; Kim et al. 1997; McCracken et al. 1997; Pennisi 1997; Patturajian et al. 1998).
The possible spatial basis for the coordination of transcription and splicing is suggested by the colocalization of both total nuclear poly(A)+ RNA (Carter et al. 1991, Carter et al. 1993) and RP IIo (Bregman et al. 1995; Mortillaro et al. 1996) with the splicing factor–rich nuclear speckles. The finding that poly(A)+ RNA is stably associated with interchromatin granules (the presumptive electron microscopic equivalent of nuclear speckles) under conditions of transcriptional inhibition led Huang et al. 1994 to propose that the speckle-associated poly(A)+ RNA is not related to mRNA. This is contrary to the results of Visa et al. 1993 who demonstrated decreases in poly(A)+ RNA–associated with interchromatin granules after actinomycin D treatment of cells. Moreover, experiments using specific gene probes demonstrate a close association of certain actively transcribing genes and/or their RNA transcripts with nuclear speckles (Huang and Spector 1991; Xing et al. 1993, Xing et al. 1995; Zhang et al. 1994; Bridge et al. 1996; Smith et al. 1999). Some gene transcripts are juxtaposed and others show internal overlap with the speckles (Xing et al. 1993, Xing et al. 1995). While not all actively transcribed genes are found closely associated, no instance has been reported of an inactive gene associated with nuclear speckles (Xing et al. 1995; Smith et al. 1999). Moreover, splicing occurs along the RNA transcript tracks with a defined polarity and is likely cotranscriptional (Xing et al. 1993, Xing et al. 1995). These findings are consistent with Wang et al. 1991 who demonstrated that pre-mRNA microinjected into the nuclei of cells preferentially associates with nuclear speckles, and that this targeting requires RNA capable of undergoing normal splicing. Spector and colleagues have further implicated nuclear speckles and their surrounding regions for the dynamic recruitment of splicing factors at sites of transcription (Jimenez-Garcia and Spector 1993; O'Keefe et al. 1994; Huang and Spector 1996a,Huang and Spector 1996b; Misteli et al. 1997).
Despite this progress, our understanding of the spatial relationships of transcriptional and RNA splicing sites in the cell nucleus remains elusive. Splicing factors and associated small nuclear ribonucleoprotein (snRNP) components are distributed in the cell nucleus in nuclear speckles with additional diffuse distribution throughout the extranucleolar compartment (Spector et al. 1991, Spector et al. 1993), whereas RP II-mediated transcription is detected in up to several thousand punctated sites (Iborra et al. 1996; Fay et al. 1997). Some studies have indicated an association of transcription sites with the diffusely staining regions of splicing factors, but the degree of association with the nuclear speckles remains unclear (Jackson et al. 1993; Wansink et al. 1993; Fay et al. 1997; Neugebauer and Roth 1997). Indeed, a popular view, based on electron microscopic autoradiography and the possible correlation of speckles with interchromatin granular clusters, is that nuclear speckles are predominantly storage sites for splicing factors rather than being active sites of coordinate transcription/splicing (Huang and Spector 1996a,Huang and Spector 1996b).
In this study, we have used the BrUTP labeling approach in permeabilized mammalian cells in conjunction with laser scanning confocal microscopy and computer image analysis to investigate the three-dimensional organization of transcription sites in the mammalian cell nucleus and their spatial relationship to nuclear speckles. An average of 2,000 individual transcription sites were measured per nucleus that are distributed as clusters in both the nucleolar (RP I-mediated) and extranucleolar (RP II-mediated) compartments. The extranucleolar transcription sites contained over 90% of the total sites and were further organized three-dimensionally into networklike arrays. A similar three-dimensional organization of transcription sites was observed after extraction for nuclear matrix or when transcription was performed on the nuclear matrix itself. Moreover, a portion of the total extranucleolar, RP II-mediated transcription sites detected in the cell nucleus are closely associated with the nuclear speckles. While the peripheral rim regions of the speckles are particularly enriched, we also detect significant levels of transcription sites inside the speckles themselves.
Materials and Methods
Labeling of DNA Replication and Transcription Sites and Immunofluorescence Procedures
DNA replication and transcription sites were labeled based on previous procedures (Nakayasu and Berezney 1989; Jackson et al. 1993; Wansink et al. 1993) and those briefly reported by Wei et al. 1998. Mouse 3T3 fibroblast cells are grown on coverslips in DME or MEM supplied with 10% FCS for 24–48 h. Cells are washed with ice-cold TBS buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2) and further washed with glycerol buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, 5 mM MgCl2, 0.5 mM EGTA, 0.5 mM PMSF) for 10 min on ice. Washed cells were permeabilized with 0.025% Triton X-100 in glycerol buffer (with 25 U/ml of RNasin; Promega Corp.) on ice for 3 min and immediately incubated at room temperature for 30 min with nucleic acid synthesis buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 150 mM NaCl, 25% glycerol, 0.5 mM PMSF, 25 U/ml of RNasin, 1.8 mM ATP) supplemented with 0.5 mM CTP, GTP, and BrUTP (Sigma Chemical Co.) for labeling transcription sites (nascent RNA), 0.1 mM dATP, dCTP, dGTP, and 25 μM digoxigenin-11-dUTP (Boehringer Mannheim) for labeling DNA replication sites (nascent DNA), or both for simultaneously labeling transcription and replication sites.
After incorporation, the cells were fixed with 3% freshly made paraformaldehyde in PBS on ice for 5 min, washed with ice-cold TBS-Tween buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.2 mM MgCl2, 0.2% Tween 20), blocked with 5% goat serum and incubated with rat anti-BrU antibodies (IgG, SeraLab) followed by biotin-conjugated goat anti–rat IgG (1:50; Jackson ImmunoResearch Laboratories, Inc.) and Texas red–conjugated streptavidin (1:100; GIBCO BRL) to detect transcription sites. Replication sites were detected with FITC-conjugated sheep antidigoxigenin Fab fragments (1:10; Boehringer Mannheim). All incubations were performed at room temperature for 30 min. Alternatively, nascent RNA was labeled in vivo, by pulsing 3T3 cells for 2 min with 30 μM BrU (Sigma Chemical Co.). The biotin-strepavidin enhancement system resulted in greatly improved sensitivity for detected individual transcription sites compared with a standard secondary antibody approach and was absolutely essential for detecting transcription sites after 2-min in vivo pulses of BrU.
Nuclear speckles were decorated with the Y12 mAb to Sm, which recognizes common core proteins of snRNPs involved in RNA processing (Lerner et al. 1981; Zieve and Sauterer 1990), followed by goat anti–mouse IgG (1:20–50; Jackson ImmunoResearch Laboratories, Inc.) conjugated to FITC or Texas red. Coiled bodies and the nuclear lamina were decorated with rabbit polyclonal anticoilin and antilamin B antibodies, respectively, and detected with FITC- or Texas red–conjugated goat anti–rabbit IgG (1:20–50; Jackson ImmunoResearch Laboratories, Inc.). Coverslips were mounted on slides in Slow-Fade (Molecular Probes, Inc.) and stored at −20°C for microscopic examination.
Laser Scanning Confocal Microscopy, Image Processing, and Quantitation
Optical sections (0.5 or 0.3 μm where indicated) were collected with a confocal microscope (MRC-1024; Bio-Rad) equipped with a Nikon Optiphot 2 microscope, a Nikon 60×, 1.4 NA objective, and a krypton argon laser to simultaneously excite FITC and Texas red at 488 and 568 nm, respectively. Emissions were collected with filters 522-DF32 for FITC and HQ598-40 for Texas red. The pixel intensity distribution was checked before image collection and adjusted so that the pixel intensities were all below saturation.
The total number of transcription sites are calculated by a new and highly improved segmentation program, presented in detail elsewhere (Samarabandu et al. 1995). In brief, by using two-dimensional image segmentation on the individual slices to obtain transcription site contours and combining this high level two-dimensional data using a modified three-dimensional connected component labeling algorithm with weak connectivity in the Z direction, we were able to reconstruct the network of three-dimensional transcription sites. Instead of using the equivalent of traditional two-dimensional 8-connectivity in three dimensions, we used a metric that evaluated the amount of overlap between successive sections to determine whether the two contours belong to the same site. After determining the three-dimensional boundary of transcription sites, the center of gravity of each site was calculated by averaging the (x, y, z) coordinates of all voxels that belong to the site (Samarabandu et al. 1995).
Segmentation of the nuclear speckles, nucleoli, the total nuclei, and measurement of the size and intensity of segregated areas was performed in IPLab (Signal Analytics Corp.). Distribution of transcription sites between the nuclear speckles and other nuclear regions was determined by direct measurement.
Fluorescence signal intensities from unprocessed confocal microscopic images were measured in IPLab. In brief, the extranucleolar regions, nucleolar regions, and the internal region of the nuclear speckles were manually selected and the mean intensities were calculated after multiple scans through the areas under analysis. The nucleolar signal was selected as background and subtracted from the signal emitting from the speckle and extranucleolar regions.
Results
Visualization of RP I and II Transcription Sites in Permeabilized Mammalian Cells
Previous studies indicated that incorporation of BrUTP into newly transcribed RNA in permeabilized cells is essentially a transcription readout system with little or no subsequent RNA processing (Jackson et al. 1993; Wansink et al. 1993; Wei et al. 1998; see introduction and Materials and Methods), and that the labeled RNA is visualized predominantly if not exclusively at sites of transcription. Both nucleolar and extranucleolar incorporation sites are detected (Fig. 1, a and b). When low levels of α-amanitin (1 μg/ml), which inhibits RP II- but not RP I- or RP III-mediated transcription (Roeder 1976), was included in the synthesis buffer, the entire extranucleolar signal is abolished (Fig. 1c and Fig. d). This demonstrates that all transcription sites visualized in the extranucleolar compartment are mediated by RP II. To confirm that nucleolar transcription is mediated by RP I, cells were treated with low concentration of actinomycin D (0.8 μg/ml), which preferentially inhibits RP I transcription of the ribosomal genes (Perry and Kelley 1970) (Fig. 1e and Fig. f). When higher concentrations of actinomycin D (3.2 μg/ml) were used in the synthesis buffer, transcription activity is completely abolished in the nucleus (Fig. 1g and Fig. h). RNase A greatly reduced transcription (Fig. 1i and Fig. j), whereas DNase I (Fig. 1k and Fig. l) and aphidicolin (Fig. 1m and Fig. n), a DNA synthesis inhibitor (Spadari et al. 1985), had no effect on transcription sites. All transcription sites are confined within the boundary of the nuclear lamina detected with antilamin B antibody (Fig. 2, E–J). Thus, transport of transcripts to the cytoplasm is undetectable, supporting the conclusion that the transcripts are still engaged on the templates in this transcription readout system (Jackson et al. 1993; Wansink et al. 1993).
Figure 1.
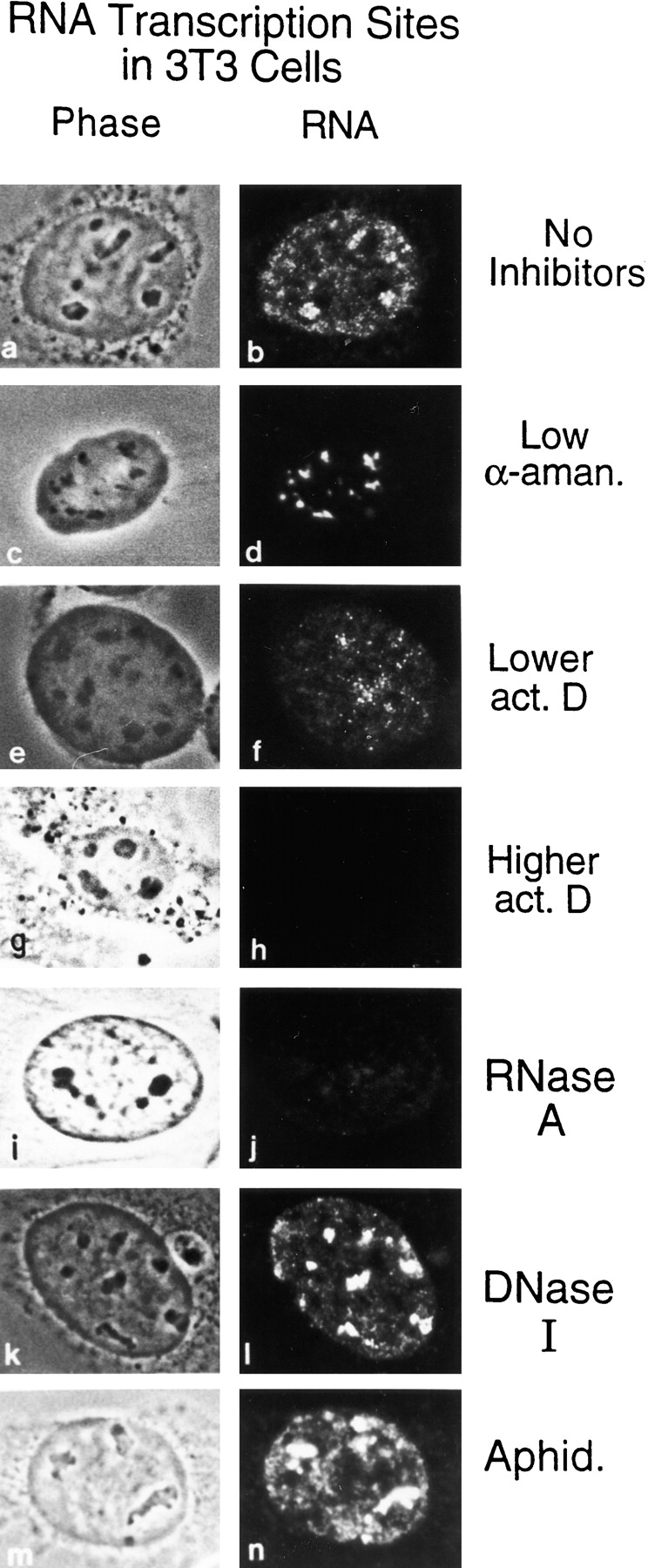
Transcription occurs at discrete sites throughout the cell nucleus. Mouse 3T3 cells were permeabilized with 0.025% Triton X-100, and then incubated with RNA synthesis buffer to label nascent transcripts with BrUTP, which was later immunodetected with anti-BrdU antibody. On the left are phase-contrast images of cells. On the right are conventional epifluorescent images of transcription sites. (a and b) Transcription sites are distributed throughout the cell nucleus. Both extranucleolar and nucleolar transcription sites are detected, with nucleolar transcription sites being more intense and discrete. (c and d) The extranucleolar transcription signal is completely abolished when 1 μg/ml of α-amanitin was included in the synthesis buffer. (e and f) Low concentration of actinomycin D (0.8 μg/ml) preferentially inhibits nucleolar transcription. (g and h) Higher concentration of actinomycin D (3.2 μg/ml) abolished all transcription sites in the cell nucleus. (i and j) Transcription signals were greatly reduced by treatment with RNase A (250 U/ml). (k and l) In contrast, DNase I digestion (50 U/ml) had no visible effects on the transcription sites. (m and n) The lack of effect of the specific DNA replication inhibitor aphidicolin (20 μg/ml) confirmed that the fluorescence labeled sites were not due to DNA synthesis.
Figure 2.
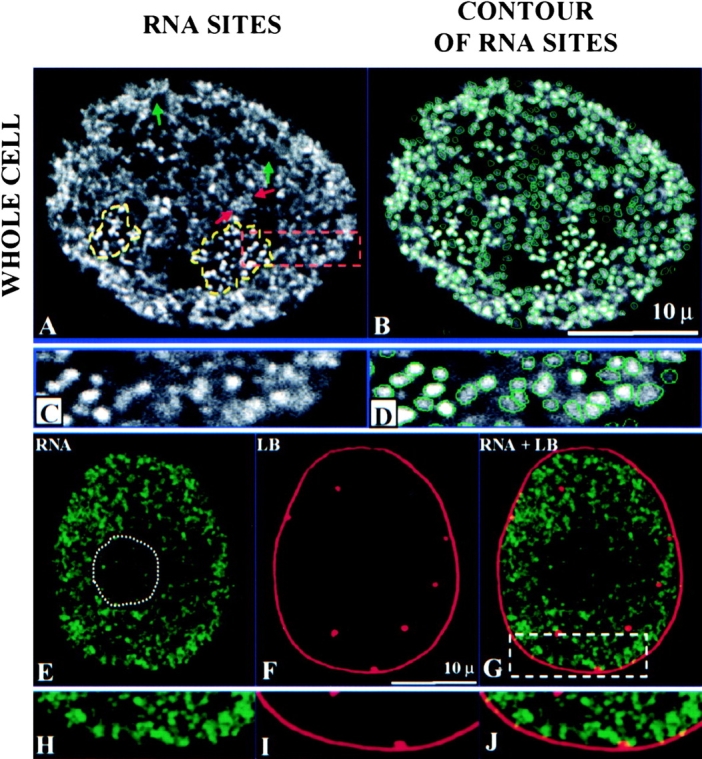
Laser scanning confocal microscopy of transcription sites, their segmentation, and spatial relationship with the nuclear lamina in permeabilized 3T3 cells. (A) Nucleolar transcription sites are encircled (yellow dashed lines). A cluster of transcription sites is indicated by red arrows. Regions with no transcription activity were indicated with green arrows. (B) A segmentation algorithm is applied to the image in A. Every site counted is contoured with a green line. (C) Higher magnification of a portion of A. (D) Higher magnification of a portion of B. (E–G) Double labeling of transcription sites and nuclear lamina. (E) Transcription sites are stained with FITC (green). (F) Nuclear lamina is detected with antilamin B antibodies and Texas red–conjugated secondary antibodies (red). (G) Merged image of transcription sites and nuclear lamina shows that transcription is occurring inside the region confined by nuclear lamina, and transcription sites are not associated with lamin B either on the lamina or with internal lamin B positive spots. (H–J) A portion of the image, indicated with a box in the merged image, is further enlarged. All images are 0.5-μm midplane optical sections.
Transcription Sites Are Organized into Clusters in the Cell Nucleus
Using a laser scanning confocal microscope we find that extranucleolar RP II transcription is organized into granular-like sites with an average x-y diameter of 0.8 ± 0.05 μm. The nucleolar RP I transcription sites (average diameter of 0.7 ± 0.04 μm) are usually more intensely labeled than the extranucleolar RP II sites and are arranged into discrete clusters. The transcription sites are not distributed evenly in the extranucleolar compartment (Fig. 2, A–D). Some regions have more sites and form clusters (active zones), whereas other regions have virtually no transcription sites (inactive zones).
To study the three-dimensional organization of transcription sites in the cell nucleus, we reconstructed a series of optical images three dimensionally and displayed them as stereo pairs. This revealed that the individual sites in the extranucleolar compartment are arranged in a higher order networklike pattern (Fig. 3 A). The individual sites that compose the overall network are readily observed at higher magnification (Fig. 3 B). x, y, z coordinates (centers of gravity) for each transcription site were calculated by a computer program and displayed in three dimensions (Fig. 3 C) with an enlarged portion shown in Fig. 3 D. Consistent with the observation of the original confocal images (Fig. 3A and Fig. B), chainlike arrays and clusters of sites were observed that extended considerable distances in three dimensions. Since each site is assigned a unique x, y, z coordinate, individual transcription sites can be easily identified among the huge population present. This procedure is illustrated by the highlighting of two individual sites (Fig. 3C and Fig. D) and has enormous potential as an approach for identifying active transcription of specific gene sequences by combining fluorescence in situ hybridization (Xing, 1993, 1995) with transcription site labeling.
Figure 3.
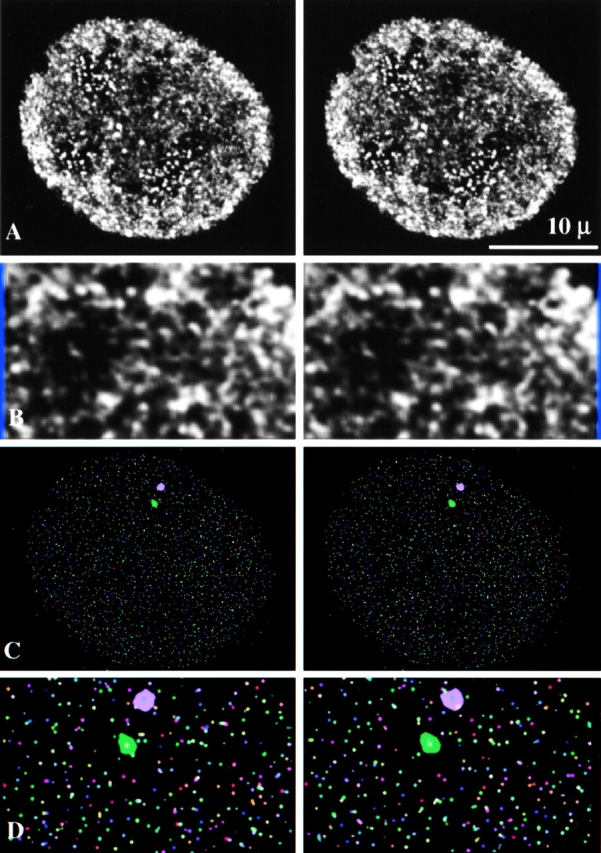
Three-dimensional visualization of transcription sites in permeabilized 3T3 cells. (A) 0.3-μm optical sections were reconstructed to form a three-dimensional image. Transcription sites are clustered in some regions of the cell nucleus, whereas others have no transcription activity. (B) A portion of A is further enlarged. Three-dimensional observations reveal clustering of individual sites into separate higher order domains. These separate higher order domains, in turn, are arranged in a pattern that resembles a networklike organization. (C) The centers of gravity (x, y, z) of transcription sites were determined by an algorithm after segmentation and displayed in three dimensions. Two specific transcription sites were pinpointed by their x, y, z coordinates and highlighted in purple and green. (D) A portion of the image of the centers of gravity is further enlarged.
Association of Transcription with the Nuclear Matrix
After transcription synthesis, the cells were subjected to DNase I digestion and 0.2- or 0.6-M ammonium sulfate (AS) extraction to obtain the nuclear matrix (Belgrader et al. 1991). Transcription sites are maintained on the nuclear matrix with an organization similar to the intact nucleus (Fig. 4A, Fig. C, Fig. E, and Fig. G). In particular, nucleolar sites maintained their high intensity and tight packing arrays, whereas the extranucleolar space was organized into clusters of sites (active zones) separated by relatively empty regions (inactive zones).
Figure 4.
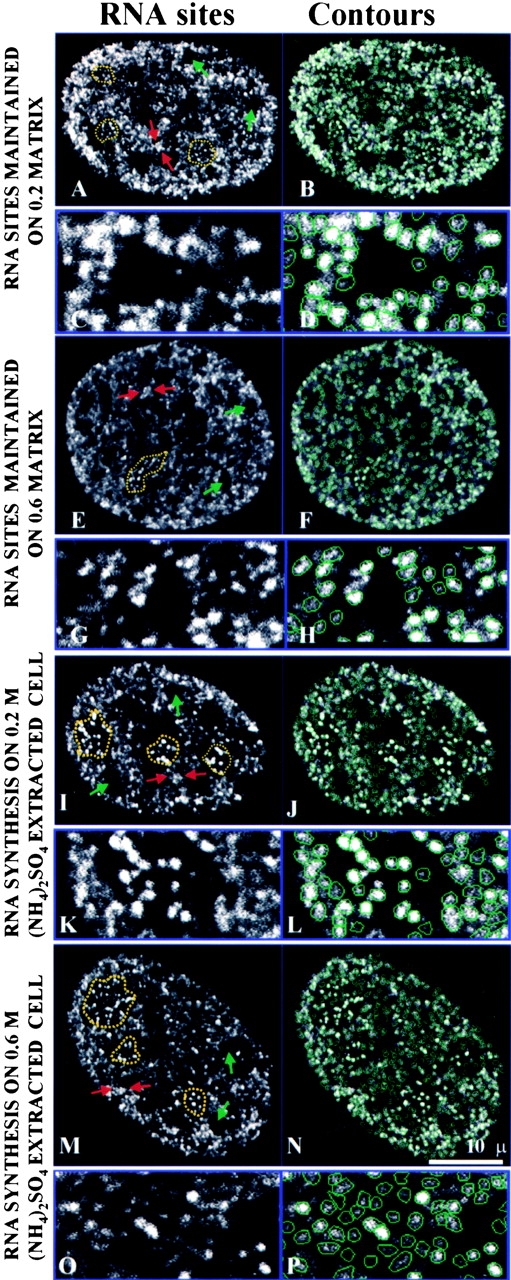
Transcription sites are maintained after preparation for nuclear matrix and the transcription activity is also preserved in salt-extracted 3T3 cells. On the left are the optical midplane images; on the right are the results after the images were subjected to the segmentation algorithm. Yellow dotted lines indicate nucleolar transcription sites. Examples of clustered transcription sites are indicated by red arrows. Nuclear regions with no transcription activity are indicated with green arrows. For all conditions, a portion of the images was further enlarged and displayed underneath. All sites that are counted are contoured with green lines. (A–D) Transcription sites are maintained after DNase I digestion followed by 0.2-M AS (ammonium sulfate) extraction for nuclear matrix. (E–H) Transcription sites are maintained after DNase I digestion followed by 0.6-M AS extraction for nuclear matrix. (I–L) Cells were first extracted with 0.2-M AS, and then subjected to RNA synthesis (M–P). Cells were first extracted with 0.6-M AS, and then subjected to RNA synthesis.
We next determined if active transcription can occur after extraction for nuclear matrix. The cells were not treated with DNase I to maintain the DNA template in an intact state. After 0.2- or 0.6-M AS extraction, the cells were subjected to transcription incorporation followed by immunodetection of Br-RNA. We found that transcription activity is maintained on the extracted cells at levels comparable to permeabilized cells. Moreover, the transcription activity was organized into sites that were indistinguishable to those detected in permeabilized cells, including the characteristic intense nucleolar clusters and the arrangement of the extranucleolar compartment into clusters of sites and other areas devoid of sites (Fig. 4I, Fig. K, Fig. M, and Fig. O).
Examination of the nuclear matrix–associated transcription sites in three dimensions revealed an organization strikingly similar to that of the intact nucleus. Clusters of sites arranged into discontinuous networklike arrays were observed after extraction of cells for nuclear matrix (Fig. 5A and Fig. B) and in transcription assays performed on the nuclear matrix (Fig. 5D and Fig. E). Chain-like arrays of transcription sites were also seen after visualization of the x, y, z coordinates (centers of gravity) of individual sites (Fig. 5C and Fig. F).
Figure 5.
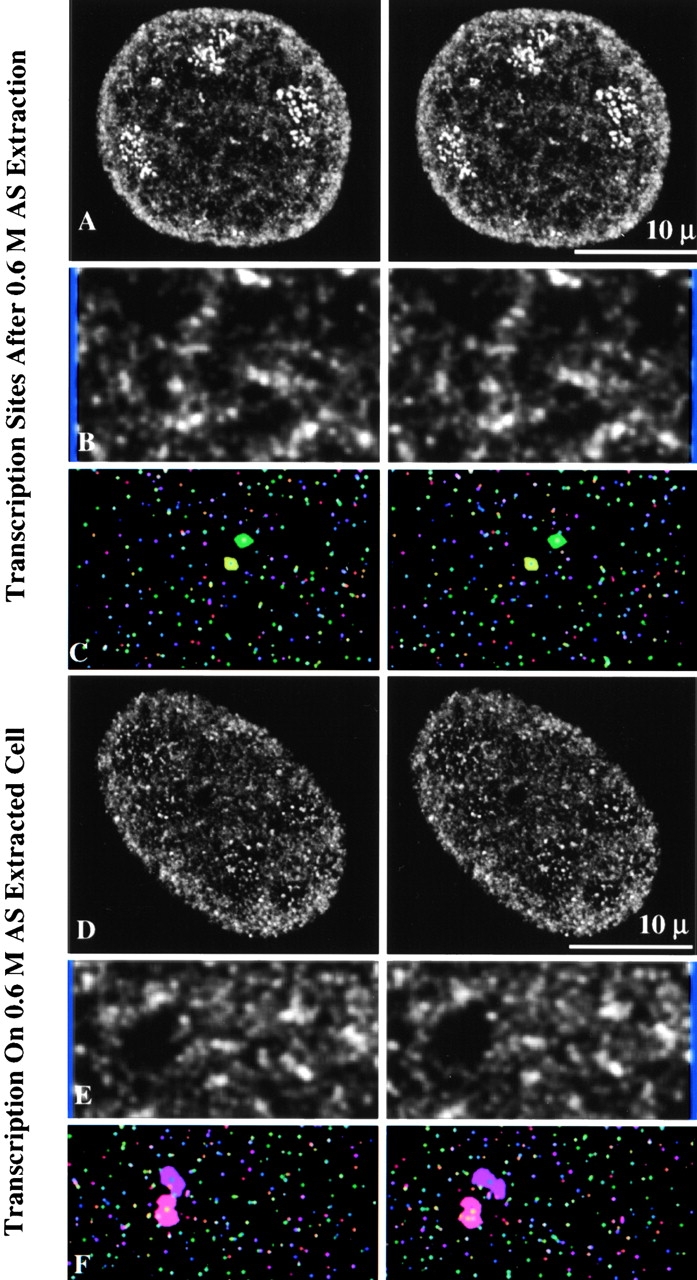
Three-dimensional visualization of transcription sites maintained on nuclear matrix and transcription activity of salt-extracted cells. 0.3-μm optical sections through 3T3 cells were reconstructed to form three-dimensional images. These results demonstrate that not only transcription sites, but also transcription activity is maintained on the nuclear matrix. (A) Transcription sites after 0.6-M AS extraction for nuclear matrix. (B) A portion of A is further enlarged. (C) Three-dimensional image of the centers of gravity of a region of A. (D) Three-dimensional visualization of transcription sites when synthesis was performed on 0.6-M AS extracted cells. (E) A portion of D is further enlarged. (F) Three-dimensional image of the centers of gravity of a region of D.
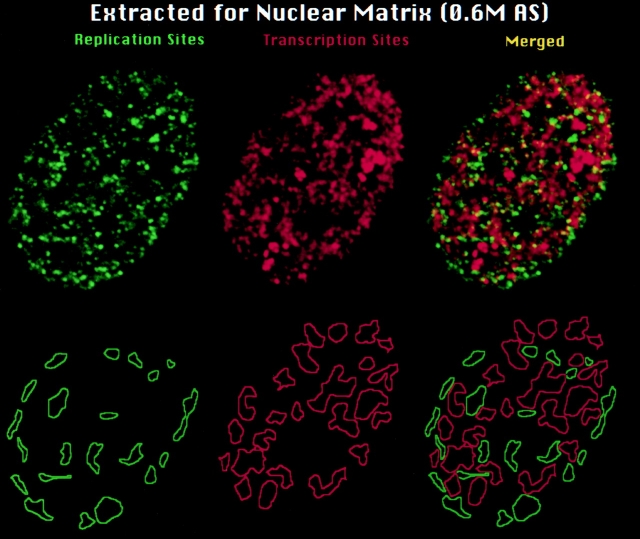
It was recently reported that replication and transcription sites simultaneously labeled during early S phase are arranged predominantly into spatially separate higher order clusters or nuclear zones (Wei et al. 1998). As illustrated in Fig. 6, this higher order arrangement is well maintained after nuclear matrix extraction. 75.2% ± 4.6 SEM (n = 10) of the total extranucleolar area containing replication and transcription sites was occupied by separate zones of replication or transcription in cells extracted for nuclear matrix compared with 79.2% ± 5.2 SEM (n = 12) for unextracted cells (Wei et al. 1998).
Figure 6.
Maintenance of DNA replication and transcription sites and their organization into higher order zones on the nuclear matrix. Replication and transcription sites were simultaneously labeled in permeabilized 3T3 cells as described in Materials and Methods, and then extracted for nuclear matrix using 0.6 M AS (Belgrader et al. 1991). Contours were drawn around regions in the nuclear matrix that contained exclusively replication (green contours lines) or transcription sites (red contour lines), respectively. Top row are the original optical sections. Bottom row are the derived contour maps. As previously demonstrated for permeabilized cells (Wei et al. 1998), the great majority of replication and transcription sites that are maintained after extraction for nuclear matrix are arranged in clusters of spatially separate zones.
There Are ∼2,000 Transcription Sites in the Cell Nucleus
A spot detection segmentation program was developed to quantify the transcription sites in the cell nucleus (Samarabandu et al. 1995). Sites that can be detected are contoured in green. The segmentation program was applied to a series of optical sections and sites detected in each section were coordinated to accurately measure the total number of transcription sites. An example of this computer algorithm applied to a typical middle section image is shown in Fig. 2B and Fig. D. A large majority of all detectable transcription sites ranging from very intense to weakly stained sites are segmented with over 90% in the extranucleolar compartment. Of the small portion of sites not segmented by this program (<10% of the total), many represent relatively weak sites positioned between relatively intensely stained ones. The values obtained for the total number of transcription sites, therefore, represent minimal estimations.
The transcription sites associated with the nuclear matrix (Fig. 4D, Fig. H, Fig. J, and Fig. N) had a similar range of intensity from strong to very weak and a similar efficiency of segmentation. From this analysis, we estimate a minimal average of 2,000 transcription sites per nucleus (Table ). The great majority of transcription sites are maintained on the nuclear matrix (Table ) after extraction of cells (1,500–1,900 sites) or transcription labeling on cells first extracted for the nuclear matrix (1,800–1,900).
Table 1.
Number of Transcription Sites Following Nuclear Matrix Preparation
| Cell preparation | Transcription sites/nucleus |
|---|---|
| Whole cell | 2,011 ± 367 |
| Transcription sites after 0.2-M AS [(NH4)2SO4] extraction | 1,871 ± 248 |
| Transcription sites after 0.6-M AS extraction | 1,475 ± 304 |
| Transcription on 0.2-M AS extracted cells | 1,909 ± 287 |
| Transcription on 0.6-M AS extracted cells | 1,786 ± 360 |
The Nuclear Speckles Have a Severalfold Higher Concentration of Splicing Factors Than Their Surroundings
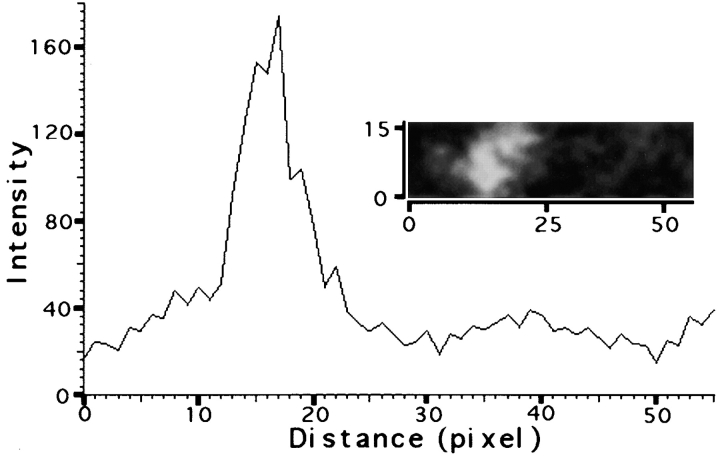
The widely held view that splicing components are highly enriched in nuclear speckles (Carter et al. 1993; Spector 1993) was recently challenged (Fay et al. 1997; Singer and Green 1997). Therefore, we measured the signal intensities of Y12 staining from unprocessed confocal microscopic images in the internal region of over 200 speckles and their surrounding nonspeckle regions in mouse 3T3 cells. An average enrichment of fivefold was found in the speckles compared with the diffuse staining in the extranucleolar regions (Fig. 7). The Y12 staining associated with the speckles is about one third of the total signal measured in the extranucleolar region based on the speckles occupying <10% of the total extranucleolar volume (Table ).
Figure 7.
Quantification of snRNP in the nuclear speckle versus other nuclear regions. snRNP was detected with mAb Y12. A typical nuclear speckle and its surroundings were selected (insert) and the fluorescent intensity was measured across 16 consecutive x-axes through this region in 3T3 cells. The average intensity of these 16 separate determinations is plotted as a function of the pixel distance in the x-plane. The pixel distances across the x- and y-axes are labeled on the insert.
Table 2.
Quantitation of Transcription Sites (TS) in the Mouse 3T3 Fibroblast Cell Nucleus
| Nuclear regions | No. of TS per confocal section | Percent distribution | Relative concentration TS/area | |
|---|---|---|---|---|
| Area | TS | |||
| Total | 418 ± 28 | 100 | 100 | 1 |
| Nucleolar | 25 ± 4.0 | 9.7 | 6 | 0.6 |
| Total extranucleolar | 393 ± 25. | 90.3 | 94 | 1 |
| Nonspeckle extranucleolar | 215 ± 25 | 81.3 | 51.4 | 0.6 |
| Speckle | 178 ± 26 | 8.7 | 42.6 | 4.9 |
| Pereipheral speckle | 114 ± 15 | — | 27.3 | — |
| Internal speckle | 64 ± 13 | — | 15.3 | — |
Values represent the average + SEM for midplane optical sections from different nuclear samples. Over 4,000 transcription sites were counted along with 214 speckles.
Transcription, but Not Replication Sites, Are Associated with Splicing Factors in Nuclear Speckles
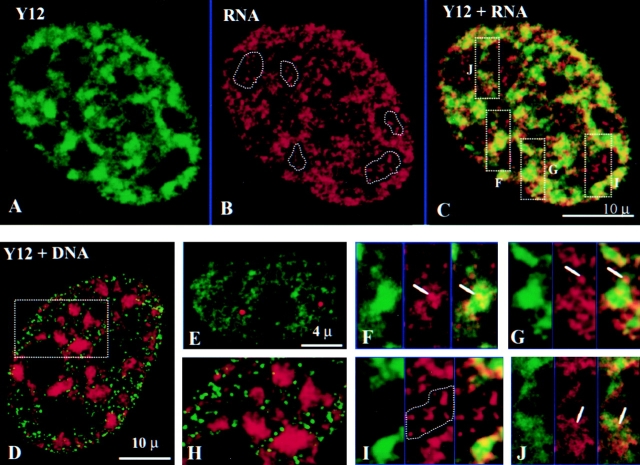
The spatial relationships of RP II transcription sites and nuclear speckles were next examined. RP II transcription sites are associated with splicing factors sites (Fig. 8, A–C) at both speckles (Fig. 8, F–G) and diffusely stained regions (Fig. 8 J). In contrast, RP I transcription sites are not associated with splicing factor sites (Fig. 8 I). Speckle-associated RP II transcription sites are located not only along the periphery (Fig. 8 G, arrows), but also in the interior regions of the speckles (Fig. 8 F, arrows). Early S phase DNA replication sites, which are distributed throughout the cell nucleus and in numbers similar to transcription sites (Nakayasu and Berezney 1989; Ma et al. 1998; Wei et al. 1998), are not associated with nuclear speckles or the more diffusely stained splicing factor sites (Fig. 8D and Fig. H). Overlap between replication and splicing factor sites was also not detected in cells of middle and late S phase (data not shown).
Figure 8.
Transcription but not replication sites are associated with nuclear speckles. (A) Y12 decorates regions where splicing factors are concentrated in nuclear speckles and more diffuse nonspeckled regions (green). (B) Transcription sites scattered throughout the cell nucleus (red). (C) Merger of A and B shows significant association of transcription sites with nuclear speckles (yellow to orange staining). (D) Replication sites (green) are not associated with splicing factors (red). (E) Coiled bodies (red) do not have transcription activity (green). (F, G, I, and J) Enlarged areas as indicated by boxes F, G, I, and J in C, with Y12 on the left, transcription sites in the middle, and merged images on the right. Some transcription sites are in the interior region of the speckles (F, arrows), whereas other sites are located along the periphery (G, arrows) or with the more diffusely stained regions of Y12 (J, arrows). Nucleolar transcription sites are not associated with splicing factors (I, encircled by a dotted line). (H) Enlarged area of D shows that replication sites are present in the regions between splicing factor speckles and do not colocalize with the more diffusely distributed splicing factors.
Since nuclear coiled bodies also contain snRNPs (Bohmann et al. 1995), we determined whether coiled bodies were also associated with nascent transcripts. We found that transcription activity was rarely associated (<10%) with the 1–3 coiled bodies per cell (Fig. 8 E; Jordan et al. 1997).
Transcription in the Internal Regions of Speckles
The resolution of the confocal microscope in the z-axis is two- to threefold less than in the x-y planes (Pawley 1995). Therefore, it is conceivable that peripherally localized transcription sites that happen to locate vertically in the center of the speckles could give the appearance of an internal location in a particular optical section (Fig. 9E and Fig. F). To investigate this possibility, we examined serial optical sections (0.3 μm) through individual speckles and found numerous examples of interior located transcription sites that occurred in the middle sections of the series, but were not present in the top or bottom sections (Fig. 9A and Fig. B, arrows). These serial sections also revealed many transcription sites located along the periphery of the speckles. Whereas the great majority of speckles had both peripheral and internally located transcription sites (Fig. 9A and Fig. B, arrows), a small percentage (<10%) displayed only peripheral ones (Fig. 9C and Fig. D, arrows).
Figure 9.
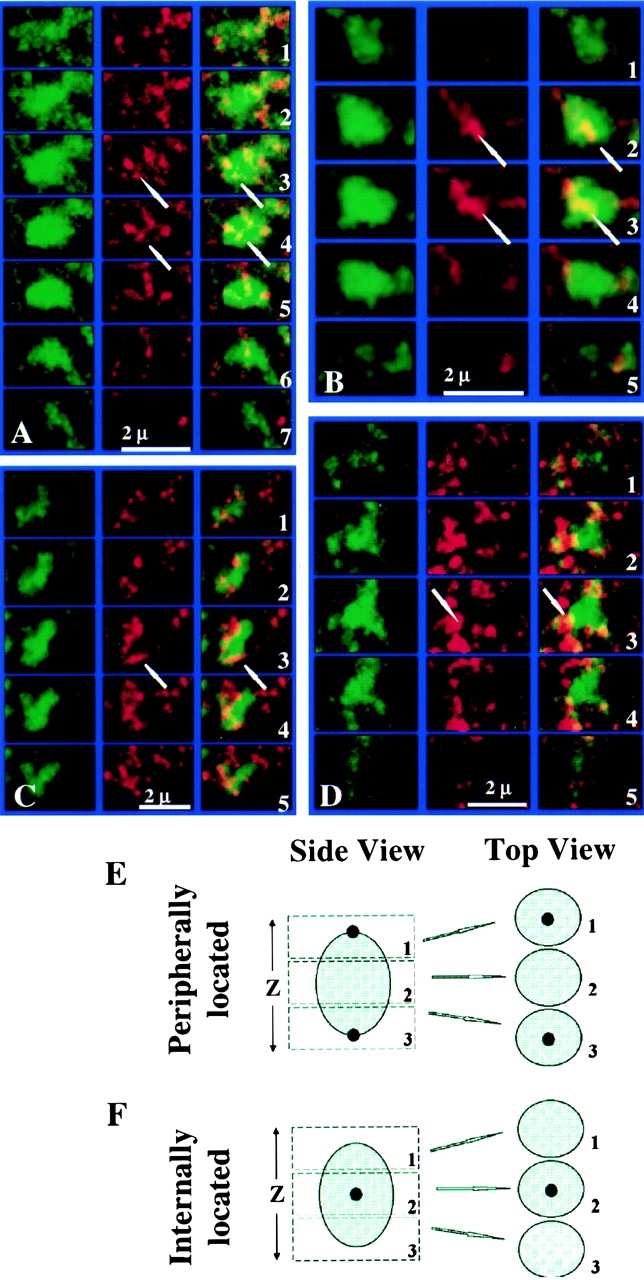
Optical section series through individual nuclear speckles. (A and B) Arrows indicate interior located transcription sites that are present in the middle sections (0.3 μm) of the series, but not in sections above or below. (C and D) Whereas the vast majority of speckles showed both internally and peripherally located transcription sites, a small population (<10%) showed only peripherally associated transcription sites (arrows). Section number for each speckle are labeled on the right. (E and F) Schematic illustration of optical sectioning method to distinguish peripheral from internal locations of transcription sites in the speckle regions. Large shadowed spheres indicate speckles. Small solid dots indicate transcription sites.
Quantitation of Transcription and Splicing Factors in the Nuclear Speckles and Nonspeckle Regions
Quantitative image analysis was performed on the middle sections from 12 randomly selected optical image sets. An average of ∼400 extranucleolar transcription sites was counted per section. 45% of these sites were speckle-associated even though the speckles contained only 9.6% of the total extranucleolar area (Table ). Whereas the nonspeckle extranucleolar regions of the nucleus contained the majority of transcription sites (55%), the speckle regions were significantly enriched in these sites (over 8-fold) compared with nonspeckle extranucleolar regions (Table ). Of the 214 speckles analyzed, the majority of speckle-associated transcription sites (64%) were concentrated along the periphery of speckles either in close juxtaposition or partially overlapping with the speckles themselves (Table ). All speckles examined had associated transcription sites and the great majority (92.2%) had a moderate to high level of associated sites (4–16 sites per speckle). Only a very small percentage of speckles (7.8%) were relatively weakly decorated with transcription sites (<4 sites per speckle).
Transcription Sites Are Associated with Nuclear Speckles In Vivo
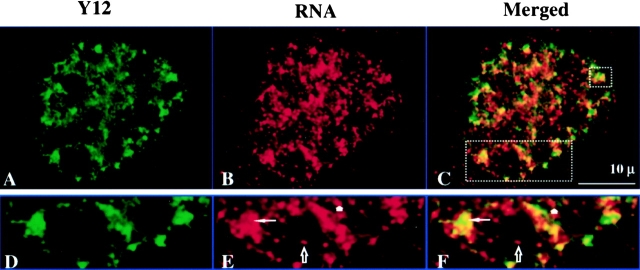
To determine whether transcription sites are also associated with speckles in vivo, 3T3 cells were pulsed with 30 μM BrU for 2 min to label nascent RNA transcripts, and then fixed immediately followed by dual color labeling for splicing factor and transcription sites. As seen in Fig. 10A–C, the double labeling pattern in the cell nucleus is virtually identical to that observed in the permeabilized cell system (Fig. 8, A–C). Transcription sites are associated with both the peripheral and interior regions of the nuclear speckles as well as with the more diffuse Y12 staining regions (Fig. 10, D–F). Nucleolar transcription sites are not associated with the Y12 staining regions (Fig. 10, D–F) as demonstrated in the permeabilized cell system (Fig. 8 I).
Figure 10.
In vivo labeled nascent RNA transcripts are associated with splicing factors. (A) Nuclear speckles are detected with the Y12 antibody (green). (B) Transcription sites are detected by incorporating BrU (2-min pulse) into nascent transcripts in vivo (red). (C) Merged image of A and B. (D–F) Enlarged portions of A–C, respectively. The thin arrow indicates a transcription site inside of a nuclear speckle. Nucleolar transcription sites are not associated with splicing factors (see hollow arrow). Transcription sites that are associated with diffusely distributed splicing factors are indicated with a short arrow.
Discussion
Identifying Transcription Sites in the Cell Nucleus and Models of Gene Organization
We have used BrUTP incorporation to label transcription sites (nascent RNA) in permeabilized cells (see introduction and Materials and Methods). By combining laser scanning confocal microscopy and a new computer segmentation algorithm (Samarabandu et al. 1995), individual sites of relatively small size (x-y diameters of 0.2–1.0 μm) are detected that are closely associated and have a wide range of fluorescence intensity. Our estimation of 2,000 transcription sites per 3T3 cell nucleus is consistent with other recent reports (Iborra et al. 1996; Fay et al. 1997; Jackson et al. 1998).
Since there are ∼30,000 genes expressed in a mammalian cell (Lewin 1975), these results can be explained in several ways. None of these explanations are mutually exclusive. Thus, more than one may be operative. First, the labeling system may only be sensitive enough to detect transcription sites for genes that transcribe at high copy number. The great majority of genes that are known to transcribe only a few copies in the cell may not be detected.
Second, it is unlikely that all genes are being transcribed simultaneously and continuously during the cell cycle. The 2,000 sites that are detected may represent one window of time for the overall transcriptional expression pattern in the cell cycle. While there is little direct data supporting this model, Wei et al. 1998 has recently discovered that nuclear zones of early S replication sites do not contain significant levels of transcription sites and vice versa. Since early S replication sites contain predominantly actively transcribed genes (Goldman et al. 1984; Hatton et al. 1988), these results indicate that at least in S phase there is a higher order control of the qualitative pattern of gene transcription (Berezney and Wei 1998).
Third, the large number of genes may be divided among the more limited number of transcription sites. In this multigene model, each site could be a transcription domain for a cluster of genes containing up to a dozen or more transcriptional units. This model has been favored by Cook and colleagues who envision a transcriptional factory where transcription of multiple genes can be regulated (Jackson et al. 1993; Hughes et al. 1995; Iborra et al. 1996). If correct, it will be of great interest to determine what kinds of genes are organized at individual transcription sites. There are several possibilities that again are not mutually exclusive. Most simply, adjacent genes along the genomic DNA sequence could be clustered together at a transcription site. Since there are growing reports of gene clusters along the genomic DNA (Calza et al. 1984; Brown et al. 1987; Hatton et al. 1988; Ben-Arie et al. 1994; Diaz et al. 1994; LaSalle and Lalande 1995), this could be an effective way of regulating expression among members of the cluster. In contrast, active genes may randomly cluster at newly assembled transcription sites. Finally, genes that produce functionally related products or that are involved in a pathway of regulation may be assembled at the same transcription site and, in turn, may be subjected to a common regulation. Those genes could be far apart along the genomic sequence and even on different chromosomes. In this regard rDNA genes, which are clustered in tandem arrays on the short arms of several chromosome pairs (Wellauer and David, 1973), are packaged together in the nucleolar structure (Henderson et al. 1972). It would be of interest to determine if genes regulated by a common activator are located on the same transcription domain. Future studies combining fluorescence in situ hybridization for specific gene localization with transcription site labeling should provide important insight into these issues.
Nucleolar Transcription Sites
Using RP I antibodies, Scheer and Rose 1984 demonstrated that the presumptive sites of rDNA transcription show a punctate staining pattern over the fibrillar center regions of the nucleolus. While no measurements of these punctate sites were reported, we have estimated a diameter range of 0.5–1.0 μm with an average of ∼0.7 μm (Fig. 1 e; Scheer and Rose 1984). This is in the same size range as the transcription sites we observed over the nucleolar regions. Similar to the number of RP I granular sites per nucleus (95–145; Haaf et al. 1991), we report an average of 110 discrete and intensely stained nucleolar transcription sites per mouse fibroblast nucleus. This is considerably less than that of rRNA genes determined by saturation hybridization (200 rRNA genes in mouse or 300 in human; Schmickel 1973; Long and Dawid, 1980). It was proposed that each RP I site contains one active rRNA gene (Scheer et al. 1984; Haaf et al. 1991) which would indicate that there are ∼115 and 106 active rRNA genes in human and mouse cells, respectively (Haaf et al. 1991).
The lower number of RP I transcription sites compared with the number of rRNA genes can also be explained by a limited clustering of rRNA genes at each transcription site (e.g., 2–3 genes per site). If this is the case, then gene clusters might represent a fundamental feature of transcription sites as units of gene expression. Jackson et al. 1993 have suggested that each RP I transcription site may contain approximately six transcriptional units, but this is based on a much lower estimation for the number of nucleolar transcription sites.
The Higher Order Arrangement of Transcription Sites and Nuclear Architecture
We consistently observe that the transcription sites, rather than being scattered throughout the extranucleolar compartment as previously suggested (Wansink et al. 1993), are arranged in nuclear zones containing clusters of sites and other regions devoid of sites. Three-dimensional reconstruction further reveals networklike arrays of the transcription sites. Contour mapping demonstrates that both transcription and replication sites in early S phase form separate nuclear zones and networklike arrays in three dimensions (Wei et al. 1998; Berezney and Wei 1998).
Since the individual transcription sites are spatially separate, some elements of higher order nuclear architecture are likely involved in maintaining these networklike arrays. Previous reports have suggested that transcription sites are maintained on the nuclear matrix (Wansink et al. 1993; Jackson et al. 1993). Our study is the first detailed analysis of the association of transcription sites with the nuclear matrix. We also observe a remarkable maintenance of RP I and II transcription sites and their higher order arrangement into separate transcription and replication zones and networklike arrays after extraction for nuclear matrix.
Association of Transcription Sites with Nuclear Speckles
The question as to whether nuclear speckles, which are generally believed to be sites for storage of splicing factors, are also sites of active transcription and/or splicing has generated considerable interest in recent years (Lawrence et al. 1993; Rosbash and Singer 1993; Visa et al. 1993; Wansink et al. 1993; Huang et al. 1994; Mattaj 1994; Moen et al. 1995; Raska 1995; Bridge et al. 1996; Clemson and Lawrence 1996; Huang and Spector 1996a,Huang and Spector 1996b; Fay et al. 1997; Neugebauer and Roth 1997; Singer and Green 1997). While studies of individual genes (Huang and Spector 1991; Xing et al. 1993, Xing et al. 1995; Bridge et al. 1996; Smith et al. 1999) and poly(A)+ RNA (Carter et al. 1991, Carter et al. 1993; Visa et al. 1993) lend support to this view (see however, Huang et al. 1994; Zhang et al. 1994), the matter is complicated by the lack of correspondence between the overall pattern of RP II-mediated transcription sites, which are scattered throughout the extranucleolar compartment in hundreds to thousands of sites, and the 20–50 much larger nuclear speckles (Wansink et al. 1993; Fay et al. 1997; Neugebauer and Roth 1997; Singer and Green 1997). Moreover, the lack of tritiated uridine labeling over the interchromatinic granule clusters (Fakan and Puvion 1980), which are believed to represent the electron microscopic equivalent of nuclear speckles (Spector et al. 1991), has led to the view that at least most of the nuclear speckles are storage sites and transcriptionally inactive. Splicing factors are proposed to be recruited from nuclear speckles to other nonspeckled sites for coordinate transcription/splicing (Jimenez-Garcia and Spector 1993; Spector et al. 1993; O'Keefe et al. 1994; Huang and Spector 1996a,Huang and Spector 1996b).
Surprisingly few studies, however, have directly measured the degree to which nascent transcripts are associated with nuclear speckles. While Wansink et al. 1993 is often quoted as providing evidence that transcription sites are not associated with nuclear speckles, merged images or quantitation of the degree of overlap between transcription sites and nuclear speckles were not presented in that study. Direct overlay of the separate channel images shown in Fig. 7 of Wansink et al. 1993, however, suggests significant levels of overlap of transcription sites with nuclear speckles (results not shown). In addition, Dundr and Raska 1993 observed labeling of nascent RNA over the interchromatin granular clusters after BrUTP incorporation in permeabilized cells.
In our study, over 200 individual nuclear speckles were examined in detail by confocal microscopic optical sectioning and quantitative image analysis. We report that significant levels of RP II transcription sites (45% of total) are associated with nuclear speckles and concentrated over eightfold compared with the remaining extranucleolar compartment. The majority of speckle-associated transcription sites are found along the periphery or juxtaposed to the speckles (approximately two thirds) with the remainder located in interior regions of the speckles. Coupled with the concentration of transcription sites in the speckled regions is an average enrichment of approximately fivefold in snRNP splicing factors.
Fay et al. 1997 previously measured the association of transcribed RNA with nuclear speckles. However, that study was limited by the relatively long in vivo pulse periods used to label the nascent RNA. Under those conditions, a significant portion of the labeled RNA would have completed transcription and might not be associated with the transcription sites. Consistent with this interpretation, the distribution of speckle-associated transcription sites in that study decreased from 21 to 10% as the in vivo labeling time increased from 9.5 to 19.5 min (Fay et al. 1997). The readout transcription approach employed in our study avoids this problem (Jackson et al. 1993; Wansink et al. 1993; see also introduction and Methods). Moreover, we observe a similar pattern of transcription sites associated with nuclear speckles after a brief in vivo pulse (2 min).
Fay et al. 1997 also reported an enrichment of only twofold of the splicing factor, SC-35 in nuclear speckles and have questioned the view that speckles are indeed sites of enrichment of splicing factors (Fay et al. 1997; Singer and Green 1997). Our measurements using a similar approach, but on original confocal images without thresholding or image enhancement, revealed a fivefold enrichment for the snRNP splicing factors and are consistent with previous studies suggesting high levels of enrichment of SC-35 and other splicing factors in nuclear speckles (Fu and Maniatis; 1990; Spector et al. 1991; Carter et al. 1993).
We propose that both nuclear speckles and the extraspeckle regions participate in coordinated transcription/RNA processing events. In this regard, Smith et al. 1999 have demonstrated that not all actively transcribed genes and their transcripts show an association with nuclear speckles. Further research is clearly needed to understand the relationship of nuclear speckles to the much smaller and diffuse sites of transcription and RNA processing in the cell nucleus. One simplistic view is that the speckles are sites of massive levels of transcription/RNA processing mediated by the limited number of genes that transcribe high copy numbers. Most genes that transcribe at low copy numbers would then be localized in the hundreds to thousands of additional sites arranged throughout the extranucleolar compartment. However, the situation is more complicated. Smith et al. 1999 recently showed that speckle association of RNA is not restricted to the limited number of highly abundant transcripts. Among four genes that are believed to have moderate levels of transcripts in the cell, two were speckle-associated and two were clearly not. These and previous studies (Xing et al. 1993, Xing et al. 1995) suggest that a significant portion of the total population of different pre-mRNA transcripts are associated with nuclear speckles, whereas another portion is not. This agrees with our finding that ∼45% of total RP II transcription sites are speckle-associated.
Several studies have stressed the dynamic nature of nuclear speckles and their role as recruitment sites for engaged transcription of activated genes (Jimenez-Garcia and Spector 1993; Spector et al. 1993; Huang and Spector 1996a,Huang and Spector 1996b). The dynamic recruitment of splicing factors from nuclear speckles to putative sites of transcription was recently observed in living cells using a GFP-expression system (Misteli et al. 1997). Moreover, massive expression of an intron-containing RNA leads to recruitment of splicing factors at sites of transcription that assemble into nuclear speckle-like structures (Huang and Spector 1996a).
These dynamic properties and studies demonstrating individual gene transcripts closely associated with nuclear speckles have led to the proposal that there are two populations of nuclear speckles in a cell: transcriptionally inactive speckles that serve as storage sites for splicing factors and transcriptionally engaged speckles, where splicing factors are massively recruited to active sites of transcription (Huang and Spector 1996a,Huang and Spector 1996b). Alternatively, it has been proposed that all nuclear speckles might be active in coordinate transcription/RNA splicing processes (Clemson and Lawrence 1996). Our results are most consistent with the latter model. All speckles examined (>4,000) had associated transcription sites. Of the 214 speckles examined in detail, the great majority (>90%) had moderate to high levels of associated transcription sites. The presence of a small population of speckles (<10%) with relatively low levels of transcription sites, however, also lends credence to the dynamic speckle model (Huang and Spector 1996a,Huang and Spector 1996b). Indeed, it is plausible that the population of speckles actively engaged in transcription and splicing may widely fluctuate depending on a variety of cellular conditions. Further research is needed to clarify these possibilities.
While our studies demonstrate that nuclear speckles participate in transcription and are likely sites of coordinated transcription/RNA processing, only limited regions of the speckles likely participate in this activity at any given time. The majority of the RP IIo and splicing factors that are concentrated in the nuclear speckles are not colocalized with the transcription sites. What leads to recruitment of active RP IIo and splicing factors remains to be elucidated, but initial biochemical studies suggest an important role of the CTD of RP II and recently identified CTD-binding proteins of the SR class (Mortillaro et al. 1996; Vincent et al. 1996; Yuryev et al. 1996; Kim et al. 1997; McCracken et al. 1997; Pennisi 1997; Steinmetz 1997; Tanner et al. 1997). One of these CTD-binding proteins, SCAF8, colocalizes with transcription sites (Patturajian et al. 1998) and another, termed matrin CYP, is a novel cyclophilin-containing SR repeat that is highly concentrated in the nuclear speckles (Bourquin et al. 1997; Mortillaro and Berezney 1998).
Association of nuclear speckles (Spector et al. 1983), RP II transcription sites (Jackson et al. 1993; Wansink et al. 1993), RP IIo (Bisotto et al. 1995; Mortillaro et al. 1996; Vincent et al. 1996), transcription factors (van Wijnon et al., 1993; Zeng et al. 1997, Zeng et al. 1998; McNeil et al. 1998), SCAF8 (Patturajian et al. 1998), and matrin CYP (Mortillaro and Berezney 1998) with the nuclear matrix (Berezney and Jeon 1995) provide an in vitro approach for elucidating the molecular associations involved in the coordination of transcription and RNA processing and their architectural integration in the mammalian cell nucleus.
Acknowledgments
Dr. Robert Summers (Confocal Microscopy & 3-D Imaging Facility of School of Medicine and Biomedical Sciences, SUNY/Buffalo) provided valuable assistance in the dual labeling confocal microscopy. We thank Alan J. Siegel (Microscopic Imaging Facility, SUNY/Buffalo) and Jim Stamos (SUNY/Buffalo) for assistance on CCD image analysis and preparation of figures, respectively. The Y12, anticoilin, and antilamin B antibodies were provided by Drs. Benjamin Blencowe (Massachusetts Institute of Technology), Robert Ochs (Scripps Research Institute), and Gunter Blobel (Rockefeller University), respectively.
This work was supported by a grant from National Institutes of Health to R. Berezney (GM-23922) and a SUNY/Buffalo Mark Diamond Graduate Student Research Grant to X. Wei (45F97).
Footnotes
1.used in this paper: AS, ammonium sulfate; CTD, carboxy-terminal domain of RNA polymerase II large subunit; RP I and II, RNA polymerase I and II; RP IIo, hyperphosphorylated RNA polymerase II; snRNP, small nuclear ribonucleoprotein; SR, serine-arginine
Dr. Samarabandu's present address is Life Imaging Systems Inc., 195 Dufferin Avenue, Suite 300, London, ON N6A 1K7, Canada. Dr. Wei's current address is Department of Ophthalmology, Harvard Medical School, 243 Charles Street, Boston, MA 02114.
References
- Bauren G., Wieslander E. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Belgrader, P., A.J. Siegel, and R. Berezney. 1991. A comprehensive study on the isolation and characterization of the HeLa S3 nuclear matrix. 98:281–291. [DOI] [PubMed]
- Ben-Arie N., Lancet D., Taylor C., Khen M., Walker N., Ledbetter D.H., Carrozzo R., Patel K., Sheer D., Lehrach H., North M.A. Olfactory receptor gene cluster on human chromosome17possible duplication of an ancestral receptor repertoire. Hum. Mol. Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- Berezney R. Visualizing DNA replication sites in the cell nucleus. Semin. Cell Biol. 1991;2:103–115. [PubMed] [Google Scholar]
- Berezney R., Jeon K.W. Nuclear MatrixStructural and Functional Organization 1995. Academic Press, Inc; Orlando, FL: pp. 1,047 [Google Scholar]
- Berezney R., Mortillaro M.J., Ma H., Wei X., Samarabandu J. The nuclear matrixa structural milieu for genomic function. Int. Rev. Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Berezney R., Wei X. The new paradigmintegrating genomic function and nuclear architecture. J. Cell. Biochem. 1998;Suppl. 30/31:238–242. [PubMed] [Google Scholar]
- Beyer A., Osheim Y. Splicing site selection, rate of splicing and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Bisotto S., Lauriault P., Duval M., Vincent M. Colocalization of a high molecular mass phosphoprotein of the nuclear matrix (p255) with spliceosomes. J. Cell Sci. 1995;108:1873–1882. doi: 10.1242/jcs.108.5.1873. [DOI] [PubMed] [Google Scholar]
- Blencowe B.J., Nickerson J.A., Issner R., Penman S., Sharp P.A. Association of nuclear matrix antigens with exon-containing splicing complexes. J. Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K., Ferreira J., Santama N., Weis K., Lamond A.I. Molecular analysis of the coiled body. J. Cell Sci. (Suppl.) 1995;19:107–113. doi: 10.1242/jcs.1995.supplement_19.16. [DOI] [PubMed] [Google Scholar]
- Bourquin J.P., Stragljar I., Meier P., Moosmann P., Silke J., Baechi T., Georgiv O., Schaffner W. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;25:2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouteille M., Laval M., Dupuy-Coin A.M. Localization of nuclear functions as revealed by ultrastructural autoradiography and cytochemistry. Vol 1. In: Busch H.P., editor. The Cell Nucleus. Academic Press; NY: 1974. pp. 3–71. [Google Scholar]
- Bregman D.B., Du L., van der Zee S., Warren S.L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge E., Riedel K.-U., Johansson B.-M., Pettersson U. Spliced exons of adenovirus late RNAs colocalize with snRNP in a specific nuclear domain. J. Cell Biol. 1996;135:303–314. doi: 10.1083/jcb.135.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.H., Iqbal M.A., Stuart S., Hatton K.S., Valinsky J., Schildkraut C.L. Rate of replication of the murine immunoglobulin heavy-chain locusevidence that the region is part of a single replicon. Mol. Cell. Biol. 1987;7:450–457. doi: 10.1128/mcb.7.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calza R.E., Eckhardt L.A., DelGiudice T., Schildkraut C.L. Changes in gene position are accompanied by a change in time of replication. Cell. 1984;36:689–696. doi: 10.1016/0092-8674(84)90349-0. [DOI] [PubMed] [Google Scholar]
- Carter K., Taneja K., Lawrence J.B. Discrete nuclear domains of poly(A)+ RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter K.C., Bowman D., Carrington W., Fogarty K., McNeil J.A., Fay F.S., Lawrence J.B. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Clemson C.M., Lawrence J.B. Multifunctional compartments in the nucleusinsights from DNA and RNA localization. J. Cell. Biochem. 1996;62:181–190. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C181::AID-JCB6%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Diaz M.O., Pomykala H., Bohlander S., Maltepe E., Malik K., Brownstein B., Olopade O.I. Structure of the human type-I interferon gene cluster determined from a YAC clone contig. Genomics. 1994;22:540–552. doi: 10.1006/geno.1994.1427. [DOI] [PubMed] [Google Scholar]
- Dundr M., Raska I. Nonisotopic ultrastructural mapping of transcription sites within the nucleolus. Exp. Cell Res. 1993;208:275–281. doi: 10.1006/excr.1993.1247. [DOI] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fakan S., Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int. Rev. Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Fay F.S., Taneja K.L., Shenoy S., Lifshitz L., Singer R. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp. Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- Fu X.-D., Maniatis T. Factor required for mammalian splicesome assembly is localized to discrete regions in the nucleus. Nature. 1990;365:82–85. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist G.P., Gray M.C., Caston L.A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Haaf T., Hayman D.L., Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Exp. Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. [DOI] [PubMed] [Google Scholar]
- Hassan A.B., Cook P.R. Visualization of replication sites in unfixed human cells. J. Cell Sci. 1993;105:541–555. doi: 10.1242/jcs.105.2.541. [DOI] [PubMed] [Google Scholar]
- Hatton K.S., Dhar V., Brown E.H., Iqbal M.A., Stuart S., Didamo V.T., Schildkraut C.L. Replication program of active and inactive multigene families in mammalian cells. Mol. Cell. Biol. 1988;5:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A.S., Warburton D., Atwood K.C. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. USA. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Spector D. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Huang S., Spector D.L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription J. Cell Biol. 133 1996. 719 732a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Spector D.L. Dynamic organization of pre-mRNA splicing factors J. Cell. Biochem 62 1996. 191 197b [DOI] [PubMed] [Google Scholar]
- Huang S., Deerinck T.J., Ellisman M.H., Spector D.L. In vivo analysis of the stability and transport of nuclear poly (A)+ RNA. J. Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.A., Pombo A., McManus J., Hozak P., Jackson D.A., Cook P.R. On the structure of replication and transcription factories. J. Cell Sci. (Suppl. 1995;) 19:59–65. doi: 10.1242/jcs.1995.supplement_19.8. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Pombo A., Jackson D.A., Cook P.R. Active RNA polymerases are localized within discrete transcription factories in human nuclei. J. Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Cook P.R. The structural basis of nuclear function. Int. Rev. Cytol. 1995;162A:125–149. doi: 10.1016/s0074-7696(08)61230-9. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Hassan B., Errington R., Cook P.R. Visualization of focal sites of transcription within human nuclei. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Iborra F.J., Manders E.M.M., Cook P.R. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia L.F., Spector D.L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Jordan P., Cunha C., Carmo-Fonseca M. The cdk7-cyclin H-MAT1 complex associated with TFIIH is localized in coiled bodies. Mol. Biol. Cell. 1997;8:1207–1217. doi: 10.1091/mbc.8.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Du L., Bregman D.B., Warren S.L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle J.M., Lalande M. Domain organization of allele-specific replication within the GABRB3 gene cluster requires a biparental 15q11-13 contribution. Nat. Genet. 1995;9:386–394. doi: 10.1038/ng0495-386. [DOI] [PubMed] [Google Scholar]
- Laskey R.A., Fairman M.P., Blow J.J. S phase of the cell cycle. Science. 1989;246:609–614. doi: 10.1126/science.2683076. [DOI] [PubMed] [Google Scholar]
- Lawrence J.B., Carter K.C., Xing X. Probing functional organization within the nucleusis genomic structure integrated with RNA metabolism? Cold Spring Harbor Symp. Quant. Biol. 1993;58:807–818. doi: 10.1101/sqb.1993.058.01.088. [DOI] [PubMed] [Google Scholar]
- LeMaire M.F., Thummel C.S. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol. Cell. Biol. 1990;10:6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E., Lerner M., Janeway C., Steitz J. Monoclonal antibodies to nucleic acid-containing cellular constituentsprobes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Units of transcription and translationsequence components of heterogeneous nuclear RNA and messenger RNA. Cell. 1975;4:77–93. doi: 10.1016/0092-8674(75)90113-0. [DOI] [PubMed] [Google Scholar]
- Long E.O., David I.B. Repeated genes in eukaryotes. Annu. Rev. Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Ma H., Samarabandu J., Devdhar R.S., Acharya R., Cheng P.-C., Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. RNA processing. Splicing in space. Nature. 1994;372:727–728. doi: 10.1038/372727a0. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S., Marvin W., Bentley D. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- McNeil S., Guo B., Stein J.L., Lian J.B., Bushmeyer S., Seto E., Atchison M.L., Penman S., van Wijnen A.J., Stein G.S. Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situthe C-terminus is a principal determinant for nuclear trafficking. J. Cell. Biochem. 1998;68:500–510. [PubMed] [Google Scholar]
- Misteli T., Caceres J.F., Spector D. The dynamics of a pre-mRNA splicing factors in living cells. Nature. 1997;387:523–526. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Moen P.T., Jr., Smith K.P., Lawrence J.B. Compartmentalization of specific pre-mRNA metabolisman emerging view. Hum. Mol. Genet. 1995;4:1779–1789. doi: 10.1093/hmg/4.suppl_1.1779. [DOI] [PubMed] [Google Scholar]
- Mortillaro M.J., Berezney R. Matrin CYP, an SR-rich cyclophilin that associates with the nuclear matrix and splicing factors. J. Biol. Chem. 1998;273:8183–8192. doi: 10.1074/jbc.273.14.8183. [DOI] [PubMed] [Google Scholar]
- Mortillaro M.J., Blencowe B.J., Wei X., Nakayasu H., Du L., Warren S.L., Sharp P.A., Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu H., Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer K.M., Roth M.B. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes. Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- Nickerson J.A., Blencowe B.J., Penman S. The architectural organization of nuclear metabolism. Int. Rev. Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- O'Keefe R.T., Henderson S.C., Spector D.L. Dynamic organization of DNA replication in mammalian cell nucleispatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe R.T., Mayeda A., Sadowski C.L., Krainer A.R., Spector D.L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajian M., Wei X., Berezney R., Corden J. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 1998;18:2406–2415. doi: 10.1128/mcb.18.4.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawley J. Fundamental Limits in Confocal Microscopy. In: Pawley J., editor. Handbook of Biological Confocal Microscopy. Plenum Press; New York: 1995. pp. 19–37. [Google Scholar]
- Pennisi E. How the nucleus gets it together. Science. 1997;276:1495–1497. doi: 10.1126/science.276.5318.1495. [DOI] [PubMed] [Google Scholar]
- Perry R.P., Kelley D.E. Inhibition of RNA synthesis by actinomycin Dcharacteristic dose-response of different RNA species. J. Cell. Physiol. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- Raska I. Nuclear ultrastructures associated with the RNA synthesis and processing. J. Cell. Biochem. 1995;59:11–26. doi: 10.1002/jcb.240590103. [DOI] [PubMed] [Google Scholar]
- Roeder R.G. Eukaryotic nuclear RNA polymerases. In: Losick R., Chamberlin M., editors. RNA Polymerase. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1976. pp. 285–329. [Google Scholar]
- Rosbash M., Singer R.H. RNA traveltracks from DNA to cytoplasm. Cell. 1993;75:399–401. doi: 10.1016/0092-8674(93)90373-x. [DOI] [PubMed] [Google Scholar]
- Samarabandu J., Ma H., Acharya R., Cheng P.C., Berezney R. Image analysis techniques for visualizing the spatial organization of DNA replication sites in the mammalian cell nucleus using multi-channel confocal microscopy. SPIE. 1995;2434:370–375. [Google Scholar]
- Scheer U., Rose K. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc. Natl. Acad. Sci. USA. 1984;81:1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Hugle B., Hazan R., Rose K.M. Drug-induced dispersal of transcribed rRNA genes and transcriptional productsimmunolocalization and silver staining of different nucleolar components in rat cells treated with 5,6-dichloro-beta-D-ribofuranosylbenzimidazole. J. Cell Biol. 1984;99:672–679. doi: 10.1083/jcb.99.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickel R.D. Quantitation of human ribosomal DNAhybridization of human DNA with ribosomal RNA for quantitation and fractionation. Pediatr. Res. 1973;7:5–12. doi: 10.1203/00006450-197301000-00002. [DOI] [PubMed] [Google Scholar]
- Singer R.H., Green M.R. Compartmentalization of eukaryotic gene expressioncauses and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- Smetana, K., and H. Busch. 1974. The nucleolar and nucleolar DNA. The Cell Nucleus. Academic Press, New York. 1:73–147.
- Smith K.P., Moen P.T., Wydner K.L., Coleman J.R., Lawrence J.B. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadari S., Focher F., Sala F., Ciarrocchi G., Koch G., Falaschi A., Pedrali-Noy G. Control of cell division by aphidicolin without adverse effects upon resting cells. Arzneim-Forsch. 1985;35:1108–1116. [PubMed] [Google Scholar]
- Spector D. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector D., Schrier W.H., Busch H. Immunoelectron microscopic localization of sn RNPs. Biol. Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Spector D., Fu X.D., Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., O'Keefe R.T., Jimenez-Garcia L.F. Dynamics of transcription and pre-mRNA splicing within the mammalian cell nucleus. Cold Spring Harbor Symp. Quant. Biol. 1993;58:799–805. doi: 10.1101/sqb.1993.058.01.087. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J. Pre-mRNA processing and the CTD of RNA polymerase IIthe tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- Tanner S., Stagljar I., Georgiev O., Schaffner W., Bourquin J.-P. A novel SR-related protein specifically interacts with the carboxy-terminal domain (CTD) of RNA polymerase II through a conserved interaction domain. Biol. Chem. 1997;378:565–571. doi: 10.1515/bchm.1997.378.6.565. [DOI] [PubMed] [Google Scholar]
- van Driel R., Wansink D.G., van Steensel B., Grande M.A., Schul W., de Jong L. Nuclear domains and the nuclear matrix. Int. Rev. Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- van Wijnen A.J., Bidwell J.P., Fey E.G., Penman S., Lian J.B., Stein J.L., Stein G.S. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- Vincent M., Lauriault P., Dubois M.F., Lavoie S., Bensaude O., Chabot B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 1996;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Puvion-Dutilleul F., Harper F., Bachellerie J.-P., Puvion E. Intranuclear distribution of poly (A) RNA determined by electron microscope in situ hybridization. Exp. Cell Res. 1993;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Wang J., Cao L., Wang Y., Pederson T. Localization of pre-messenger RNA at discrete nuclear sites. Proc. Natl. Acad. Sci. USA. 1991;88:7391–7395. doi: 10.1073/pnas.88.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink D.G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Samarabandu J., Devdhar R.S., Siegel A.J., Acharya R., Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1504. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Wellauer P.K., Dawid I.B. Secondary structure maps of RNAprocessing of HeLa ribosomal RNA. Proc. Natl. Acad. Sci. USA. 1973;70:2827–2831. doi: 10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J., Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase IIevidence for cotranscriptional splicing. Mol. Cell. Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Johnson C.V., Dobner P., Lawrence J.B. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y., Johnson C.V., Moen P.T., Jr., McNeil J.A., Lawrence J.B. Nonrandom gene organizationstructural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J. Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A., Patturajan M., Litingtung Y., Joshi R.V., Gentile C., Gebara M., Corden J.L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., van Wijnen A.J., Stein J.L., Meyers S., Sun W., Shopland L., Lawrence J.B., Penman S., Lian J.B., Stein G.S., Hiebert S.W. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-alpha transcription factors. Proc. Natl. Acad. Sci. USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., McNeil S., Pockwinse S., Nickerson J., Shopland L., Lawrence J.B., Penman S., Hiebert S., Lian J.B., van Wijnen A.J., Stein J.L, Stein G.S. Intranuclear targeting of AML/CBF alpha regulatory factors to nuclear matrix-associated transcriptional domains. Proc. Natl. Acad. Sci. USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Taneja K.L., Singer R.H., Green M.R. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- Zieve G., Sauterer R. Cell biology of the snRNP particles. Crit. Rev. Biochem. Mol. Bio. 1990;25:1–46. doi: 10.3109/10409239009090604. [DOI] [PubMed] [Google Scholar]