Abstract
The establishment and maintenance of polarity is of fundamental importance for the function of epithelial and neuronal cells. In Drosophila, the multi-PDZ domain protein Bazooka (Baz) is required for establishment of apico-basal polarity in epithelia and in neuroblasts, the stem cells of the central nervous system. In the latter, Baz anchors Inscuteable in the apical cytocortex, which is essential for asymmetric localization of cell fate determinants and for proper orientation of the mitotic spindle. Here we show that Baz directly binds to the Drosophila atypical isoform of protein kinase C and that both proteins are mutually dependent on each other for correct apical localization. Loss-of-function mutants of the Drosophila atypical isoform of PKC show loss of apico-basal polarity, multilayering of epithelia, mislocalization of Inscuteable and abnormal spindle orientation in neuroblasts. Together, these data provide strong evidence for the existence of an evolutionary conserved mechanism that controls apico-basal polarity in epithelia and neuronal stem cells. This study is the first functional analysis of an atypical protein kinase C isoform using a loss-of-function allele in a genetically tractable organism.
Keywords: cell polarity, atypical PKC, Bazooka, tight junction, asymmetric cell division
Introduction
Polarity is a common feature of many different cell types and is the prerequisite for various processes including vectorial transport of molecules, directed cell migration, and asymmetric cell division. Epithelial cells, for example, possess apical and basolateral plasma membrane domains that are distinguished from each other by their different lipid and protein composition (Rodriguez-Boulan and Nelson 1989; Drubin and Nelson 1996). Similar differences exist between axonal and somatodendritic membrane domains of neurons (Foletti et al. 1999; Winckler and Mellman 1999). Polarity is not restricted to the plasma membrane but can also be observed in the underlying cytocortex. Examples of cell types with pronounced cortical polarity are the blastomeres of the early Caenorhabditis elegans embryo (Bowerman and Shelton 1999) and Drosophila neuroblasts (Lu et al. 1998).
The mechanisms leading to the generation of cell polarity are just beginning to emerge. Membrane proteins and lipids are sorted in the trans Golgi network and are subsequently delivered to distinct regions of the plasma membrane via vesicular transport (Ikonen and Simons 1998). The cytocortex may acquire polarity by interaction of cortical proteins with certain membrane lipids; e.g., by binding of proteins containing pleckstrin homology domains to phosphatidylinositol (3,4,5) trisphosphate or by binding to the cytoplasmic tails of transmembrane proteins. Vice versa, the correct localization of many transmembrane proteins depends on binding to cortical proteins. So far, it is not known whether membrane polarity can be established in the absence of cortical polarity or whether both processes are strictly dependent on each other.
How are the different membrane domains separated from each other in polarized cells? In vertebrate epithelia, the tight junction (TJ) is considered to be the boundary between apical and basolateral plasma membrane domains (Stevenson and Keon 1998; Tsukita et al. 1999). In addition, TJs create a paracellular seal that prevents the free diffusion of macromolecules in the extracellular space between cells. At the ultrastructural level, TJs are characterized by the fusion of the outer leaflet of the plasma membrane of adjacent cells. The formation of TJs depends on transmembrane proteins of the claudin family (Furuse et al. 1998; Tsukita and Furuse 1999). Claudins and occludin, another transmembrane protein of the TJ, associate with several cortical proteins (ZO-1, ZO-2, ZO-3, and cingulin) that form a link with the actin cytoskeleton (Stevenson and Keon 1998; Tsukita and Furuse 1999; Tsukita et al. 1999).
In contrast to vertebrate epithelial cells, neurons do not possess TJs, but nonetheless establish and maintain separate axonal and somatodendritic plasma membrane domains. The border between these membrane domains is located at the initial axonal segment (Winckler and Mellman 1999). Measurements of the force required to move transmembrane and GPI-linked proteins in the plane of the membrane revealed that the lateral mobility of both classes of proteins is strongly reduced in the initial segment (Winckler et al. 1999). Treatment with the actin depolymerizing drug latrunculin B disrupts the diffusion barrier in this region (Winckler et al. 1999), suggesting that the submembraneous cytoskeleton plays an important role in maintenance of neuronal cell surface polarity.
Like neurons, most epithelial tissues in arthropods lack TJs, despite being highly polarized (Tepass and Hartenstein 1994; Müller 2000). It has been proposed that the septate junction (SJ) could be the functional equivalent of TJs in arthropods. This may be true at least in some cases for the sealing of the paracellular space (Skaer et al. 1987; Baumgartner et al. 1996), but the SJ is probably not responsible for the formation of a diffusion barrier in the plasma membrane. In Drosophila, the separation of apical and basolateral membrane domains can already be observed at the time of gastrulation (3 h after egg laying) (Müller and Wieschaus 1996), while SJs form much later (11 h after egg laying) (Tepass and Hartenstein 1994). Moreover, the septate junction forms basally of the zonula adherens (ZA), a belt-like adhesive junction around the apex of epithelial cells, whereas the border between the apical and the basolateral plasma membrane domain is located at the apical end of the ZA (Tepass 1996). So how is the separation of apical and basolateral plasma membrane domains achieved in Drosophila epithelia?
Double mutants lacking zygotic expression of the genes stardust (sdt) and bazooka (baz) fail to establish plasma membrane polarity after cellularization of the Drosophila embryo (Müller and Wieschaus 1996). This phenotype is characterized by expression of the basolateral marker Neurotactin (Nrt) on the whole cell surface and mislocalization of the ZA component Armadillo (Arm). Moreover, in sdt, baz double mutants, the monolayered organization of the blastoderm epithelium is lost and cells acquire irregular shapes. These morphological changes are reminiscent of those seen during epithelial-mesenchymal transitions. Essentially, the same phenotype as in sdt, baz double mutants is observed in baz mutants lacking maternal and zygotic Bazooka (Baz), whereas zygotic sdt and baz single mutants show a weaker phenotype later in development (Tepass and Knust 1993; Grawe et al. 1996; Müller and Wieschaus 1996; Kuchinke et al. 1998). These data suggest that baz is absolutely required for establishment of plasma membrane polarity and epithelial morphology, whereas the early function of sdt may be partially redundant with that of baz.
baz is also required for establishment of apico-basal polarity and asymmetric division of neuroblasts in the developing central nervous system (CNS). Neuroblasts delaminate from the neuroectodermal epithelium and undergo several rounds of asymmetric cell division, generating a ganglion mother cell and another neuroblast in each division. Before division, the mitotic spindle rotates by 90° and localization of the cell fate determinants Prospero and Numb becomes restricted to the basal cortex of the neuroblast. These events are prerequisites for proper segregation of Prospero and Numb into the ganglion mother cell. From delamination to early anaphase, Baz is localized in the apical cortex of neuroblasts, where it forms a complex with Inscuteable (Insc) (Schober et al. 1999; Wodarz et al. 1999), a protein required for rotation of the mitotic spindle and correct localization of Prospero and Numb (Kraut and Campos-Ortega 1996; Kraut et al. 1996; Kaltschmidt et al. 2000; reviewed in Jan and Jan 2000). In the absence of Baz, asymmetric cortical localization of Insc is abolished, leading to randomized spindle orientation and mislocalization of cell fate determinants (Schober et al. 1999; Wodarz et al. 1999). These data led to the conclusion that apico-basal polarity in neuroblasts depends on maintenance of apical Baz expression and is thus inherited from the neuroectodermal epithelium.
baz encodes a protein with three PDZ domains which shows significant sequence similarity along its entire length to Par-3 (Caenorhabditis elegans) and ASIP (rat) (Etemad-Moghadam et al. 1995; Izumi et al. 1998; Kuchinke et al. 1998). In the early C. elegans embryo, Par-3 is asymmetrically localized in the anterior cortex of the zygote and of blastomeres that undergo asymmetric cell divisions (Etemad-Moghadam et al. 1995). In these cells, Par-3 controls spindle orientation and asymmetric localization of cell fate determinants (Etemad-Moghadam et al. 1995; reviewed in Rose and Kemphues 1998). Later on, Par-3 is also expressed in the apical cortex of the embryonic gut epithelium (O. Bossinger, personal communication). Par-3 binds to PKC-3, an atypical protein kinase C (aPKC) isoform (Tabuse et al. 1998; Wu et al. 1998). Both proteins are mutually dependent on each other for correct cortical localization. Moreover, embryos depleted of PKC-3 by RNA interference show a very similar phenotype to par-3 mutant embryos (Tabuse et al. 1998). ASIP was isolated as a binding partner of the mammalian aPKC isoforms, PKCλ and PKCζ (Izumi et al. 1998). Intriguingly, ASIP and PKCλ colocalize at the TJ in vertebrate epithelial cells (Izumi et al. 1998). These observations suggest that the association of ASIP/Par-3 with aPKCs is functionally important and evolutionarily conserved.
We have identified an aPKC from Drosophila (DaPKC) that shows very high sequence similarity to PKCλ and PKCζ from vertebrates and PKC-3 from C. elegans. DaPKC and Baz coimmunoprecipitate and directly bind to each other in a yeast two-hybrid assay. In embryos, both proteins colocalize in the apical cortex of almost all epithelial tissues and in neuroblasts. Apical localization of DaPKC in epithelia and neuroblasts is abolished in baz mutants, and vice versa, Baz is delocalized in DaPKC mutants. We show that the phenotype of DaPKC loss-of-function mutants resembles that of baz mutants, consistent with a close functional interdependence of both proteins. Together, our data provide the first in vivo evidence for an essential role of an atypical protein kinase C isoform in establishment and maintenance of epithelial and neuronal polarity.
Materials and Methods
Identification of DaPKC
BLAST searches of the Berkeley Drosophila genome database revealed that EST clone HL05754 (obtained from Research Genetics) contains a cDNA insert with high similarity to atypical PKCs from vertebrates and C. elegans. HL05754 was fully sequenced and contains bases 1–1917 of the full length DaPKC cDNA sequence plus additional sequences derived from a transcript unrelated to DaPKC. Additional 3′ cDNA sequence including the 3′ end of the DaPKC coding region was amplified by 3′ RACE from embryonic mRNA with the 5′/3′ RACE Kit (Roche) using the following primers: 5′-CGCGTTCATGGATATCGTCAGC-3′ (forward) and 5′-GTTGATATTGCTGCTGCTGCTGC-3′ (reverse). The remaining part of the 3′ noncoding region has been deduced from the genomic sequence 3′ of the RACE product, which does not contain a splice donor site but contains a consensus polyadenylation motif (AATAAA).
Fly Stocks and Genetics
Oregon R was used as wild-type stock. Df(2R)Jp1 removes the cytological interval 51C3;52F5-9, which contains the complete coding region of DaPKC. The DaPKCk06403 P-element insertion allele was obtained as l(2)k06403 from the Bloomington Drosophila stock center. Revertants of DaPKCk06403 were generated using P[Δ2-3] (Robertson et al. 1988) as a transposase source. bazXi106germ line clones were produced using the FLP/DFS technique (Chou and Perrimon 1992; Müller and Wieschaus 1996). Baz overexpression in embryos was achieved with the UAS-GAL4 system (Brand and Perrimon 1993) using a UAS-Baz transgene (Kuchinke et al. 1998) and the maternal GAL4 driver mat67G4 (D. St. Johnston and J.P. Vincent, unpublished observations). For analysis of DaPKC localization in insc mutants, inscP49 and inscP72 (Kraut and Campos-Ortega 1996) were used in combination with a CyO P[ftz::lacZ] balancer. To generate hemizygous DaPKCk06403 embryos with the wild-type maternal dosage of DaPKC, we crossed females with a compound second chromosome, C(2)v (Merrill et al. 1988), to DaPKCk06403/CyO males. Compound autosomes have both left arms attached to one centromere and both right arms attached to the other, instead of the normal arrangement in which the left and right arms of a given chromosome are attached to the same centromere. C(2)v females produce gametes that contain either two left or two right arms of the attached second chromosome. Thus, one quarter of the progeny from the cross C(2)v × DaPKCk06403/CyO lack zygotic expression of DaPKC. DaPKC mutant embryos were identified by lack of staining with anti–aPKC antibody C20. Staging of embryos was according to Campos-Ortega and Hartenstein 1997.
Immunohistochemistry and In Situ Hybridization
For antibody stainings, embryos were fixed either in 4% formaldehyde, 100 mM phosphate buffer, pH 7.4 (for stainings with antibodies against PKCζ (C20), Insc, β-galactosidase, and Baz in neuroblasts) or according to the heat-methanol fixation procedure described in Müller and Wieschaus 1996. The following primary antibodies were used: rabbit anti–PKCζ C20 (Santa Cruz Biotechnology, Inc.), 1:1,000; mouse anti-Nrt (Hortsch et al. 1990), 1:5; rat anti-Baz (Wodarz et al. 1999), 1:300; mouse anti-Arm N2-7A1 (Peifer et al. 1994) 1:10; rabbit anti-Insc (Kraut and Campos-Ortega 1996), 1:1,000; mouse anti-Sex-lethal (Bopp et al. 1991), 1:5; mouse anti–β-galactosidase (Promega), 1:5,000. DNA was stained with YoYo-1 (Molecular Probes). Cy2-, Cy3-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1:400. In situ hybridization was performed using an antisense RNA probe of DaPKC labeled with digoxigenin-UTP (Roche) according to standard procedures. Images were taken on a TCSNT confocal microscope (Leica) or on an Axioplan microscope (Carl Zeiss, Inc.) and processed using Photoshop (Adobe) and Canvas (Deneba) software.
Immunoprecipitation and Western Blotting
Immunoprecipitation and western blotting were carried out as described (Willert et al. 1997; Wodarz et al. 1999). For IPs, 3 μl of mouse anti-Baz or 30 μl of monoclonal mouse anti-Nrt were added to a cell lysate containing 500 μg of total protein from S2 cells overexpressing Baz (Wodarz et al. 1999). For Western analysis, rabbit anti-PKCζ C20 was diluted 1:2,000.
Yeast Two-Hybrid Interaction Assays
Full-length DaPKC was cloned in frame with the GAL4 transactivation domain in pACT2 (CLONTECH Laboratories, Inc.). For generation of bait constructs, the regions of Baz shown in Fig. 2 b (below) were cloned in frame with the GAL4 DNA binding domain in pGBT9 (CLONTECH Laboratories, Inc.). Two hybrid assays were performed according to the manufacturers instructions. Details on the generation of two-hybrid constructs will be provided on request.
Figure 2.
DaPKC and Baz directly bind to each other. (a) Anti-Baz antibody (COOH-terminal, mouse) was used for immunoprecipitation (IP) of an extract from S2 cells overexpressing Baz. Anti-Nrt antibody was used as a negative control. Immunoprecipitates underwent SDS-PAGE and Western analysis with anti-aPKC antibody C20. The Western blot of the cell lysate used for immunoprecipitation (input) is shown on the right. The single band detected by antibody C20 migrates with a molecular weight of ∼75 kD. (b) DaPKC binds directly to Baz. The structure of Baz and the position of the three PDZ domains are shown schematically at the top. The regions of Baz fused in frame with the GAL4 DNA-binding domain in the bait constructs are indicated by bars below the schematic drawing of Baz. In the column on the right: (+) interaction of the bait constructs with full length DaPKC, and (−) the lack of interaction. Numbering of amino acid residues is according to Kuchinke et al. 1998.
Results
Molecular Characterization of the DaPKC Gene
To identify an atypical protein kinase C isoform from Drosophila, we screened the Berkeley Drosophila genome database (http://www.fruitfly.org) with sequences from mouse PKCλ and C. elegans PKC-3 (Tabuse et al. 1998) using the BLAST algorithm (Altschul et al. 1997). One EST clone (HL05754) showed significant sequence similarity to the NH2 termini of both PKCλ and PKC-3. Further sequencing of HL05754 revealed that it contains most of the coding region of DaPKC, except for a few hundred basepairs that are missing at the 3′ end. BLAST searches with the HL05754 cDNA fragment showed that the DaPKC gene is located in genomic region 51D on the right arm of chromosome 2, a region that had been sequenced by the Berkeley Drosophila genome project (Adams et al. 2000). Based on sequence similarity to mouse and C. elegans aPKCs and on sequence analysis tools predicting exon–intron boundaries, we identified three additional putative 3′ exons that are missing in HL05754. The existence of the predicted transcript was confirmed by 3′ RACE analysis of embryonic mRNA. Comparison of the DaPKC cDNA sequence to the genomic sequence of the DaPKC locus revealed the existence of at least 10 exons (Fig. 1 a). Both the first and the last exon are noncoding and the last exon contains a canonical polyadenylation signal (AATAAA). Conceptual translation of the cDNA gives rise to a protein of 606 amino acids with a predicted molecular weight of 69.5 kD. DaPKC shows the highest sequence similarity to mouse PKCλ (68% identity), rat PKCζ (63% identity), and C. elegans PKC-3 (58% identity) (Fig. 1 b). In comparison, DaPKC shows significantly lower sequence similarity to two conventional PKC isoforms from Drosophila (Schaeffer et al. 1989), PKC 53E (29% identity) and PKC 98F (36% identity). BLAST searches of the completed genome sequence of Drosophila (Adams et al. 2000) revealed that DaPKC is the only aPKC in Drosophila.
Figure 1.

Structure of the DaPKC locus. (a) The DaPKC gene is located on the right arm of the second chromosome at position 51D. The gene comprises 10 exons (boxes). Translated regions are filled in black and untranslated regions in gray. ATG and TGA mark the translation initiation and termination codons, respectively. In the lethal P-element insertion line l(2)k06403 the PlacW element (hatched box) is inserted into the third intron, which causes a loss-of-function phenotype. (b) Alignment of DaPKC with aPKC isoforms of mouse, rat, and C. elegans. The DaPKC amino acid sequence (aPKC Dm) shows 68% identity to mouse PKCλ (PKCλ Mm), 63% identity to rat PKCζ (PKCζ Rn) and 58% identity to C. elegans PKC-3 (PKC-3 Ce). Amino acid residues identical in at least two of the four kinases are shown in reversed font. The region comprising the kinase domain is underlined. These sequence data are available from GenBank/EMBL/DDBJ under accession number AF288482.
For detection of DaPKC on Western blots and in whole-mount immunofluorescence stainings, we tested an antibody raised against a peptide corresponding to the COOH-terminal 20 amino acids of rat PKCζ (C20) for cross-reactivity with DaPKC. At the very COOH terminus, DaPKC and rat PKCζ show 75% identity and 95% similarity at the amino acid level (Fig. 1 b). Antibody C20 recognizes a protein of ∼75 kD in Western Blots of Drosophila embryos and Schneider S2 cells (Fig. 2 a), which is in good agreement with the predicted molecular weight of DaPKC. In whole-mount immunofluorescence stainings of embryos, antibody C20 recognizes an epitope that is expressed in the same spatial and temporal pattern as the DaPKC mRNA (see below). The staining intensity is reduced to background level in embryos homozygous for a deficiency that removes the DaPKC locus (data not shown) and in embryos homozygous for the lethal P-element insertion l(2)k06403, which is located in the third intron of the DaPKC locus (Fig. 1 a, 9 a, below). From these observations, we conclude that antibody C20 specifically recognizes the DaPKC protein.
DaPKC Binds Directly to Baz
To test whether DaPKC and Baz are associated in a protein complex, we performed coimmunoprecipitation experiments. Baz was immunoprecipitated from extracts of S2 cells overexpressing Baz (Wodarz et al. 1999). S2 cells express endogenous DaPKC. The immune complex was subjected to SDS-PAGE and Western analysis with anti-aPKC antibody C20 (Fig. 2 a). Anti-Nrt antibody (Hortsch et al. 1990) was used for immunoprecipitation as a negative control. While a significant amount of DaPKC coimmunoprecipitated with Baz, no signal was detected in the Nrt control (Fig. 2 a).
To determine whether binding of DaPKC to Baz is direct, we performed interaction studies with the yeast two-hybrid system. A construct containing full-length DaPKC fused to the transactivation domain of GAL4 was cotransformed into yeast with bait constructs containing different regions of Baz fused to the GAL4 DNA-binding domain (Fig. 2 b). Interaction of the bait constructs with DaPKC was assayed by X-Gal filter assays. Bait constructs containing the second and third PDZ domain of Baz showed interaction with full length DaPKC, whereas all constructs lacking the second or third PDZ domain gave negative results (Fig. 2 b). We conclude that binding of DaPKC to Baz is direct and that the region from amino acid 401 to 737 of Baz is sufficient for binding to DaPKC.
Expression Pattern of DaPKC
To see where DaPKC is expressed during embryonic development, we analyzed the mRNA distribution by RNA in situ hybridization. DaPKC mRNA is already detectable in freshly laid eggs before the onset of zygotic transcription (Fig. 3 a) and thus must be deposited in the egg during oogenesis. At the cellular blastoderm stage, DaPKC mRNA is present in all cells except for the pole cells (Fig. 3 b). During gastrulation, strong expression of DaPKC is detectable in tissues that undergo morphogenetic movements; e.g., the invaginating mesoderm, the proctodeum, and the cephalic furrow (Fig. 3 c). In embryos at the extended germ band stage, prominent DaPKC expression is detectable in neuroblasts (Fig. 3d and Fig. e). In several epithelial tissues, in particular in the fore- and hindgut and in the Malpighian tubules, DaPKC mRNA is highly enriched in the apical cytocortex (Fig. 3d and Fig. f), reminiscent of the polarized localization of baz and crumbs (crb) mRNAs (Tepass et al. 1990; Kuchinke et al. 1998).
Figure 3.
mRNA expression pattern of DaPKC in wild-type embryos. (a) DaPKC mRNA is already present in freshly laid eggs and early embryos before the onset of zygotic transcription and thus must be deposited in the egg during oogenesis. (b) In embryos at the cellular blastoderm stage, DaPKC is expressed in all cells except for the pole cells (arrow). (c) During gastrulation, DaPKC expression is detectable in ectoderm and mesoderm and is elevated in cells that undergo morphogenetic movements; e.g., in the cephalic furrow (cf), in the proctodeal (pr), and the stomodeal (st) invaginations and in the mesoderm (ms). (d) At the extended germ band stage (stage 11), DaPKC is strongly expressed in neuroblasts (nb, arrows) and in most ectodermally derived epithelia, except for the amnioserosa. Note that DaPKC mRNA is highly enriched in the apical cytocortex of several epithelial tissues, including the primordia of the foregut (fg) and hindgut (hg). (e) Higher magnification of a stage 11 embryo showing strong expression of DaPKC in neuroblasts (arrows). (f) Higher magnification of a stage 11 embryo showing apical localization of DaPKC mRNA in foregut (left) and hindgut (right). The apical cell surfaces of fore- and hindgut are marked by arrows and the basal cell surfaces by arrowheads, respectively. Except for f, which is a dorsal view, all embryos are shown in a lateral view. Dorsal is up and anterior to the left. Bar in a–d = 100 μm; bar in e and f = 50 μm.
The distribution of DaPKC protein was analyzed using anti-PKCζ antibody C20. During cellularization of the embryo, DaPKC becomes localized to the apical cytocortex of all cells except for the pole cells, which do not contain detectable amounts of DaPKC (Fig. 4, a and b). Already at this stage DaPKC is enriched at apico-lateral cell borders, giving rise to a honeycomb pattern in en face views of the blastoderm (Fig. 4 c). After completion of cellularization, DaPKC is highly concentrated in the apico-lateral cortex (Fig. 4d and Fig. e) and shows little overlap with the basolateral marker Nrt (Fig. 4d′ and d″) (de la Escalera et al. 1990; Hortsch et al. 1990). Apical localization is maintained throughout embryonic development in most epithelia that are derived from the ectoderm (e. g. epidermis, fore-, and hindgut), Malpighian tubules, and the tracheal system (Fig. 4, f–i). The only ectodermal epithelium devoid of DaPKC expression is the amnioserosa.
Figure 4.

Expression pattern and subcellular localization of DaPKC protein in wild-type embryos. (a–c) During cellularization (cell cycle 14), DaPKC is already enriched in the apical cytocortex of the newly forming blastoderm cells (a), but is not detectable in the pole cells (pc) (b). Cortical staining of DaPKC is concentrated at cell borders (c). At this stage, plasma membrane polarity has not been fully established yet, as can be seen by the nonpolar distribution of Nrt, an integral membrane protein that becomes later confined to the basolateral membrane (a′). (d–e) In embryos at gastrulation, DaPKC localization is confined to the apicalmost region of the lateral cortex and, at lower levels, to the cortex underlying the free apical surface (d). This localization pattern is complementary to that of Nrt (d′), which is strictly basolateral after completion of cellularization (merged image in d″). In en face views, DaPKC forms a contiguous belt around the apical circumference of each blastoderm cell (e). (f and g) At the extended germ band stage (stage 12), DaPKC is detectable in the apical cortex of all epithelia derived from the ectoderm, except for the amnioserosa. (h and i) Epithelial expression of DaPKC is maintained after germ band retraction (stage 14) and remains confined to the apical cytocortex. sns, stomatogastric nervous system; fg, foregut; hg, hindgut; ep, epidermis; tp, tracheal pits; tr, tracheae; mt, Malpighian tubules. In all panels, DaPKC is in red and Nrt is in green. a and a′, and d, d′, and d″, are double labelings of the same embryo, respectively. f and g show different focal planes of the same embryo. Anterior is to the left and dorsal up in all panels. Bars in a–e and i = 10 μm. Bars in f–h = 100 μm.
Strong expression of DaPKC was also detected in neuroblasts, the stem cells of the embryonic CNS. During delamination of neuroblasts, DaPKC is localized in the apical stalk that is wedged between adjacent cells of the neuroectodermal epithelium (Fig. 5, a–c). In pro- and metaphase, DaPKC forms apical cortical crescents (Fig. 5, d–i). In anaphase, DaPKC staining is strongly diminished and expands over a broader region of the neuroblast cortex, but is clearly excluded from the budding ganglion mother cell (Fig. 5, j–l). Thus, from delamination through pro- and metaphase, localization of DaPKC in neuroblasts is very similar to that of Baz and Insc (Kraut and Campos-Ortega 1996; Kraut et al. 1996; Schober et al. 1999; Wodarz et al. 1999). Simultaneously with DaPKC, cell outlines of epithelial cells and neuroblasts were visualized with the Nrt antibody (Fig. 5b, Fig. e, Fig. h, and Fig. k). Similar to epithelial cells, Nrt expression is clearly polarized in neuroblasts. Strong staining for Nrt is detectable in the basal and lateral membrane of neuroblasts, whereas staining is strongly reduced in apical regions where DaPKC is expressed (Fig. 5, arrowheads).
Figure 5.

Expression of DaPKC in neuroblasts. Wild-type embryos at the extended germ band stage (stage 10) were triple labeled for DaPKC (red), Nrt (blue), and DNA (YoYo-1, green). (a–c) During delamination of neuroblasts, DaPKC is expressed in the apical stalk that is wedged between adjacent epithelial cells. In prophase (d–f) and metaphase (g–i), DaPKC forms a crescent in the apical cortex of neuroblasts. In anaphase (j–l), DaPKC staining in neuroblasts is strongly reduced but is still detectable in an enlarged region of the apical cortex. No staining is detectable in budding ganglion mother cells (GMCs). Note that Nrt staining is reduced in regions of the plasma membrane where DaPKC is present in the cortex (regions between arrowheads in b and e). Neuroblasts are marked by asterisks and the budding GMCs in j and k by a circle. Merged images of all three stainings are shown in the right column. Apical is up in all panels. Bar = 10 μm.
To test whether DaPKC and Baz are colocalized, we performed double-label immunofluorescence stainings of embryos (Fig. 6, a–c). DaPKC (Fig. 6 a, red) and Baz (Fig. 6 b, green) are clearly colocalized in the epidermis (Fig. 6 c, yellow) and in neuroblasts (data not shown). To determine the precise subcellular localization of DaPKC and Baz with respect to the ZA, we performed double-label immunofluorescence stainings with antibodies against Arm, a component of the ZA (Fig. 6 d, red) and Baz (Fig. 6 e, green). The merged image (Fig. 6 f) shows that Baz is localized apically to Arm. The same is true for DaPKC (data not shown). At the resolution of the confocal microscope, we cannot rule out the possibility that the localization of Baz and DaPKC partially overlaps with Arm in the ZA.
Figure 6.
Apical localization of DaPKC depends on Baz. (a–c) DaPKC and Baz are colocalized in epithelia. A wild-type embryo at stage 14 was doubly stained for DaPKC (a, red) and Baz (b, green). Both proteins show extensive colocalization (c). (d–f) Baz is localized apically to the ZA marker Arm. A wild-type embryo at stage 14 was doubly stained for Arm (d, red) and Baz (e, green). In the merged image (f) there is very little overlap and Baz is expressed apically to Arm. The indentations of the epidermis in a–f represent segment boundaries. (g–i) DaPKC localization is lost and Nrt localization is not restricted to the basolateral membrane in baz mutant embryos. In a bazXi106 null embryo at gastrulation, DaPKC is not localized in the cortex (g) and Nrt is present on the whole-cell surface (h), merged image in i. Note also that the blastoderm epithelium is multilayered and cells show a very irregular shape (compare Fig. 4d, Fig. d′, and d″). (j–l) Overexpression of Baz is sufficient for ectopic localization of DaPKC. When overexpressed with the GAL4 system, Baz is not restricted to the apical cortex of epithelial cells and neuroblasts, but becomes localized to ectopic sites in the cortex (k, arrows). Concomitantly, DaPKC is also detectable at ectopic sites in the cortex (j, arrows), where it colocalizes with Baz (l, arrows). Apical is up in all panels. Bars = 10 μm.
Localization of DaPKC Depends on Baz
Binding and colocalization of DaPKC and Baz suggested that both proteins may functionally interact with each other. In stainings of baz mutant embryos derived from germ line clones (baz null embryos) with anti-aPKC antibody, we could not detect any apical localization of DaPKC in epithelia (Fig. 6g and Fig. i) and in neuroblasts (Fig. 7g and Fig. h). Instead, DaPKC was distributed in a diffuse fashion in the cytoplasm. baz null embryos also showed a loss of membrane polarity that became evident by mislocalization of the basolateral transmembrane protein Nrt. In contrast to wild type (Fig. 4 d′), Nrt was not excluded from the apical plasma membrane (Fig. 6 h). Moreover, the monolayered structure of the epidermis was lost and cells piled up on top of each other (Fig. 6 h), as has been described before for sdt, baz double mutants (Müller and Wieschaus 1996).
Figure 7.
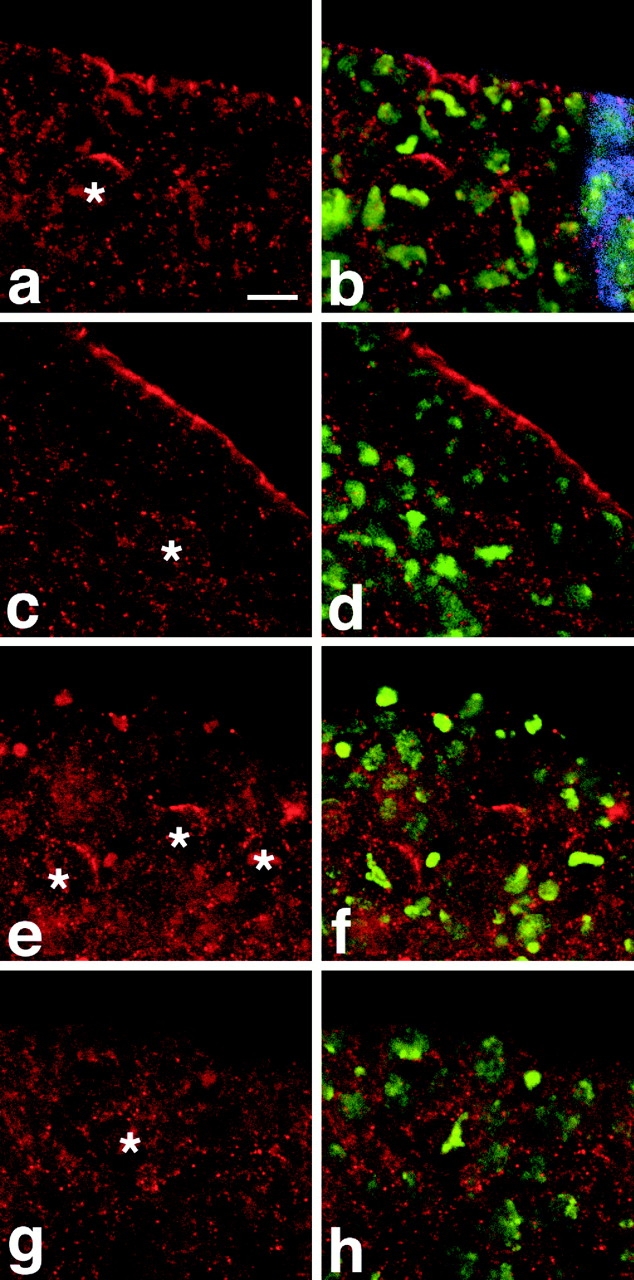
Apical localization of DaPKC in neuroblasts depends on Baz and Insc. (a and b) In a heterozygous inscP49/CyO embryo at stage 10, DaPKC (red) forms apical crescents in metaphase neuroblasts, as in wild type. (c and d) In inscP49 homozygous mutant embryos, apical localization of DaPKC in neuroblasts is lost, while apical localization of DaPKC in the neuroectodermal epithelium is unaffected. (e and f) In bazXi106/+ embryos that are derived from a homozygous mutant germ line (germ line clones with a paternal wild-type copy), asymmetric DaPKC crescents are detectable in neuroblasts, although the neuroectodermal epithelium has almost completely disintegrated. (g and h) In bazXi106/Y embryos derived from germ line clones (baz null), DaPKC localization is completely diffuse and neither cortical nor asymmetric in neuroblasts and epithelia. Note also the abnormal orientation of metaphase plates in d, f, and h. Homozygous mutant inscP49 embryos were identified by the absence of β-galactosidase expression derived from the CyO P[ftz::lacZ] balancer (blue stripe in b). Hemizygous mutant bazXi106/Y embryos were identified by absence of Sex-lethal expression (not shown). DNA was stained with YoYo-1 (green). Apical is up in all panels. Bar = 10 μm.
To test whether mislocalization of Baz is sufficient to induce mislocalization of DaPKC, we overexpressed Baz by means of the GAL4 system (Brand and Perrimon 1993; Wodarz et al. 1999). Under these conditions, Baz was not confined to the apical cytocortex anymore and was found in more lateral and basal positions in epithelia and neuroblasts (Fig. 6 k). Concomitantly, DaPKC was also mislocalized (Fig. 6 j) and colocalized in ectopic positions with ectopic Baz (Fig. 6 l), confirming that ectopic Baz can recruit DaPKC to ectopic sites in the cytocortex.
It has been shown before that Baz is required for apical localization of Insc in neuroblasts and that Insc is required for stabilization of Baz in neuroblasts after delamination (Schober et al. 1999; Wodarz et al. 1999; Yu et al. 2000). We tested whether Baz and Insc are also required for localization of DaPKC in neuroblasts. DaPKC localization was indistinguishable from wild type in neuroblasts of inscP49/CyO heterozygous embryos (Fig. 7, a and b), but was neither cortical nor apical in neuroblasts of inscP49 homozygous mutant embryos (Fig. 7c and Fig. d). In embryos lacking maternal Baz but carrying a paternal wild-type allele of baz (partial paternal rescue), asymmetric cortical localization of DaPKC was detected in most neuroblasts at metaphase (Fig. 7e and Fig. f). However, DaPKC crescents and metaphase plates were often misoriented with respect to the surface of the embryo (Fig. 7e and Fig. f), a phenotype that has also been observed at low penetrance in embryos lacking only zygotic expression of Baz (Kuchinke et al. 1998). In embryos lacking both maternal and zygotic expression of Baz (baz null), DaPKC was completely delocalized in neuroblasts and epithelial tissues (Fig. 6 g and 7, g and h). These results indicate that Baz is absolutely required for apical localization of DaPKC in neuroblasts and epithelial tissues, while Insc is required for localization of DaPKC only in neuroblasts.
Genetic Analysis of DaPKC Loss-of-Function Mutants
To study the consequences of DaPKC loss-of-function on epithelial polarity and asymmetric division of neuroblasts, we analyzed the phenotype of mutants in the DaPKC locus. BLAST searches with the genomic sequence of the DaPKC gene revealed that the P-element insertion l(2)k06403 is located in the third intron of the DaPKC gene (Fig. 1 a). This insertion line is homozygous lethal, contains a single PlacW P-element, and does not complement Df(2R)Jp1, which removes the cytological interval 51C3;52F5-9. Staining intensity with anti-PKCζ antibody C20 was reduced to background level in embryos homozygous mutant for l(2)k06403 (Fig. 9 a, below). We mobilized the P-element using the transposase source P[Δ2-3] (Robertson et al. 1988) and recovered several revertants lacking the w + marker of the original P-element insertion. Of seven revertant lines we tested, 3 were homozygous viable, demonstrating that insertion of the P-element into the DaPKC locus was the cause of lethality and of the observed phenotype (see below). We thus conclude that l(2)k06403 is an allele of DaPKC and renamed it DaPKCk06403.
Figure 9.

DaPKC mutants show defects in epithelial polarity and morphology. (a and b) Homozygous DaPKCk06403 mutant embryos do not stain with anti-PKCζ antibody C20. A mutant embryo at gastrulation was doubly labeled with anti-PKCζ antibody C20 (a) and anti-Nrt (b). Note the absence of specific staining in a and the multilayering of the blastoderm epithelium in b. (c–f) Homozygous DaPKCk06403 mutant embryos at gastrulation show abnormal localization of Baz and Nrt. Baz (c and e) is not localized to the apical cortex of the blastoderm cells, but instead shows a diffuse, nonpolarized cortical staining. Except for some cells in the outermost cell layer, Nrt (d and f) is localized on the whole plasma membrane and is not restricted to the basolateral membrane, as in wild type. Note the multilayering of the epithelium and the extremely irregular cell shape. e and f are higher magnification views of the boxed area in d. A region devoid of cells due to cell death is marked by an arrow in c and d. (g and h) Embryos derived from heterozygous DaPKCk06403/CyO mothers that carry a zygotic wild-type allele of DaPKC frequently develop characteristic head defects. At the anterior tip (region between arrowheads), the epithelium is multilayered, shows nonpolarized Baz staining (g) and expression of Nrt on the whole cell surface [h, merged channels for Nrt (green) and Baz (red)]. (i and j) Hemizygous DaPKCk06403 embryos derived from C(2)v mothers show loss of Baz (i) and Arm (j) staining in the ventral neuroectoderm. The embryo in g and h is at stage 8 and the embryo in i and j is at stage 10. a–h are optical cross sections and i and j are a view on the ventral surface of the embryo. In all panels, anterior is to the left. Bar in a–d = 100 μm. Bars in e–j = 20 μm.
Homozygous mutant DaPKCk06403 embryos do not produce any cuticle (Fig. 8 b). The reason for this phenotype is early embryonic lethality before the onset of cuticle secretion. Transheterozygous DaPKCk06403/Df(2R)Jp1 embryos show exactly the same cuticle phenotype (Fig. 8 c), suggesting that DaPKCk06403 is a strong hypomorphic or null allele. The analysis of the DaPKC loss-of-function phenotype was complicated by the fact that DaPKC mutants show maternal haploinsufficiency with incomplete penetrance. The evidence for this conclusion comes from the following observations: we noticed that, in egg collections from the DaPKCk06403/CyO stock, significantly more than 50% of embryos (including CyO/CyO homozygotes) failed to hatch (63.2%, n = 644), which means that even embryos with a zygotic wild-type allele of DaPKC frequently show developmental defects (Fig. 8 e and 9, g and h). This finding was confirmed in crosses of heterozygous DaPKCk06403/CyO females to wild-type males, where we also observed considerable embryonic lethality (25%, n = 244). The reciprocal cross did not show an increase of embryonic lethality compared with wild-type controls. We therefore conclude that the decreased viability of the progeny from DaPKCk06403/CyO mothers is caused by the reduction of the maternal DaPKC level during oogenesis.
Figure 8.

Cuticle phenotypes of DaPKC and baz mutants are very similar. (a) Wild-type cuticle. (b) Homozygous mutant DaPKCk06403 embryos derived from DaPKCk06403/CyO mothers do not form cuticle because they die before cuticle secretion begins. (c) The same phenotype is observed in embryos from DaPKCk06403/CyO mothers that are transheterozygous for DaPKCk06403 and Df(2R)Jp1 that removes the whole DaPKC coding region. (d) Many bazXi106 null embryos also do not produce cuticle and resemble embryos lacking DaPKC. (e) Embryos from heterozygous DaPKCk06403/CyO mothers that possess a zygotic wild-type allele of DaPKC frequently die with characteristic head defects. (f and g) Embryos derived from C(2)v mothers that are hemizygous for DaPKCk06403 form cuticles with severe head defects and large ventral holes (f) or lack ventral cuticle altogether (g). (h) Zygotic bazXi106 mutants also form cuticles with large ventral holes and head defects, reminiscent of the one shown in f. Anterior is to the left in all panels. Dorsal is up in a–e and g, while f and h are ventral views. Bar = 100 μm.
To produce embryos with the wild-type maternal contribution of DaPKC, but lacking any zygotic DaPKC expression, we crossed C(2)v females (Merrill et al. 1988) to DaPKCk06403/CyO males and analyzed the cuticle phenotype of their progeny (for details, see Materials and Methods). One quarter of the embryos derived from that cross completely lacked zygotic expression of DaPKC and produced cuticles with characteristic defects. In most cases, head structures were missing and the ventral cuticle either showed large holes (Fig. 8 f) or was missing altogether (g). These phenotypes are strikingly similar to those of baz mutants. baz null embryos derived from germ-line clones produce very little or no cuticle (Fig. 8 d) and resemble DaPKCk06403 mutant embryos derived from heterozygous mothers (b and c). baz mutant embryos lacking only the zygotic expression produce cuticles with characteristic head defects and ventral holes (Fig. 8 h) that are very similar to DaPKCk06403 mutant embryos derived from C(2)v mothers (f and g).
DaPKC Mutants Show Loss of Polarity in Epithelia and Neuroblasts
To investigate the role of DaPKC in the control of epithelial organization and polarity, we stained DaPKCk06403mutant embryos with antibodies against Baz, Nrt, and Arm, the Drosophila β-catenin homologue. Most homozygous DaPKCk06403 embryos from heterozygous mothers arrested very early in development and died before or during cellularization (data not shown). Those that developed further showed dramatic defects in epithelial organization and polarity. The blastoderm epithelium of these embryos was multilayered, cell shapes were extremely irregular and apico-basal polarity of the epithelium was lost (Fig. 9, a–f). Instead of being localized to the apical cortex, Baz was found in randomly scattered aggregates (Fig. 9c and Fig. e). The basolateral marker Nrt was localized on the whole-cell surface in most cells (Fig. 9d and Fig. f; compare Fig. 4 d′).
A significant fraction of embryos derived from DaPKCk06403/CyO heterozygous mothers that possessed at least one zygotic wild-type allele of DaPKC showed characteristic defects in the head region (Fig. 9g and Fig. h). While epithelial structure and distribution of Baz and Nrt was normal in the trunk region of these embryos, the epithelium at the anterior tip of the embryos was multilayered, showed a delocalized distribution of Baz (Fig. 9g and Fig. h) and expression of Nrt on the whole cell surface (h). Thus, the defects observed in the head region of these embryos were very similar to the defects observed in the whole blastoderm epithelium of homozygous DaPKCk06403 embryos from heterozygous mothers (Fig. 9, a–f). Most likely, these defects reflect an early requirement for DaPKC before the onset of zygotic transcription and are caused by insufficient maternal supply of DaPKC. Consistent with this interpretation, homozygous DaPKCk06403 embryos with the wild-type maternal contribution of DaPKC (see Materials and Methods) developed further than homozygous mutant embryos derived from heterozygous mothers and did not show obvious defects before germ band extension. At this stage, patches devoid of apical Baz (Fig. 9 i) and Arm (j) staining appeared, especially in the ventral neuroectoderm and in the head. Optical cross sections of these regions revealed defects in epithelial organization and polarity very similar to those shown in Fig. 9, a–h (data not shown).
To study the effect of DaPKC loss-of-function on asymmetric division of neuroblasts in the embryonic CNS, we stained DaPKCk06403 mutant embryos that received the full maternal dosage of DaPKC with antibodies against Baz and Insc (Fig. 10). In most metaphase neuroblasts of these embryos, Baz was not detectable (Fig. 10, a and b) and Insc staining was diffuse, instead of forming a tight apical crescent (Fig. 10c and Fig. d). In addition, the orientation of metaphase plates often deviated from the normal orientation parallel to the surface of the embryo (Fig. 10b and Fig. d), reflecting abnormal orientation of the mitotic spindle.
Figure 10.

DaPKC is required for localization of Baz and Insc and for correct orientation of the metaphase plate in neuroblasts. Hemizygous DaPKCk06403 embryos derived from C(2)v mothers were stained for Baz and DNA (a and b) or for Insc and DNA (c and d). Baz staining is strongly reduced and is not localized to the apical cortex in metaphase neuroblasts of DaPKCk06403 hemizygous embryos (a and b), while Insc is still detectable in the cytoplasm but fails to form a tight apical crescent (c and d). Note also that orientation of metaphase plates with respect to the surface of the embryo is abnormal in DaPKCk06403 mutants (b and d). Neuroblasts are marked by asterisks. Apical is up in all panels. b and d are merged images of the Baz and DNA channels (b) or the Insc and DNA channels (d). Bar = 10 μm.
Discussion
We have identified the gene encoding the only atypical PKC isoform in Drosophila and show that DaPKC binds directly to Baz and is colocalized with Baz in epithelia and neuroblasts. Apical localization of DaPKC is lost in baz mutants and vice versa, demonstrating that both proteins are mutually dependent on each other for correct localization. In neuroblasts, localization of DaPKC is not only dependent on Baz, but also on Insc. Baz levels are strongly reduced in neuroblasts of insc mutant embryos, most likely because Insc is required for stabilization of Baz (Schober et al. 1999; Wodarz et al. 1999; Yu et al. 2000). Thus, the effect of Insc on DaPKC localization is probably indirect and can be explained by the loss of Baz in insc mutant neuroblasts. Another protein, partner of Insc (Pins), has recently been identified as a binding partner of Insc (Schaefer et al. 2000; Yu et al. 2000). Pins, Baz, and Insc are mutually dependent on each other for correct localization and/or stability (Schaefer et al. 2000; Yu et al. 2000), indicating that loss of any single known component of this complex compromises stability and/or localization of the other components in the apical cortex of neuroblasts.
These findings are reminiscent of the situation in the early C. elegans embryo, where PKC-3, Par-3 and another PDZ domain protein, Par-6, are mutually dependent on each other for correct localization in the anterior cytocortex (Watts et al. 1996; Tabuse et al. 1998; Hung and Kemphues 1999). Consistent with these results, the phenotype of embryos depleted of PKC-3 by RNA interference is very similar to the phenotype of par-3 and par-6 mutants (Etemad-Moghadam et al. 1995; Watts et al. 1996; Tabuse et al. 1998; Hung and Kemphues 1999). Interestingly, a Drosophila homologue of par-6 does exist (Tabuse et al. 1998), raising the possibility that the interaction of Par-3/Baz, PKC-3/DaPKC and Par-6 has been evolutionarily conserved.
Another example for a close functional interaction between a PDZ domain protein and protein kinase C has recently been uncovered in Drosophila. The multi-PDZ domain protein InaD binds to the eye-specific, conventional isoform of PKC and is required for its proper localization in photoreceptors (Tsunoda et al. 1997; Xu et al. 1998). InaD contains five PDZ domains and distinct binding partners have been identified for each of them. Intriguingly, all of the proteins that bind to InaD are part of the phototransduction cascade in the Drosophila eye (Tsunoda et al. 1997; Xu et al. 1998). Thus, it has been proposed that InaD provides a scaffold for the assembly of a signaling complex, a so called “transducisome.”
In the case of DaPKC and Baz, the situation is more complicated. Consistent with a function as a scaffold, Baz is required for localization of the signaling protein DaPKC. However, Baz itself is not properly localized in the absence of DaPKC. It is easy to imagine how a structural multi-PDZ domain protein like InaD or Baz can localize a protein kinase, but how can DaPKC be responsible for localization and stabilization of Baz? Baz possesses a PKC consensus phosphorylation site that is conserved between Baz, Par-3, and ASIP. Phosphorylation of this site by DaPKC could be important to regulate binding of Baz to other proteins or to protect Baz from proteolytic degradation. It is also possible that DaPKC binds simultaneously to Baz and another protein that may be required for localization of Baz. A detailed structure–function analysis of both Baz and DaPKC will be necessary to clarify this issue.
Analysis of the DaPKC loss-of-function phenotype revealed that DaPKC is already required very early during embryogenesis, before the onset of zygotic transcription. Most homozygous DaPKCk06403 embryos with a reduced maternal dosage of DaPKC die before cellularization is completed. What could be the reason for this early death? aPKCs have been implicated in the control of apoptotic cell death in vertebrate tissue culture cells. Inhibition of aPKCs induces apoptosis (Diaz-Meco et al. 1996). Treatment of cells with UV irradiation also triggers apoptosis and rapidly inhibits aPKC kinase activity, suggesting that inhibition of aPKCs is an early event in the apoptotic signaling cascade (Berra et al. 1997). In accordance with these data, aPKCs have been implicated in the transduction of survival signals downstream of growth factor receptors. In contrast to conventional and novel PKC isoforms, aPKCs can be activated by phosphatidylinositol(3,4,5)trisphosphate and ceramide, two second messengers that are generated in response to inflammatory cytokines and growth factors (Nakanishi et al. 1993; Lozano et al. 1994; Akimoto et al. 1996). Our observation that DaPKC mutant embryos show premature cell death and strongly increased TUNEL labeling (A. Wodarz, unpublished observations), which is a hallmark of apoptosis, is consistent with a function of DaPKC in the transmission of survival signals. A detailed analysis of the role of DaPKC in the control of cell death in Drosophila is beyond the scope of this work and will be addressed in a future publication.
The loss-of-function phenotype of DaPKC mutants in epithelia is very similar to the phenotypes described for baz null mutants and zygotic sdt, baz double mutants (Müller and Wieschaus 1996). The most striking abnormalities in these mutants are loss of the monolayered epithelial organization, irregular cell shapes, and loss of plasma membrane polarity. Multilayering of epithelia and abnormal cell shapes are most likely caused by defects in cell adhesion. Indeed, formation of the ZA, a region of intense, cadherin-mediated cell contact, is defective in DaPKC, baz, and sdt mutants (Grawe et al. 1996; Müller and Wieschaus 1996). Another gene, crb, is also required for correct positioning and maintenance of the ZA (Wodarz et al. 1995; Grawe et al. 1996; Tepass 1996; reviewed in Müller 2000). DaPKC, Baz, and Crb are all localized apically of the ZA (Tepass 1996; this work), so how can they control formation of the ZA? Crb, a transmembrane protein (Tepass et al. 1990) binds via its cytoplasmic tail to the cortical multi-PDZ domain protein Discs Lost (Bhat et al. 1999; Klebes and Knust 2000), which probably associates with additional proteins. This complex could be involved in the formation of a protein scaffold in the apical cytocortex that prevents ZA components from moving further apically. A similar function can be envisioned for Baz, since it is also a multi-PDZ domain protein with the capacity to interact with several partners at the same time.
How does DaPKC fit into this model? As discussed above, DaPKC is required for localization and stabilization of Baz, but this may not be its only function in ZA formation. Several reports show that PKCs are involved in the assembly of adherens junctions and TJs (Lewis et al. 1994; Citi and Denisenko 1995; Stuart and Nigam 1995; van Hengel et al. 1997). The majority of these studies used cultured cell lines and analyzed the effects of different inhibitors and agonists of PKCs on localization and phosphorylation of junctional proteins, cell adhesion, and cell morphology. Although these studies provided compelling evidence for an involvement of PKCs in junction formation, in most cases neither the specific PKC isoforms responsible for the observed phenotypes nor the targets of these PKCs have been unambiguously identified. In one interesting study, inhibition of aPKCs induced epithelial-mesenchymal transformation in quail neural tube explants, while inhibitors of conventional or novel PKCs had little or no effect in this assay (Minichiello et al. 1999).
In addition to their effects on epithelial organization and cell shape, mutations in DaPKC, baz, sdt, and crb also affect plasma membrane polarity (Wodarz et al. 1993, Wodarz et al. 1995; Müller and Wieschaus 1996; Klebes and Knust 2000). As discussed in the Introduction, establishment and maintenance of plasma membrane polarity requires the separation of apical and basolateral membrane domains by a diffusion barrier in the plane of the membrane. In vertebrate epithelia, this diffusion barrier is created by the TJ. In arthropod epithelia, TJs have not been found by ultrastructural analysis (Tepass and Hartenstein 1994). We note, however, that the vertebrate homologues of DaPKC and Baz, PKCλ, PKCζ, and ASIP, are localized at the TJ in epithelial cells (Izumi et al. 1998). Moreover, DaPKC and Baz are localized apically to the ZA in Drosophila epithelia, which corresponds to the position of the TJ in vertebrate epithelia. Thus, based on their localization and their mutant phenotypes, we propose that DaPKC and Baz are components of an evolutionarily conserved protein complex that may serve similar functions as the TJ in vertebrates.
Neuroblasts do not possess elaborate cell junctions but clearly show cortical and, at least to some extent, plasma membrane polarity. DaPKC and Baz are required for anchoring Insc in the apical neuroblast cortex (Schober et al. 1999; Wodarz et al. 1999; this work) and it is conceivable that DaPKC and Baz may also be involved in the formation of a submembraneous protein scaffold analogous to the model we have proposed for epithelia. Consistent with this idea is the finding that Nrt staining is reduced precisely in those regions of the neuroblast plasma membrane where DaPKC and Baz are localized beneath the membrane. Thus, DaPKC and Baz may be generally responsible for the separation of membrane domains by preventing diffusion of basolateral proteins into the apical domain.
From the available data, it is impossible to decide whether the primary function of DaPKC in neuroblasts is the stabilization of Baz or whether DaPKC phosphorylates additional targets involved in asymmetric division of neuroblasts. One candidate for phosphorylation by DaPKC is Miranda, an adaptor protein with six consensus PKC phosphorylation sites that binds to Prospero and Insc (Ikeshima-Kataoka et al. 1997; Shen et al. 1997, Shen et al. 1998; Schuldt et al. 1998). Miranda colocalizes with Insc only briefly in late interphase, and then moves together with Prospero to the basal cortex of the neuroblast during prophase (Shen et al. 1998). It is an attractive possibility that phosphorylation of Miranda by DaPKC regulates binding of Miranda to Insc and its release from the apical complex later in the cell cycle.
In conclusion, we have shown that DaPKC is an essential binding partner of Baz in epithelia and neuroblasts. Surprisingly, Baz does not simply function as a scaffold to anchor DaPKC in the apical cytocortex, but is itself dependent on DaPKC for proper localization and stability. This mutual dependence is indicative of an intimate cross-talk between structural proteins like Baz and the signaling protein DaPKC. The link between signal transduction components and structural components of the cytocortex may be important to allow rapid rearrangement of cellular junctions and cell shape changes such as those occurring during delamination of neuroblasts. To fully understand the role of DaPKC in the generation of cellular asymmetry, it will be essential to identify the physiological activators, inhibitors, and downstream targets of this important protein kinase.
Acknowledgments
We thank Bill Chia, Shigeo Ohno, Girish Deshpande, and the Developmental Studies Hybridoma Bank for antibodies, and Arno Müller, Eric Wieschaus, Daniel St. Johnston, José Campos-Ortega, and Kathy Matthews from the Bloomington Drosophila stock center for sending numerous stocks. We also thank Arno Müller, Kevin Johnson, and Olaf Bossinger for critical reading of the manuscript, Soya Kim for performing TUNEL stainings, and Olaf Bossinger for communication of results before publication. Special thanks to Arno Müller for giving advice on the use of compound stocks.
This work was supported by grants from the Deutsche Forschungsgemeinschaft.
Footnotes
Abbreviations used in this paper: Arm, Armadillo; Baz, Bazooka; CNS, central nervous system; Crb, Crumbs; DaPKC, Drosophila atypical PKC; Insc, Inscuteable; Nrt, Neurotactin; sdt, stardust; TJ, tight junction; ZA, zonula adherens.
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F. The genomic sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Akimoto K., Takahashi R., Moriya S., Nishioka N., Takayanagi J., Kimura K., Fukui Y., Osada S., Mizuno K., Hirai S. EGF or PDGF receptors activate atypical PKClambda through phosphatidylinositol 3-kinase. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:788–798. [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLASTa new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S., Littleton J.T., Broadie K., Bhat M.A., Harbecke R., Lengyel J.A., Chiquet-Ehrismann R., Prokop A., Bellen H.J. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87:1059–1068. doi: 10.1016/s0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- Berra E., Municio M.M., Sanz L., Frutos S., Diaz-Meco M.T., Moscat J. Positioning atypical protein kinase C isoforms in the UV-induced apoptotic signaling cascade. Mol. Cell. Biol. 1997;17:4346–4354. doi: 10.1128/mcb.17.8.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M.A., Izaddoost S., Lu Y., Cho K.O., Choi K.W., Bellen H.J. Discs lost, a novel multi-PDZ domain protein, establishes and maintains epithelial polarity. Cell. 1999;96:833–845. doi: 10.1016/s0092-8674(00)80593-0. [DOI] [PubMed] [Google Scholar]
- Bopp D., Bell L.R., Cline T.W., Schedl P. Developmental distribution of female-specific sex-lethal proteins in Drosophila melanogaster . Genes Dev. 1991;5:403–415. doi: 10.1101/gad.5.3.403. [DOI] [PubMed] [Google Scholar]
- Bowerman B., Shelton C.A. Cell polarity in the early Caenorhabditis elegans embryo. Curr. Opin. Genet. Dev. 1999;9:390–395. doi: 10.1016/S0959-437X(99)80059-8. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J.A., Hartenstein V. The embryonic development of Drosophila melanogaster. Springer Verlag; Berlin, Heidelberg, Germany: 1997. [Google Scholar]
- Chou T.B., Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila . Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citi S., Denisenko N. Phosphorylation of the tight junction protein cingulin and the effects of protein kinase inhibitors and activators in MDCK epithelial cells. J. Cell Sci. 1995;108:2917–2926. doi: 10.1242/jcs.108.8.2917. [DOI] [PubMed] [Google Scholar]
- de la Escalera S., Bockamp E.O., Moya F., Piovant M., Jimenez F. Characterization and gene cloning of neurotactin, a Drosophila transmembrane protein related to cholinesterases. EMBO (Eur. Mol. Biol. Organ.) J. 1990;9:3593–3601. doi: 10.1002/j.1460-2075.1990.tb07570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Meco M.T., Municio M.M., Frutos S., Sanchez P., Lozano J., Sanz L., Moscat J. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell. 1996;86:777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]
- Drubin D.G., Nelson W.J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo S., Kemphues K.J. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Foletti D.L., Prekeris R., Scheller R.H. Generation and maintenance of neuronal polaritymechanisms of transport and targeting. Neuron. 1999;23:641–644. doi: 10.1016/s0896-6273(01)80022-2. [DOI] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Fujimoto K., Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawe F., Wodarz A., Lee B., Knust E., Skaer H. The Drosophila genes crumbs and stardust are involved in the biogenesis of adherens junctions. Development (Camb.) 1996;122:951–959. doi: 10.1242/dev.122.3.951. [DOI] [PubMed] [Google Scholar]
- Hortsch M., Patel N.H., Bieber A.J., Traquina Z.R., Goodman C.S. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development (Camb.) 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Hung T.J., Kemphues K.J. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development (Camb.) 1999;126:127–135. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J.B., Nabeshima Y., Doe C.Q., Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Ikonen E., Simons K. Protein and lipid sorting from the trans-Golgi network to the plasma membrane in polarized cells. Semin. Cell. Dev. Biol. 1998;9:503–509. doi: 10.1006/scdb.1998.0258. [DOI] [PubMed] [Google Scholar]
- Izumi Y., Hirose T., Tamai Y., Hirai S., Nagashima Y., Fujimoto T., Tabuse Y., Kemphues K.J., Ohno S. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J. Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan Y.N., Jan L.Y. Polarity in cell divisionwhat frames thy fearful asymmetry? Cell. 2000;100:599–602. doi: 10.1016/s0092-8674(00)80695-9. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt J.A., Davidson C.M., Brown N.H., Brand A.H. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat. Cell. Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- Klebes A., Knust E. A conserved motif in Crumbs is required for E-cadherin localisation and zonula adherens formation in Drosophila . Curr. Biol. 2000;10:76–85. doi: 10.1016/s0960-9822(99)00277-8. [DOI] [PubMed] [Google Scholar]
- Kraut R., Campos-Ortega J.A. Inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev. Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- Kraut R., Chia W., Jan L.Y., Jan Y.N., Knoblich J.A. Role of inscuteable in orienting asymmetric cell divisions in Drosophila . Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Kuchinke U., Grawe F., Knust E. Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol. 1998;8:1357–1365. doi: 10.1016/s0960-9822(98)00016-5. [DOI] [PubMed] [Google Scholar]
- Lewis J.E., Jensen P.J., Johnson K.R., Wheelock M.J. E-cadherin mediates adherens junction organization through protein kinase C. J. Cell Sci. 1994;107:3615–3621. doi: 10.1242/jcs.107.12.3615. [DOI] [PubMed] [Google Scholar]
- Lozano J., Berra E., Municio M.M., Diaz-Meco M.T., Dominguez I., Sanz L., Moscat J. Protein kinase C zeta isoform is critical for kappa B-dependent promoter activation by sphingomyelinase. J. Biol. Chem. 1994;269:19200–19202. [PubMed] [Google Scholar]
- Lu B., Jan L.Y., Jan Y.N. Asymmetric cell divisionlessons from flies and worms. Curr. Opin. Gene. Dev. 1998;8:392–399. doi: 10.1016/s0959-437x(98)80108-1. [DOI] [PubMed] [Google Scholar]
- Merrill P.T., Sweeton D., Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster . Development (Camb.) 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- Minichiello J., Ben-Ya'acov A., Hearn C.J., Needham B., Newgreen D.F. Induction of epithelio-mesenchymal transformation of quail embryonic neural cells by inhibition of atypical protein kinase-C. Cell Tissue Res. 1999;295:195–206. doi: 10.1007/s004410051225. [DOI] [PubMed] [Google Scholar]
- Müller H.A.J., Wieschaus E. Armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila . J. Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.A.J. Genetic control of epithelial cell polaritylessons from Drosophila . Dev. Dyn. 2000;218:52–67. doi: 10.1002/(SICI)1097-0177(200005)218:1<52::AID-DVDY5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nakanishi H., Brewer K.A., Exton J.H. Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1993;268:13–16. [PubMed] [Google Scholar]
- Peifer M., Sweeton D., Casey M., Wieschaus E. Wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development (Camb.) 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- Robertson H.M., Preston C.R., Phillis R.W., Johnson-Schlitz D.M., Benz W.K., Engels W.R. A stable genomic source of P element transposase in Drosophila melanogaster . Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Nelson W.J. Morphogenesis of the polarized epithelial cell phenotype. Science. 1989;245:718–725. doi: 10.1126/science.2672330. [DOI] [PubMed] [Google Scholar]
- Rose L.S., Kemphues K.J. Early patterning of the C. elegans embryo. Annu. Rev. Genet. 1998;32:521–545. doi: 10.1146/annurev.genet.32.1.521. [DOI] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko A., Knoblich J.A. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila . Curr. Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Schaeffer E., Smith D., Mardon G., Quinn W., Zuker C. Isolation and characterization of two new Drosophila protein kinase C genes, including one specifically expressed in photoreceptor cells. Cell. 1989;57:403–412. doi: 10.1016/0092-8674(89)90915-x. [DOI] [PubMed] [Google Scholar]
- Schober M., Schaefer M., Knoblich J.A. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams J.H., Davidson C.M., Micklem D.R., Haseloff J., St D., Johnston, Brand A.H. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C.P., Jan L.Y., Jan Y.N. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila . Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Shen C.P., Knoblich J.A., Chan Y.M., Jiang M.M., Jan L.Y., Jan Y.N. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila . Genes Dev. 1998;12:1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaer H.B., Maddrell S.H., Harrison J.B. The permeability properties of septate junctions in Malpighian tubules of Rhodnius. J. Cell Sci. 1987;88:251–265. doi: 10.1242/jcs.88.2.251. [DOI] [PubMed] [Google Scholar]
- Stevenson B.R., Keon B.H. The tight junctionmorphology to molecules. Annu. Rev. Cell. Dev. Biol. 1998;14:89–109. doi: 10.1146/annurev.cellbio.14.1.89. [DOI] [PubMed] [Google Scholar]
- Stuart R.O., Nigam S.K. Regulated assembly of tight junctions by protein kinase C. Proc. Natl. Acad. Sci. USA. 1995;92:6072–6076. doi: 10.1073/pnas.92.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuse Y., Izumi Y., Piano F., Kemphues K.J., Miwa J., Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans . Development (Camb.) 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Tepass U. Crumbs, a component of the apical membrane, is required for zonula adherens formation in primary epithelia of Drosophila . Dev. Biol. 1996;177:217–225. doi: 10.1006/dbio.1996.0157. [DOI] [PubMed] [Google Scholar]
- Tepass U., Hartenstein V. The development of cellular junctions in the Drosophila embryo. Dev. Biol. 1994;161:563–596. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- Tepass U., Knust E. Crumbs and stardust act in a genetic pathway that controls the organization of epithelia in Drosophila melanogaster . Dev. Biol. 1993;159:311–326. doi: 10.1006/dbio.1993.1243. [DOI] [PubMed] [Google Scholar]
- Tepass U., Theres C., Knust E. Crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–799. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Furuse M. Occludin and claudins in tight-junction strandsleading or supporting players? Trends Cell. Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Furuse M., Itoh M. Structural and signalling molecules come together at tight junctions. Curr. Opin. Cell Biol. 1999;11:628–633. doi: 10.1016/s0955-0674(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Tsunoda S., Sierralta J., Sun Y., Bodner R., Suzuki E., Becker A., Socolich M., Zuker C.S. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein–coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- van Hengel J., Gohon L., Bruyneel E., Vermeulen S., Cornelissen M., Mareel M., von Roy F. Protein kinase C activation upregulates intercellular adhesion of alpha-catenin–negative human colon cancer cell variants via induction of desmosomes. J. Cell Biol. 1997;137:1103–1116. doi: 10.1083/jcb.137.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J.L., Etemad-Moghadam B., Guo S., Boyd L., Draper B.W., Mello C.C., Priess J.R., Kemphues K.J. Par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development (Camb.) 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Willert K., Brink M., Wodarz A., Varmus H., Nusse R. Casein kinase 2 associates with and phosphorylates dishevelled. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:3089–3096. doi: 10.1093/emboj/16.11.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler B., Forscher P., Mellman I. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature. 1999;397:698–701. doi: 10.1038/17806. [DOI] [PubMed] [Google Scholar]
- Winckler B., Mellman I. Neuronal polaritycontrolling the sorting and diffusion of membrane components. Neuron. 1999;23:637–640. doi: 10.1016/s0896-6273(01)80021-0. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Grawe F., Knust E. Crumbs is involved in the control of apical protein targeting during Drosophila epithelial development. Mech. Dev. 1993;44:175–187. doi: 10.1016/0925-4773(93)90066-7. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Hinz U., Engelbert M., Knust E. Expression of Crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila . Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- Wu S.L., Staudinger J., Olson E.N., Rubin C.S. Structure, expression, and properties of an atypical protein kinase C (PKC3) from Caenorhabditis elegans. PKC3 is required for the normal progression of embryogenesis and viability of the organism. J. Biol. Chem. 1998;273:1130–1143. doi: 10.1074/jbc.273.2.1130. [DOI] [PubMed] [Google Scholar]
- Xu X.Z., Choudhury A., Li X., Montell C. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J. Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X., Chia W. Analysis of partner of Inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in Inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]





