Abstract
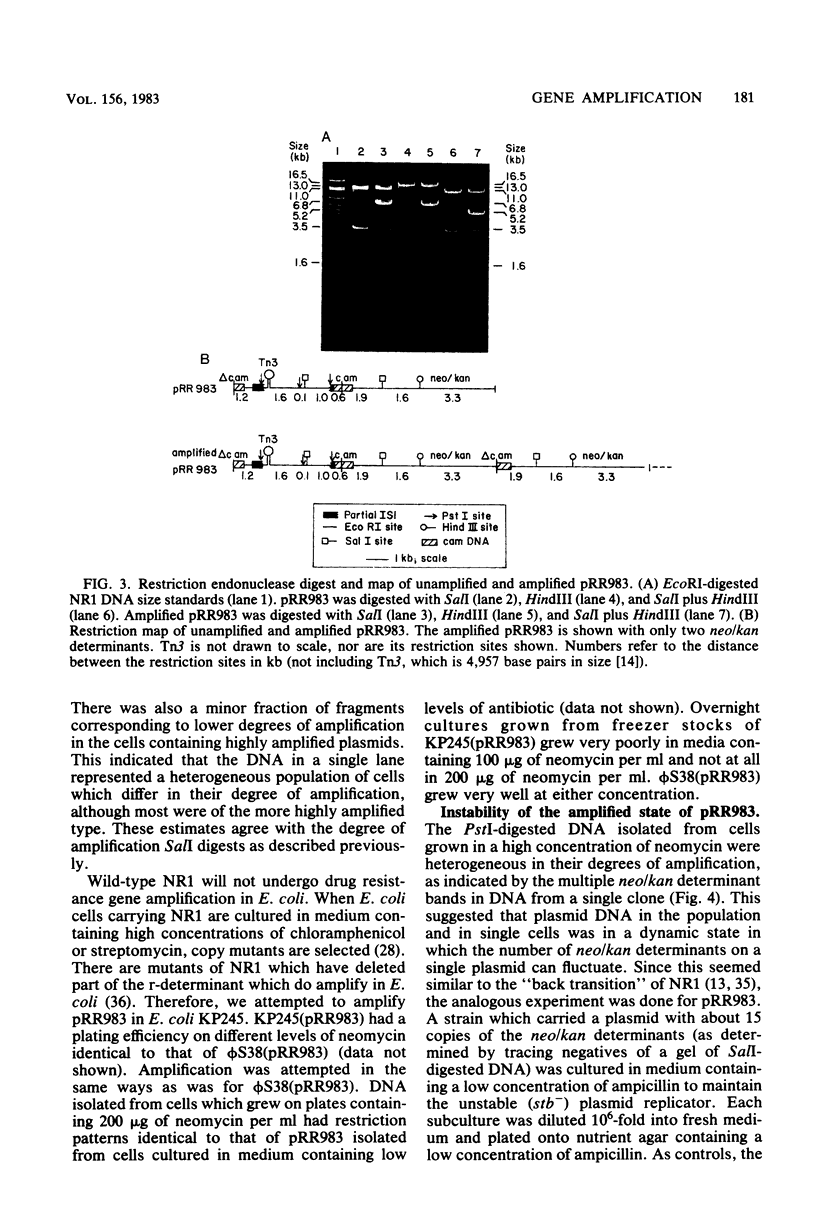
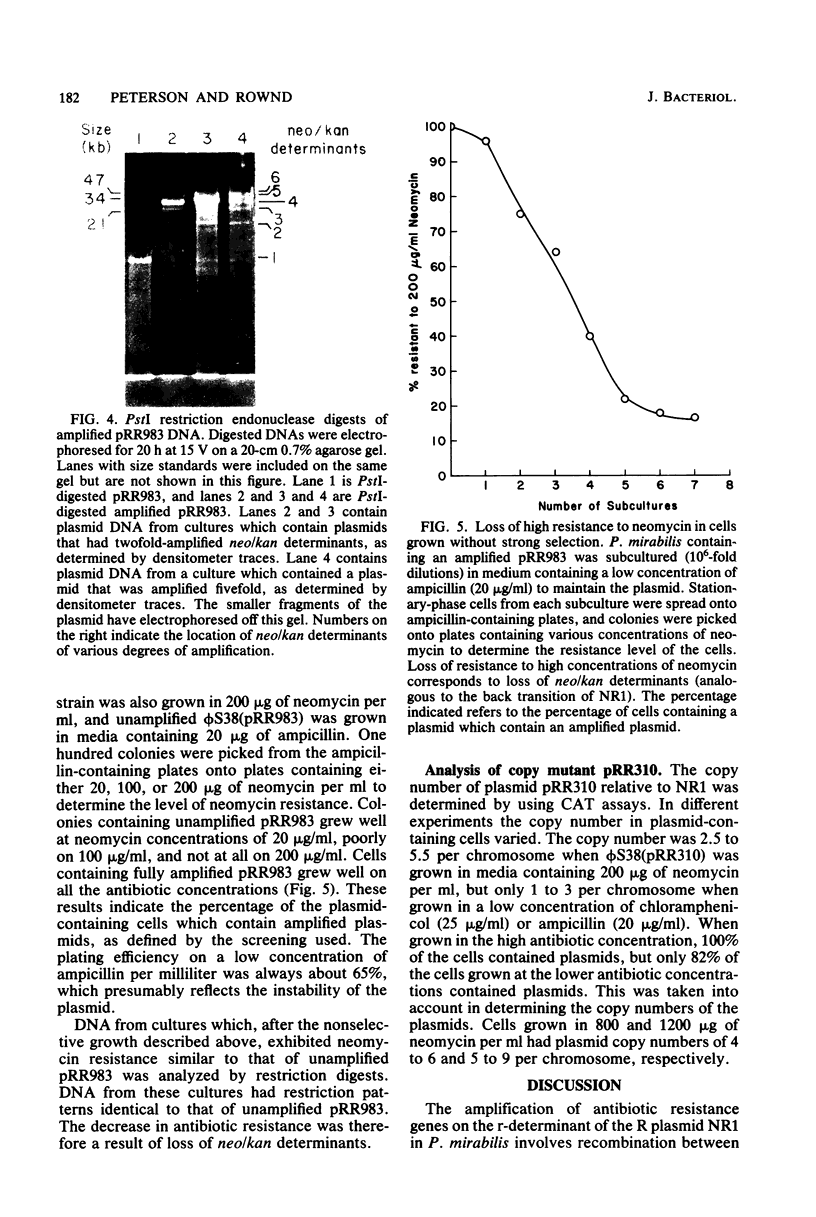
A plasmid (pRR983) was constructed which has a gene coding for neomycin and kanamycin resistance flanked by direct repeats of regions of homology which contain no known insertion sequences. pRR983 does not have any homologous IS1 sequences. Growth of Proteus mirabilis harboring pRR983 in medium containing high concentration of neomycin resulted in cells which were highly resistant to both neomycin and kanamycin. Plasmid DNA was analyzed by using restriction endonucleases. In most cases the neomycin resistance gene had been tandemly duplicated by using the homologous DNA sequences flanking the resistance gene as recombination sites. This is analogous to tandem duplication of drug resistance genes on NR1 using the two direct repeats of IS1 as recombination sites. The amplified plasmid DNA returned to its original structure by the deletion of amplified neomycin resistance determinants when the host cells were cultured without selection for high resistance to neomycin.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem chromosomal duplications in Salmonella typhimurium: fusion of histidine genes to novel promoters. J Mol Biol. 1978 Feb 15;119(1):147–166. doi: 10.1016/0022-2836(78)90274-7. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chandler M., de la Tour E. B., Willems D., Caro L. Some properties of the chloramphenicol resistance transposon Tn9. Mol Gen Genet. 1979 Oct 3;176(2):221–231. doi: 10.1007/BF00273216. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R., Wong-Staal F., Gallo R. C. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature. 1982 Sep 2;299(5878):61–63. doi: 10.1038/299061a0. [DOI] [PubMed] [Google Scholar]
- Edlund T., Grundström T., Normark S. Isolation and characterization of DNA repetitions carrying the chromosomal beta-lactamase gene of Escherichia coli K-12. Mol Gen Genet. 1979 Jun 7;173(2):115–125. doi: 10.1007/BF00330301. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Shaw W. V. Chloramphenicol acetyltransferases specified by fi minus R factors. Antimicrob Agents Chemother. 1973 Jan;3(1):99–104. doi: 10.1128/aac.3.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J., Brown P. C., Schimke R. T. Amplified dihydrofolate reductase genes in unstably methotrexate-resistant cells are associated with double minute chromosomes. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5669–5673. doi: 10.1073/pnas.76.11.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupersztoch-Portnoy Y. M., Lovett M. A., Helinski D. R. Strand and site specificity of the relaxation event for the relaxation complex of the antibiotic resistance plasmid R6K. Biochemistry. 1974 Dec 31;13(27):5484–5490. doi: 10.1021/bi00724a005. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo H., Ohtsubo E. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc Natl Acad Sci U S A. 1982 Jan;79(2):277–281. doi: 10.1073/pnas.79.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoli R., Iida S., Bickle T. A. The DNA sequence of an IS/-flanked transposon coding for resistance to chloramphenicol and fusidic acid. FEBS Lett. 1980 Jan 28;110(1):11–14. doi: 10.1016/0014-5793(80)80011-1. [DOI] [PubMed] [Google Scholar]
- Mattes R., Burkardt H. J., Schmitt R. Repetition of tetracycline resistance determinant genes on R plasmid pRSD1 in Escherichia coli. Mol Gen Genet. 1979 Jan 10;168(2):173–184. doi: 10.1007/BF00431443. [DOI] [PubMed] [Google Scholar]
- Melera P. W., Lewis J. A., Biedler J. L., Hession C. Antifolate-resistant Chinese hamster cells. Evidence for dihydrofolate reductase gene amplification among independently derived sublines overproducing different dihydrofolate reductases. J Biol Chem. 1980 Jul 25;255(14):7024–7028. [PubMed] [Google Scholar]
- Meyer J., Iida S. Amplification of chloramphenicol resistance transposons carried by phage P1Cm in Escherichia coli. Mol Gen Genet. 1979 Oct 3;176(2):209–219. doi: 10.1007/BF00273215. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Hashimoto H., Mickel S., Rownd R. Round of replication mutant of a drug resistance factor. J Bacteriol. 1974 Jun;118(3):855–866. doi: 10.1128/jb.118.3.855-866.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya R., Rownd R. Transduction of R factors by a Proteus mirabilis bacteriophage. J Bacteriol. 1971 Jun;106(3):773–783. doi: 10.1128/jb.106.3.773-783.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Schimke R. T., Urlaub G., Chasin L. A. Amplified dihydrofolate reductase genes are localized to a homogeneously staining region of a single chromosome in a methotrexate-resistant Chinese hamster ovary cell line. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5553–5556. doi: 10.1073/pnas.75.11.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Nyman K., Doroszkiewicz W., Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981 Aug 13;292(5824):640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff W. S. Tn5 transposition and its regulation. Cell. 1982 Dec;31(2 Pt 1):307–308. doi: 10.1016/0092-8674(82)90123-4. [DOI] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Brodsky R. F. Characterization of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol. 1968 Jan;95(1):28–36. doi: 10.1128/jb.95.1.28-36.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Mahowald A. P. Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1096–1100. doi: 10.1073/pnas.77.2.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. F., Murray N. E. Convergent transcription in bacteriophage lambda: interference with gene expression. J Mol Biol. 1979 Sep 15;133(2):249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Ogata Y. Genetic stability of various resistance factors in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1970 May;102(2):363–368. doi: 10.1128/jb.102.2.363-368.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K., Schraml S., Shales S. W., Schmitt R. Tetracycline resistance transposon Tn1721: recA-dependent gene amplification and expression of tetracycline resistance. J Bacteriol. 1981 Sep;147(3):851–859. doi: 10.1128/jb.147.3.851-859.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Taylor D. P., Rownd R. H. Method for obtaining more-accurate covalently closed circular plasmid-to-chromosome ratios from bacterial lysates by dye-buoyant density centrifugation. J Bacteriol. 1977 Apr;130(1):148–153. doi: 10.1128/jb.130.1.148-153.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Identification and characterization of a small sequence located at two sites on the amplifiable tetracycline resistance plasmid pAMalpha1 in Streptococcus faecalis. J Bacteriol. 1977 Jan;129(1):400–406. doi: 10.1128/jb.129.1.400-406.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Plasmid-determined tetracycline resistance in Streptococcus faecalis: tandemly repeated resistance determinants in amplified forms of pAMalpha1 DNA. J Mol Biol. 1976 Apr 15;102(3):583–600. doi: 10.1016/0022-2836(76)90336-3. [DOI] [PubMed] [Google Scholar]