Abstract
BACKGROUND
Clinical decision support systems can improve medical diagnosis and reduce diagnostic errors. Older systems, however, were cumbersome to use and had limited success in identifying the correct diagnosis in complicated cases.
OBJECTIVE
To measure the sensitivity and speed of “Isabel” (Isabel Healthcare Inc., USA), a new web-based clinical decision support system designed to suggest the correct diagnosis in complex medical cases involving adults.
METHODS
We tested 50 consecutive Internal Medicine case records published in the New England Journal of Medicine. We first either manually entered 3 to 6 key clinical findings from the case (recommended approach) or pasted in the entire case history. The investigator entering key words was aware of the correct diagnosis. We then determined how often the correct diagnosis was suggested in the list of 30 differential diagnoses generated by the clinical decision support system. We also evaluated the speed of data entry and results recovery.
RESULTS
The clinical decision support system suggested the correct diagnosis in 48 of 50 cases (96%) with key findings entry, and in 37 of the 50 cases (74%) if the entire case history was pasted in. Pasting took seconds, manual entry less than a minute, and results were provided within 2–3 seconds with either approach.
CONCLUSIONS
The Isabel clinical decision support system quickly suggested the correct diagnosis in almost all of these complex cases, particularly with key finding entry. The system performed well in this experimental setting and merits evaluation in more natural settings and clinical practice.
KEY WORDS: clinical diagnosis support systems, Isabel, internal medicine, diagnostic error, Google, decision support
INTRODUCTION
The best clinicians excel in their ability to discern the correct diagnosis in perplexing cases. This skill requires an extensive knowledge base, keen interviewing and examination skills, and the ability to synthesize coherently all of the available information. Unfortunately, the level of expertise varies among clinicians, and even the most expert can sometimes fail. There is also a growing appreciation that diagnostic errors can be made just as easily in simple cases as in the most complex. Given this dilemma and the fact that diagnostic error rates are not trivial, clinicians are well-advised to explore tools that can help them establish correct diagnoses.
Clinical diagnosis support systems (CDSS) can direct physicians to the correct diagnosis and have the potential to reduce the rate of diagnostic errors in medicine.1,2 The first-generation computer-based products (e.g., QMR—First Databank, Inc, CA; Iliad—University of Utah; DXplain—Massachusetts General Hospital, Boston, MA) used precompiled knowledge bases of syndromes and diseases with their characteristic symptoms, signs, and laboratory findings. The user would enter findings from their own patients selected from a menu of choices, and the programs would use Bayesian logic or pattern-matching algorithms to suggest diagnostic possibilities. Typically, the suggestions were judged to be helpful in clinical settings, even when used by expert clinicians.3 These diagnosis support systems were also useful in teaching clinical reasoning.4,5 Surprisingly and despite their demonstrated utility in experimental settings, none of these earlier systems gained widespread acceptance for clinical use, apparently related to the considerable time needed to input clinical data and their somewhat limited sensitivity and specificity.3,6 A study of Iliad and QMR in an emergency department setting, for example, found that the final impression of the attending physician was found among the suggested diagnoses only 72% and 52% of the time, respectively, and data input required 20 to 40 minutes for each case.7
In this study, we evaluated the clinical performance of “Isabel” (Isabel Healthcare Inc, USA), a new, second generation, web-based CDSS that accepts either key findings or whole-text entry and uses a novel search strategy to identify candidate diagnoses from the clinical findings. The clinician first enters the key findings from the case using free-text entry (see Fig 1). There is no limit on the number of terms entered, although excellent results are typically obtained with entering just a few key findings. The program includes a thesaurus that facilitates recognition of terms. The program then uses natural language processing and search algorithms to compare these terms to those used in a selected reference library. For Internal Medicine cases, the library includes 6 key textbooks anchored around the Oxford Textbooks of Medicine, 4th Edition (2003) and the Oxford Textbook of Geriatric Medicine and 46 major journals in general and subspecialty medicine and toxicology. The search domain and results are filtered to take into account the patient’s age, sex, geographic location, pregnancy status, and other clinical parameters that are either preselected by the clinician or automatically entered if the system is integrated with the clinician’s electronic medical record. The system then displays a total of 30 suggested diagnoses, with 10 diagnoses presented on each web page (see Fig. 2). The order of listing reflects an indication of the matching between the findings selected and the reference materials searched but is not meant to suggest a ranked order of clinical probabilities. As in the first generation systems, more detailed information on each diagnosis can be obtained by links to authoritative texts.
Figure 1.
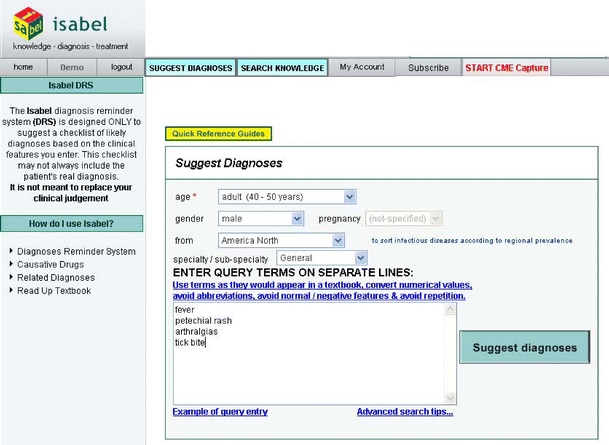
The data-entry screen of the Isabel diagnosis support software. The clinician manually enters the age, gender, locality, and specialty. The query terms can be entered manually from the key findings or findings can be pasted from an existing write-up. This patient was ultimately found to have a B-cell lymphoma secreting a cryo-paraprotein
Figure 2.
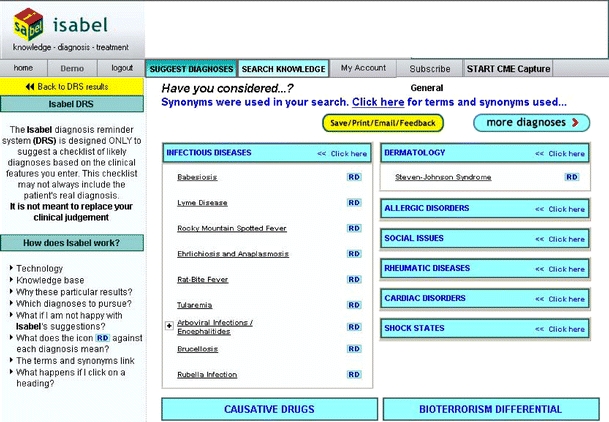
The first page of results of the Isabel diagnosis support software. Additional diagnoses are presented by selecting the ‘more diagnoses’ box
The Isabel CDSS was originally developed for use in pediatrics. In an initial evaluation, 13 clinicians (trainees and staff) at St Mary’s Hospital, London submitted a total of 99 case scenarios of hypothetical case presentations for different diagnoses, and Isabel displayed the expected diagnosis in 91% of these cases.8 Out of 100 real case scenarios gathered from 4 major teaching hospitals in the UK, Isabel suggested the correct diagnosis in 83 of 87 cases (95%). In a separate evaluation of 24 case scenarios in which the gold standard differential diagnosis was established by two senior academic pediatricians, Isabel decreased the chance of clinicians (a mix of trainees and staff clinicians) omitting a key diagnosis by suggesting a highly relevant new diagnosis in 1 of every 8 cases. In this study, the time to enter data and obtain diagnostic suggestions averaged less than 1 minute.9
The Isabel clinical diagnosis support system has now been adapted for adult medicine. The goal of this study was to evaluate the speed and accuracy of this product in suggesting the correct diagnosis in a series of complex cases in Internal Medicine.
METHOD
We considered 61 consecutive “Case Records of Massachusetts General Hospital” (New England Journal of Medicine, vol. 350:166–176, 2004 and 353:189–198, 2005). Each case had an anatomical or final diagnosis, which was considered to be correct by the discussants. We excluded 11 cases (patients under the age of 10 and cases that focused solely on management issues). The 50 remaining case histories were copied and pasted into the Isabel data-entry field. The pasted material typically included the history, physical examination findings, and laboratory test results, but data from tables and figures were not submitted. Beyond entering the patient’s age, sex, and nationality, the investigators did not attempt to otherwise tailor the search strategy. Findings were compared to the recommended (but slightly more time consuming) strategy of entering discrete key findings, as compiled by a senior internist (MLG). Because the correct diagnosis is presented at the end of each case, data entry was not blinded.
RESULTS
Using the recommended method of manually entering key findings, the list of diagnoses suggested by Isabel contained the correct diagnosis in 48 of the 50 cases (96%). Typically 3–6 key findings from each case were used. The 2 diagnoses that were not suggested (progressive multifocal encephalopathy and nephrogenic fibrosing dermopathy) were not included in the Isabel database at the time of the study; thus, these 2 cases would never have been suggested, even with different keywords.
Using the copy/paste method for entering the whole text, the list of diagnoses suggested by Isabel contained the correct diagnosis in 37 of the 50 cases (76%). Isabel presented 10 diagnoses on the first web page and 10 additional diagnoses on subsequent pages up to a total of 30 diagnoses. Because users may tend to disregard suggestions not shown on later web pages, we tracked this parameter for the copy/paste method of data entry: The correct diagnosis was presented on the first page in 19 of the 37 cases (51%) or first two pages in 28 of the 37 cases (77%). Similar data were not collected for manual data entry because the order of presentation depended on which key findings were entered.
Both data entry approaches were fast: Manually entering data and obtaining diagnostic suggestions typically required less than 1 minute per case, and the copy/paste method typically required less than 5 seconds.
DISCUSSION
Diagnostic errors are an underappreciated cause of medical error,10 and any intervention that has the potential to produce correct and timely medical diagnosis is worthy of serious consideration. Our recent analysis of diagnostic errors in Internal Medicine found that clinicians often stop thinking after arriving at a preliminary diagnosis that explains all the key findings, leading to context errors and ‘premature closure’, where further possibilities are not considered.11 These and other errors contribute to diagnoses that are wrong or delayed, causing substantial harm in the patients affected. Systems that help clinicians explore a more complete range of diagnostic possibilities could conceivably reduce these types of error.
Many different CDSSs have been developed over the years, and these typically matched the manually entered features of the case in question to a database of key findings abstracted from experts or the clinical literature. The sensitivity of these systems was in the range of 50%–60%, and the time needed to access and query the database was often several minutes.3 More recently, the possibility of using Google to search for clinical diagnoses has been suggested. However, a formal evaluation of this approach on a subset of the same “Case Records” cases used in our study found a sensitivity of 58%,12 in the range of the first-generation CDSSs and unacceptably low for clinical use.
The findings of our study indicate that CDSS products have evolved substantially. Using the Isabel CDSS, we found that data entry takes under 1 minute, and the sensitivity in a series of highly complex cases approached 100% using entry of key findings. Entry of entire case histories using copy/paste functionality allowed even faster data entry but reduced sensitivity. The loss of sensitivity seemed primarily related to negative findings included in the pasted history and physical (e.g., “the patient denies chest pain”), which are treated as positive findings (chest pain) by the search algorithm.
There are several relevant limitations of this study that make it difficult to predict how Isabel might perform as a diagnostic aid in clinical practice. First, the results obtained here reflect the theoretical upper limit of performance, given that an investigator who was aware of the correct diagnosis entered the key findings. Further, clinicians in real life seldom have the wealth of reliable and organized information that is presented in the Case Records or the time needed to use a CDSS in every case. To the extent that Isabel functions as a ‘learned intermediary’,13 success in using the program will also clearly depend on the clinical expertise of the user and their facility in working with Isabel. A serious existential concern is whether presenting a clinician with dozens of diagnostic suggestions might be a distraction or lead to unnecessary testing. We have previously identified these trade-offs as an unavoidable cost of improving patient safety: The price of improving the odds of reaching a correct diagnosis is the extra time and resources consumed in using the CDSS and considering alternative diagnoses that might turn out to be irrelevant.14
In summary, the Isabel CDSS performed quickly and accurately in suggesting correct diagnoses for complex adult medicine cases. However, the test setting was artificial, and the CDSS should be evaluated in more natural environments for its potential to support clinical diagnosis and reduce the rate of diagnostic error in medicine.
Acknowledgements
The administrative and library-related support of Ms. Grace Garey and Ms. Mary Lou Glazer is gratefully acknowledged. Funding was provided by the National Patient Safety Foundation. This study was approved by the Institutional Review Board (IRB).
Conflict of interest statement None disclosed.
References
- 1.Berner ES. Clinical Decision Support Systems. New York: Springer; 1999.
- 2.Garg AX, Adhikari NKJ, McDonald H, et al. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes. JAMA. 2005;293:1223–38. [DOI] [PubMed]
- 3.Berner ES, Webster GD, Shugerman AA, et al. Performance of four computer-based diagnostic systems. N Engl J Med. 1994;330:1792–6. [DOI] [PubMed]
- 4.Friedman CP, Elstein AS, Wolf FM, et al. Enhancement of clinicians’ diagnostic reasoning by computer-based consultation: a multisite study of 2 systems. JAMA. 1999;282:1851–6. [DOI] [PubMed]
- 5.Lincoln MJ, Turner CW, Haug PJ, et al. Iliad’s role in the generalization of learning across a medical domain. Proc Annu Symp Comput Appl Med Care. 1992;174–8. [PMC free article] [PubMed]
- 6.Kassirer JP. A report card on computer-assisted diagnosis—the grade: C. N Engl J Med. 1994;330:1824–5. [DOI] [PubMed]
- 7.Graber MA, VanScoy D. How well does decision support software perform in the emergency department? Emerg Med J. 2003;20:426–8. [DOI] [PMC free article] [PubMed]
- 8.Ramnarayan P, Tomlinson A, Rao A, Coren M, Winrow A, Britto J. ISABEL: a web-based differential diagnosis aid for paediatrics: results from an initial performance evaluation. Arch Dis Child. 2003;88:408–13. [DOI] [PMC free article] [PubMed]
- 9.Ramnarayan P, Roberts GC, Coren M, et al. Assessment of the potential impact of a reminder system on the reduction of diagnostic errors: a quasi-experimental study. BMC Med Inform Decis Mak. 2006;6:22. [DOI] [PMC free article] [PubMed]
- 10.Graber ML. Diagnostic error in medicine—a case of neglect. Joint Comm J Saf Qual Improv. 2004;31:112–19. [DOI] [PubMed]
- 11.Graber ML, Franklin N, Gordon RR. Diagnostic error in internal medicine. Arch Int Med. 2005;165:1493–9. [DOI] [PubMed]
- 12.Tang H, Ng JHK. Googling for a diagnosis—use of Google as a diagnostic aid: internet based study. Br Med J. 2006. [DOI] [PMC free article] [PubMed]
- 13.Kantor G. Guest software review: Isabel diagnosis software. 2006;1–31.
- 14.Graber ML, Franklin N, Gordon R. Reducing diagnostic error in medicine: what’s the goal? Acad Med. 2002;77:981–92. [DOI] [PubMed]


