Abstract
Autophagy is a membrane trafficking to vacuole/lysosome induced by nutrient starvation. In Saccharomyces cerevisiae, Tor protein, a phosphatidylinositol kinase-related kinase, is involved in the repression of autophagy induction by a largely unknown mechanism. Here, we show that the protein kinase activity of Apg1 is enhanced by starvation or rapamycin treatment. In addition, we have also found that Apg13, which binds to and activates Apg1, is hyperphosphorylated in a Tor-dependent manner, reducing its affinity to Apg1. This Apg1–Apg13 association is required for autophagy, but not for the cytoplasm-to-vacuole targeting (Cvt) pathway, another vesicular transport mechanism in which factors essential for autophagy (Apg proteins) are also employed under vegetative growth conditions. Finally, other Apg1-associating proteins, such as Apg17 and Cvt9, are shown to function specifically in autophagy or the Cvt pathway, respectively, suggesting that the Apg1 complex plays an important role in switching between two distinct vesicular transport systems in a nutrient-dependent manner.
Keywords: starvation, Cvt, yeast, rapamycin, phosphorylation
Introduction
Cell growth is tightly linked with the cell's perception of its nutritional environment. In particular, microorganisms, such as the yeast Saccharomyces cerevisiae, that spend most of their time in the stationary phase in the wild, require accurate responses to varying nutrient conditions for their survival. Therefore, yeast is a good model organism for the study of nutrient detection and response. Autophagy is one of several responses to nutrient starvation (Klionsky and Ohsumi 1999). A large number of cytoplasmic components are nonselectively enclosed within a double-membrane structure called an autophagosome, in which they are transported into the vacuole/lysosome to be degraded by resident hydrolases. Such turnover of a large amount of cytoplasm mediated by autophagy is essential for survival under nutrient-depleted conditions. We have isolated several genes essential for autophagy (termed APG) and have been investigating the function of the gene products (Tsukada and Ohsumi 1993; Funakoshi et al. 1997; Matsuura et al. 1997; Mizushima et al. 1998). Genetic and morphological analyses revealed that the degradative process of autophagy shares mechanistic components with the cytoplasm-to-vacuole targeting (Cvt) pathway (Harding et al. 1996; Scott et al. 1996; Baba et al. 1997), which is biosynthetic, delivering a resident hydrolase, aminopeptidase I (API), to the vacuole (Klionsky and Ohsumi 1999). On the other hand, autophagy and the Cvt pathway are distinct in many aspects. The two pathways appear to be regulated separately; the Cvt pathway is mainly observed under growing conditions, whereas autophagy is induced by starvation (Baba et al. 1997; Klionsky and Ohsumi 1999). Furthermore, Cvt vesicles and autophagosomes, the vesicles formed in the Cvt pathway and autophagy, respectively, are clearly different in size (Baba et al. 1997). Recently, the t-SNARE Tlg2 and Sec1-homologue Vps45 were found to be required for the Cvt pathway, but dispensable for autophagy, suggesting that these two pathways are mechanistically distinct (Abeliovich et al. 1999).
Tor is a phosphatidylinositol kinase-related kinase that promotes cell cycle progression in response to nutrient availability (Thomas and Hall 1997). The function of Tor appears to be conserved from yeast to mammals. Treatment with the immunosuppressant rapamycin, a specific inhibitor of Tor, induced cell cycle arrest at G0. Rapamycin-induced Tor inhibition also induces several characteristic phenotypes of starved cells. For example, accumulation of glycogen, repression of genes that are stimulated in growing cells, and stimulation of genes that are induced during starvation, all result from inhibition of Tor (Hardwick et al. 1999). Rapamycin mimics the starvation response even under nutrient rich conditions. Our laboratory has found that rapamycin induces autophagy in yeast (Noda and Ohsumi 1998). However, the molecular mechanism by which Tor negatively regulates autophagy remains to be determined. Here, we present evidence that Apg1, a protein kinase essential for autophagy, plays a pivotal role in induction of Tor-regulated autophagy.
Materials and Methods
Standard techniques were used for yeast manipulation (Kaiser et al. 1994). The strains used are listed in Table . Two-hybrid screening was performed as described previously (James et al. 1996). An NH2-terminally truncated (8 amino acid) form of APG1 open reading frame, subcloned into pGBD-C2 vector, was used as bait to screen a yeast genomic library, and interacting proteins were identified by DNA sequencing. A DNA fragment including the entire APG17 gene was cloned from yeast genomic DNA using PCR. The kinase-negative apg1 K54A mutation was obtained using the QuikChange site-directed mutagenesis kit (Stratagene).
Table 1.
Yeast Strains Used in This Study
| Strain | Genotype | Source |
|---|---|---|
| KA311A | Mata, his3 leu2 trp1 ura3 | K. Irie (Nagoya University, Nagoya, Japan) |
| BJ2168 | Mat a leu2 trp1 ura3 pep4-3 prb1-1122 prc1-407 | Y.G.S.C. (Yeast Genetic Stock Center) |
| TN125 | Mat a ade2 his3 leu2 lys2 trp1 ura3 pho8::pho8Δ60 | Noda et al. 1995 |
| CY5754 | Mat a ade2 his3 leu2 trp1 ura3 can1-100 tap42Δ::TRP1 pCEN(LEU2)[TAP42] | K. Arndt (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) |
| CY5755 | Mat a ade2 his3 leu2 trp1 ura3 can1-100 tap42Δ::TRP1 pCEN(LEU2)[tap42-11] | K. Arndt |
| JK9-3da | Mat a his4 leu2-3,112 trp1 ura3-52 rme1 GAL+ HMLa | M.N. Hall (University of Basel, Basel, Switzerland) |
| JH11-1c | JK9-3da TOR1-1 | M.N. Hall |
| YYK36 | KA311A apg1Δ::LEU2 | This study |
| YYK126 | TN125 apg1Δ::LEU2 | This study |
| YYK130 | TN125 apg13Δ::TRP1 | This study |
| YYK121 | TN125 apg17Δ::HIS3 | This study |
| YYK119 | KA311A apg13Δ::TRP1 | This study |
| YYK111 | KA311A apg17Δ::HIS3 | This study |
| YYK107 | KA311A cvt9Δ::URA3 | This study |
| PJ69-4A | Mat a his3-200 leu2-3,112 trp1-901 ura3-52 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | P. James (University of Wisconsin, Madison,WI) |
Antibody against Apg13 protein was raised against a glutathione S–transferase (GST)-Apg13 fusion protein. Specifically, a 1.9-kb BglII-XbaI fragment of the APG13 gene coding for 396 amino acids of Apg13p was subcloned into the vector pGEX-2T. Escherichia coli strain BL21 was transformed with the resulting plasmid. The expressed fusion protein was purified with glutathione-Sepharose 4B (Amersham Pharmacia Biotech). Expression and purification of the fusion protein was carried out according to the manufacturer's instructions. The purified fusion protein was used to immunize a rabbit by Shibayagi.
Immunoprecipitation, kinase assay, and immunodetection of NH2-terminally hemagglutinin (HA)-tagged Apg1 (HAApg1) were performed as described previously (Kamada et al. 1995). HAApg1 was immunoprecipitated with anti-HA mAb (16B12; BabCO), and an in vitro protein kinase assay was performed in the presence of γ[32P]ATP and myelin basic protein (MBP, substrate). The amount of immunoprecipitated HAApg1 was monitored by immunoblot. For coimmunoprecipitation experiments, yeast cells exponentially grown in YEPD medium were treated with zymolyase 100T (Seikagaku Kogyo) to generate spheroplasts. The resultant spheroplasts were treated with or without 0.2 μg/ml of rapamycin and broken by resuspending in lysis buffer (PBS, pH 7.4, 1 mM EDTA, 1 mM EGTA, 2 mM Na3VO4, 50 mM KF, 15 mM Na2H2P2O7, 15 mM P-nitrophenylphosphate, 20 μg/ml leupeptin, 20 μg/ml benzamidine, 10 μg/ml pepstatin A, 40 μg/ml aprotinin, 1 mM PMSF, and 0.5% Tween-20). Cell lysate was cleared by 10-min centrifugation at 6,500 g and 30-min incubation with protein G–Sepharose (Amersham Pharmacia Biotech). HAApg1 in the cleared cell lysate was bound to anti-HA mAb, and Apg13 was detected with anti-Apg13 antibody. The resultant immunoprecipitates were also analyzed by protein kinase assay and immunoblot with anti-HA.
For in vivo labeling of Apg13, cells (TFD13-W3) expressing APG13 were in vivo-labeled with 50 μCi of 35S (Tran35S, ICN) for 10 min, or 50 μCi of 32Pi overnight in SD medium, and transferred to YEPD or nitrogen-depleted medium SD(−N) for 1 h. Apg13 protein was immunoprecipitated following TCA precipitation. Immunoprecipitated Apg13 was treated with 5 U of alkaline phosphatase for 1 h at 30°C. Immunoprecipitated protein was analyzed by SDS-PAGE, followed by autoradiography.
Progression of autophagy was estimated by the increase of alkaline phosphatase activity in the cells expressing a cytosolic proform of the phosphatase protein (pho8Δ60p; Noda et al. 1995) with α-naphtyl phosphate as a substrate. Results were shown as means and errors calculated from three independent experiments. Maturation of vacuole-targeted precursor API was detected by immunoblot.
Results
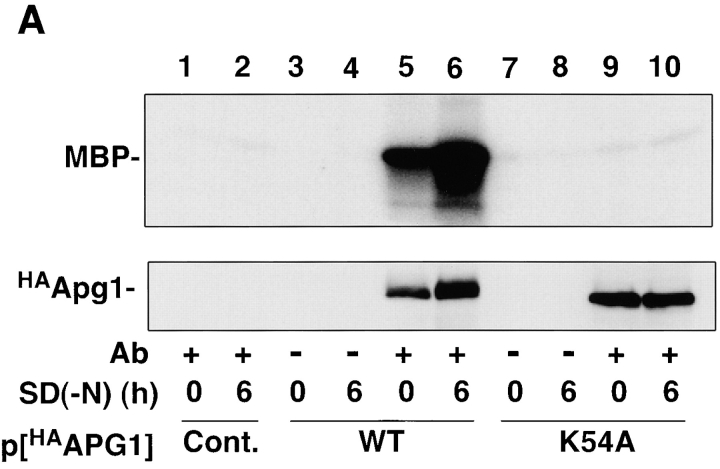
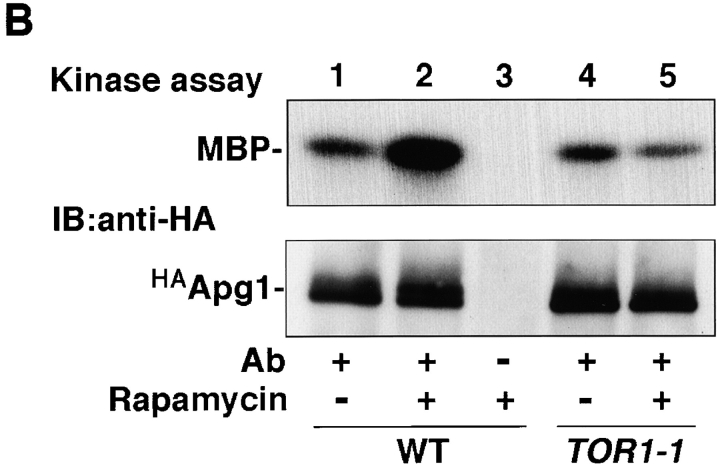
In an effort to study the mechanism of autophagy induction, we focused on the APG1 gene, which encodes a protein kinase whose activity is essential for autophagy (Matsuura et al. 1997). NH2-terminally HA-tagged Apg1 (HAApg1) was immunoprecipitated with anti-HA ascite and the resultant immunocomplex was analyzed using an in vitro kinase assay. Apg1 kinase activity was found to be highly elevated in cells grown under starvation conditions (Fig. 1 A). After a 6-h incubation in nitrogen-depleted medium, SD(−N), the amount of activated Apg1 had apparently increased, and was accompanied by slower gel migration, presumably because of autophosphorylation (Fig. 1 A, lane 6, bottom). The increase in Apg1 kinase activity is not due to this apparent increase in the protein amount, because shorter treatments with rapamycin (for example, see Fig. 1 B) resulted in Apg1 activation without an increase in the amount detected. Apg1 activity was also increased by rapamycin treatment, but the effect of rapamycin was abolished in a rapamycin resistant TOR1 mutant (TOR1-1; Kunz et al. 1993; Fig. 1 B). These results suggest that Apg1 activation is required for the induction of autophagy and that it is mediated by Tor proteins.
Figure 1.
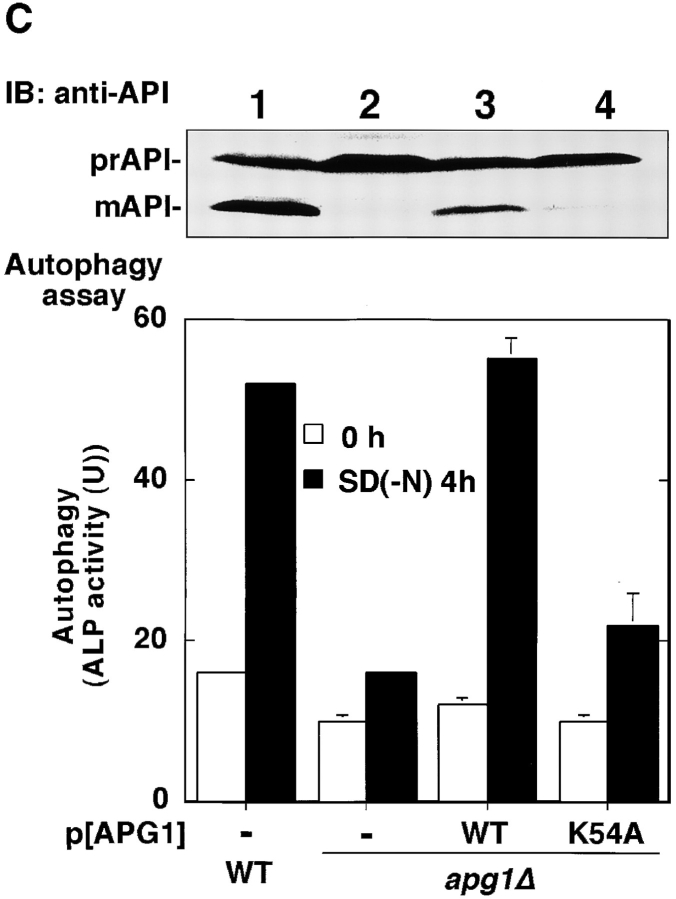
Apg1 is activated by starvation and rapamycin treatment. A, Apg1 is activated by starvation. Cells (KA311A, Cont.) expressing HA-tagged Apg1 (HAApg1, lanes 3–6) or kinase negative HAApg1K54A (lanes 7–10) grown in YEPD at 30°C were incubated in SD(−N) medium for 6 h. Immunoprecipitation of HAApg1 from the cell lysate and protein kinase assay were carried out as described in Materials and Methods (kinase assay, top). Immunoprecipitated HAApg1 was monitored by immunoblot (IB, bottom). B, Apg1 activation is mediated by Tor. Wild-type (WT, JK9-3da, lanes 1–3) or rapamycin-resistant TOR1-1 (JH11-1c, lanes 4 and 5) cells expressing HAApg1 grown in YEPD were treated with or without 0.4 μg/ml of rapamycin for 1.5 h. Apg1 kinase assay and immunodetection were performed. C, Kinase activity of Apg1 is required for both the Cvt pathway and autophagy. Wild-type (TN 125, lane 1) or apg1Δ cells (YYK126, lanes 2–4) transformed with wild-type APG1 (WT, lane 3) or kinase negative apg1 K54A (K54A, lane 4) plasmids were grown in YEPD and subjected to immunoblot (IB) with anti-API antibody (top). The precursor form of API in the cytosol (prAPI) and vacuolar-targeted and processed mature API (mAPI) are indicated in the panel. Progression of autophagy was estimated by monitoring the increase of alkaline phosphatase (ALP) activity (bottom).
A kinase-negative apg1 mutant (K54A; see Fig. 1 A) was defective in autophagy and the Cvt pathway (Fig. 1 C). This indicates not only that the enhanced Apg1 kinase activity is required for autophagy, but that basal Apg1 activity in growing cells (Fig. 1 A, lane 5) is essential for the Cvt pathway.
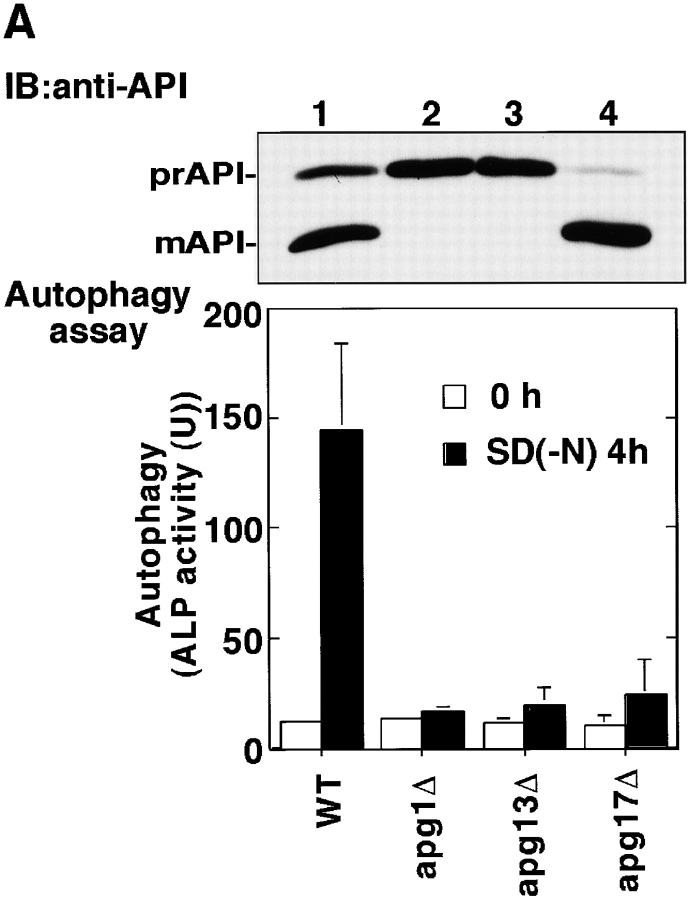
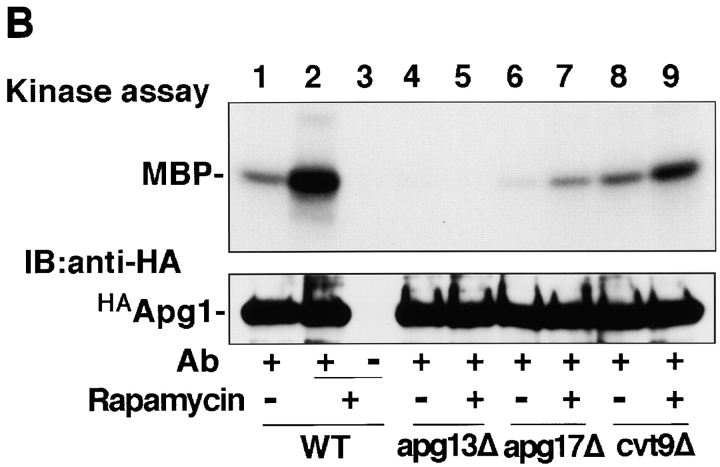
Next, we performed a two-hybrid screening with APG1 as bait to identify Apg1-associating proteins, which may regulate Apg1 activity. The following three genes were obtained from the screen: APG13 (Funakoshi et al. 1997) and two novel genes, which were subsequently found to be essential for either autophagy or the Cvt pathway, or both. One gene, designated as APG17 (YLR423c), was essential for only autophagy and was not required for the Cvt pathway (Fig. 2 A). The other, CVT9 (Harding et al. 1996; D.J. Klionsky, personal communication), was required for the Cvt pathway, but not for autophagy. Among the 16 APG genes discovered so far, APG17 is the first one identified whose function is restricted to autophagy. It is interesting to note that Apg1 binds to proteins whose function is specific to either autophagy (Apg17) or the Cvt pathway (Cvt9). Overexpression of APG1 in an apg13Δ mutant partially rescues the autophagy defect (Funakoshi et al. 1997). Similarly, the apg17Δ mutant was also rescued by overexpression of APG1 (data not shown), indicating that these three genes interact functionally. We tested whether these Apg1-associated proteins are involved in Apg1 activation. As shown in Fig. 2 B, deletion of each gene affected Apg1 activation by rapamycin treatment. In the apg13Δ mutant, attenuated Apg1 activity was observed. In rapamycin-treated apg17Δ cells, Apg1 activity also was found to be largely impaired (∼20% of the wild-type). On the other hand, deletion of CVT9, which is not needed for autophagy, resulted in rapamycin-induced activation of Apg1 to 50% of wild-type. The effects of deleting APG13 and APG17 on Apg1 activity are not the result of a general autophagy defect, because deletion of other APG genes, such as APG5 (Mizushima et al. 1998), does not affect the activation of Apg1 (data not shown). These results indicate that the activated state of Apg1 is required for autophagy induction, and that Apg13 and Apg17 play a key role in the activation of Apg1 in response to Tor inhibition.
Figure 2.
Apg1-associating proteins are required for Apg1 activation. A, Wild-type (TN125, lane 1), apg1Δ (YYK126, lane 2), apg13Δ (YYK130, lane 3), and apg17Δ (YYK121, lane 4) cells were analyzed with anti-API blot (top) and ALP assay (bottom). B, Wild-type (KA311A), apg13Δ (YYK119), apg17Δ (YYK111), and cvt9Δ (YYK107) cells expressing HAApg1 were treated with rapamycin (0.2 μg/ml, 1 h) and analyzed by kinase assay.
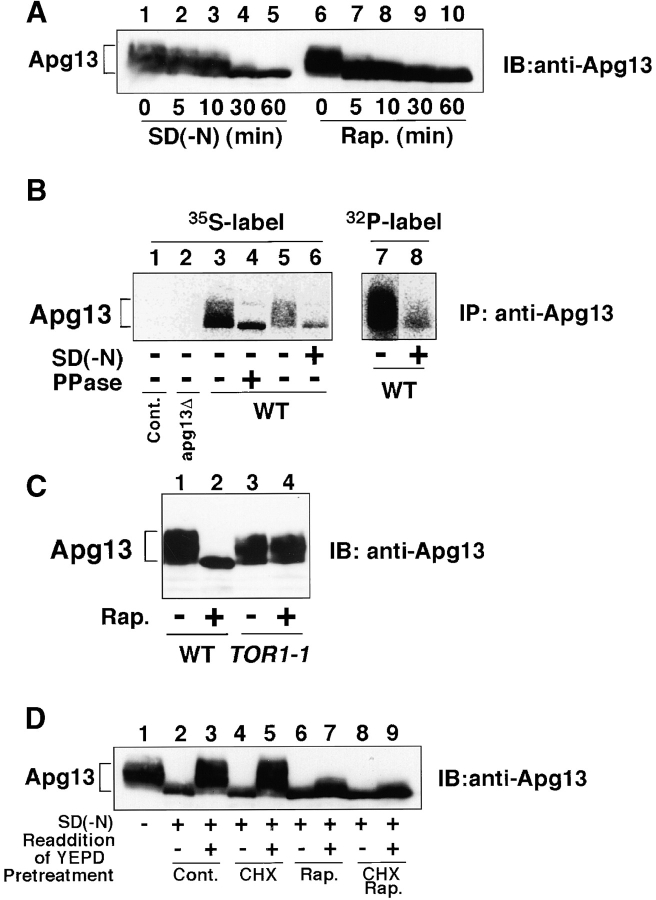
The next question we addressed was how these Apg1-associating proteins transmit the starvation signal from Tor to Apg1. To this end, the biochemical character of the Apg13 protein was investigated. Overexpression of APG13 resulted in a smeared Apg13 band on the immunoblot caused by retarded migration, indicating that it was modified in some way (Fig. 3 A). This modified form was observed only in growing cells, and after starvation or rapamycin treatment, the slower-migrating form disappeared. In particular, it was noted that the disappearance of the slower-migrating form occurred within five minutes after rapamycin treatment. In vivo labeling and in vitro phosphatase treatment revealed that the Apg13 bandshift was due to phosphorylation (Fig. 3 B). These results suggest that Apg13 is phosphorylated in a Tor-dependent manner, which was confirmed by the observation that dephosphorylation of Apg13 in response to rapamycin was not seen in TOR1-1 cells (Fig. 3 C). The dephosphorylated, faster-migrating form of Apg13 in starved cells was rapidly phosphorylated after readdition of YEPD medium (Fig. 3 D), suggesting that the phosphorylation state of Apg13 is extremely sensitive to nutrient conditions. This rephosphorylation is attenuated significantly by rapamycin, but is not affected by cycloheximide. This excludes the possibility that the phosphorylated form of Apg13 is degraded upon starvation and Apg13 is de novo synthesized after medium addition. The faster-migrating form of Apg13 (as well as the slower-migrating form) was found to be labeled with 32Pi (Fig. 3 B, lane 8), suggesting that Apg13 remains partially phosphorylated under starvation conditions.
Figure 3.
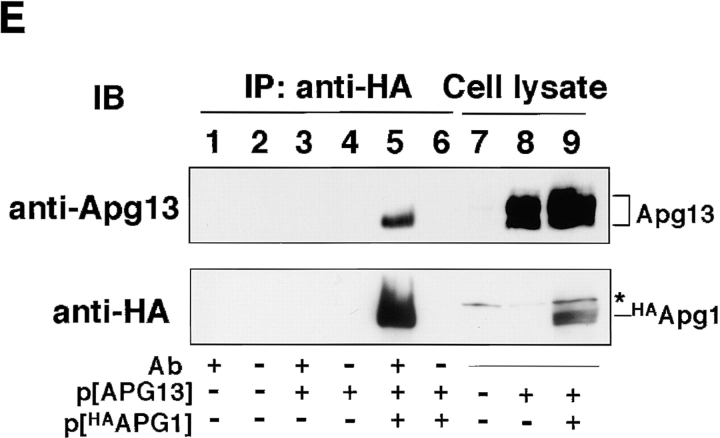
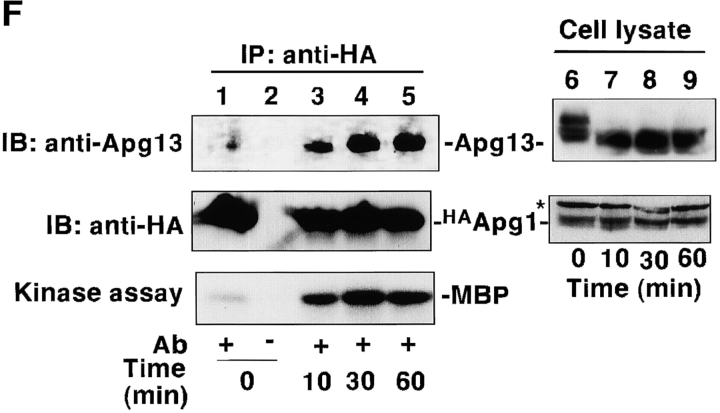
Tor-dependent phosphorylation of Apg13 inhibits Apg1–Apg13 association. A, Bandshift of Apg13 in response to starvation and rapamycin. YEPD-grown cells (BJ2168) expressing APG13 with YEp352[APG13] (lanes 1 and 6) were transferred to SD(−N) medium (lanes 2–5) or treated with 0.2 μg/ml rapamycin (lanes 7–10) and incubated for the indicated times. Total protein was analyzed by immunoblot using anti-Apg13 serum. B, Apg13 is hyperphosphorylated. Cells (TFD13-W3) overexpressing APG13 were labeled in vivo with 35S (lanes 1–6) or 32Pi (lanes 7 and 8), and shifted to YEPD (lane 5) or SD(−N) (lanes 6 and 8) for 1 h. Apg13 protein was immunoprecipitated and treated with alkaline phosphatase (PPase; lane 4). Immunoprecipitated protein was subjected to SDS-PAGE, followed by autoradiography. Cont, immunoprecipitation was carried out with preimmune serum. C, Tor-mediated phosphorylation/dephosphorylation of Apg13. Wild-type (WT, JK9-3da) or TOR1-1 (JH11-1c) cells expressing Apg13 grown in YEPD were treated with or without 0.2 μg/ml of rapamycin for 1 h. An immunoblot using anti-Apg13 serum was carried out. D, Phosphorylation/dephosphorylation cycle of Apg13 in response to nutrient conditions. YEPD-grown cells (BJ2168) overexpressing Apg13 (lane 1) were incubated in SD(−N) for 1.5 h (lane 2) with or without 30-min treatment with 10 μg/ml cycloheximide (lanes 4 and 5), 0.2 μg/ml of rapamycin (lanes 6 and 7), or both (lanes 8 and 9). Then, half a sample volume of 2× YEPD was added to the cells, after which they were incubated for 10 min (lanes 3, 5, 7, and 9). Apg13 protein was detected by immunoblot. E, Coimmunoprecipitation of Apg13 with Apg1. YEPD grown cells (BJ2168) overexpressing HAApg1 and/or Apg13 with high-copy plasmids pRS426[HA APG1] (p[HAAPG1]) and YEp351[APG13] (p[APG13]) were lysed, and HAApg1 was immunoprecipitated (IP) from the cell lysate. Apg13 and HAApg1 were detected by immunoblot (lanes 1–6). Apg13 and HAApg1 detected in total cell lysate are also shown (lanes 7–9). The hyperphosphorylated form of Apg13 remaining in the supernatant of the immunoprecipitate was still detected (data not shown), excluding the possibility that Apg1-bound Apg13 was dephosphorylated during the experiment. The asterisk shows a band that anti-HA ascite nonspecifically recognized in total lysate. F, Apg1–Apg13 association is promoted by rapamycin treatment. YEPD-grown cells (BJ2168) expressing HAApg1 and Apg13 with low-copy plasmids pRS316[HA APG1] and pRS315[APG13] were enzymatically converted to spheroplasts and treated with 0.2 μg/ml of rapamycin (at time 0). Spheroplasts were harvested at the indicated times, and coimmunoprecipitation was performed. Apg1 protein kinase assay of the immunoprecipitates was also performed (bottom).
To confirm that there is a physical association between Apg1 and Apg13, we carried out a coimmunoprecipitation experiment. A vacuolar protease-deficient strain was used for these experiments because Apg13 is quite labile in cell lysate. When HA APG1 and APG13 were expressed from a high-copy plasmid, only the faster-migrating form of Apg13 was detected in the Apg1 immunocomplex (Fig. 3 E), indicating that the hyperphosphorylated form of Apg13 has little affinity for Apg1. Next, we performed the experiment using a low-copy plasmid to more closely approximate physiological cellular conditions. The amount of Apg13 bound to Apg1 increased rapidly (as quickly as 10 min) after rapamycin treatment (Fig. 3 F), which corresponds well to the time course of Apg13 dephosphorylation. Apg1 activation was also observed to be tightly linked with this Apg1–Apg13 association. These results strongly indicate that Tor negatively regulates Apg1 kinase activity by means of (hyper)phosphorylation of Apg13, which reduces the affinity of Apg13 for Apg1.
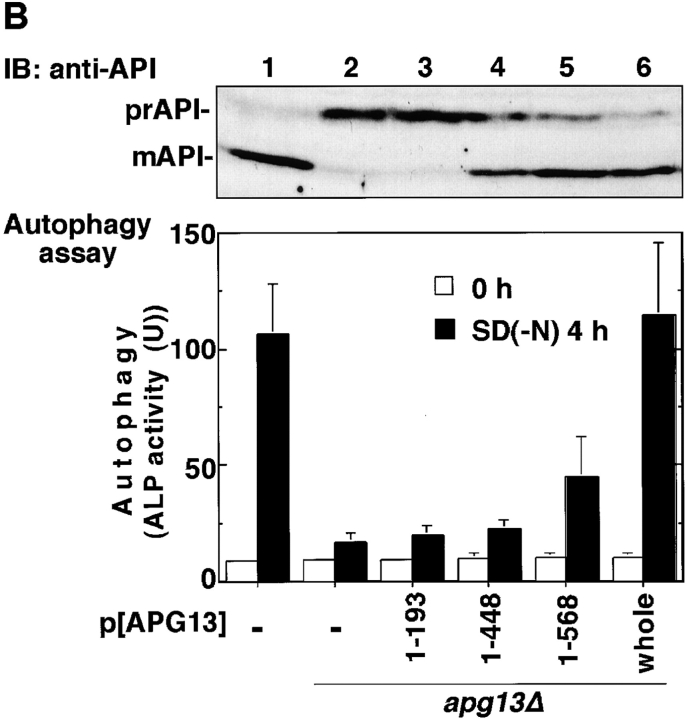
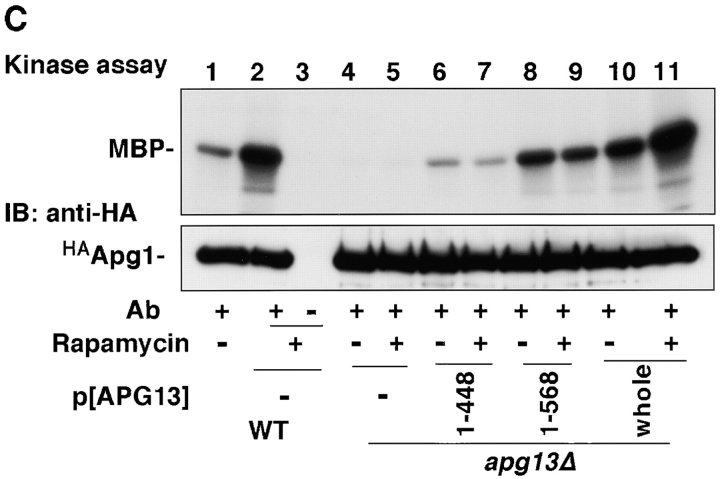
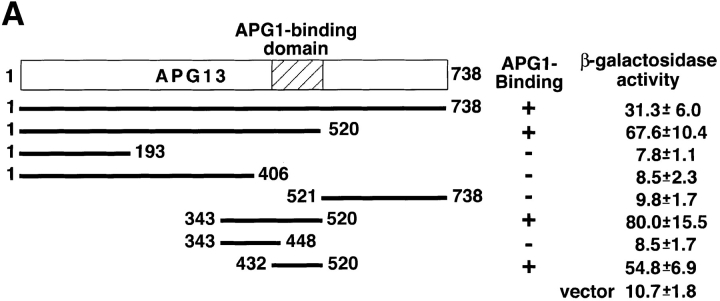
From these results, we hypothesized that Apg13 binding to Apg1 is required for the induction of autophagy, but not for the Cvt pathway. Therefore, we examined autophagy using apg13 mutants. The two-hybrid assay was used to determine the Apg1-binding site on Apg13, using various APG13 open reading frame fragments as prey. A central 89-amino acid region (432–520) was found to be responsible for Apg1 binding (Fig. 4 A). We also isolated a COOH-terminal truncated form of Apg13, Apg13(1–448), whose Ser449 was mutated to a stop codon using in vitro mutagenesis (Kaiser et al. 1994). This mutant has a mutation within the putative Apg1-binding site, and is unable to associate with Apg1, as confirmed by the two-hybrid assay and coimmunoprecipitation (Fig. 4 A, and data not shown). This result indicates a role for a domain of Apg13 around amino acid 448 in binding Apg1. We tested several COOH-terminal truncated Apg13 mutants including Apg13(1–448) for autophagy activity. Apg13(1–448) did not rescue the autophagic defect at all (Fig. 4 B, bottom), presumably due to the absence of the Apg1-binding domain. Another COOH-terminal truncated form, Apg13 (1–568), which contains the entire putative Apg1-binding domain, displayed partial, but significant, autophagic activity, when compared with Apg13(1–448). Apg13(1–568) was less competent for autophagic import than full-length Apg13, suggesting that additional sequences downstream of the Apg1 binding site are important for maximal activity. On the other hand, Apg13(1–448) partially, but significantly, rescued vacuolar-targeting of precursor API (Fig. 4 B, top), suggesting that this truncated protein is still functional for the Cvt pathway. Apg1 activity in the apg13Δ mutant was partially restored in the presence of either Apg13(1–448) or Apg13(1–568), and was fully recovered in the presence of the whole APG13 construct (Fig. 4 C). The kinase activity of the Apg13(1–568) transformant was clearly higher than that of the Apg13(1–448) transformant. These results confirmed the hypothesis that the Apg1–Apg13 association and subsequent activation of Apg1 are required for autophagy induction in response to starvation.
Figure 4.
Apg1–Apg13 association is required for autophagy, but not for the Cvt pathway. A, Apg1-binding site of Apg13. Interaction of the indicated Apg13 fragments (prey) with Apg1 (bait) in the yeast two-hybrid system. Binding activity was estimated by β-galactosidase activity (Kaiser et al. 1994, shown as means and errors calculated from three independent experiments). B, Apg1–Apg13 association is required specifically for autophagy. Wild-type cells (TN125) or apg13Δ cells (YYK130) harboring the indicated APG13 constructs grown in YEPD were analyzed by anti-API immunoblot (top). An ALP assay was performed after 4 h SD(−N) incubation (bottom). C, Apg1 kinase assay was performed using the above APG13 transformants before or after rapamycin (0.2 μg/ml, 1 h) treatment.
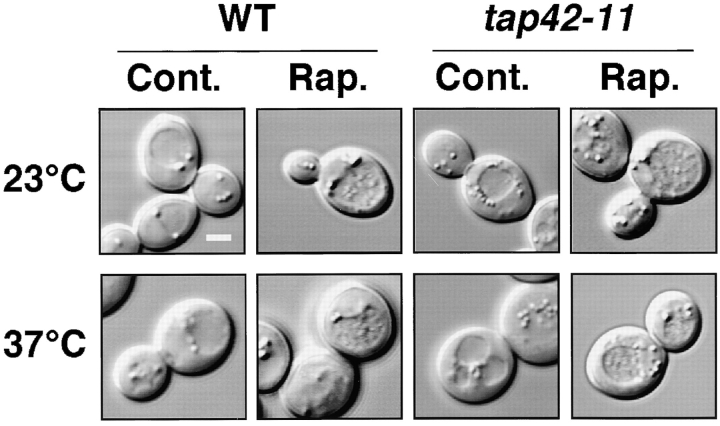
It is currently thought that Tor signaling in yeast is bifurcated (Thomas and Hall 1997). One pathway involves the small GTPase Rho1, which is responsible for actin organization and is not affected by rapamycin. The other involves Tap42, a rapamycin-sensitive phosphatase-associating protein that is necessary for the initiation of protein translation and amino acid permease turnover (Di Como and Arndt 1996). TAP42 is known to be located directly downstream of Tor, and is required for Tor-mediated signaling, especially the rapamycin-sensitive branch (Jiang and Broach 1999). To investigate this issue further, we tested the ability of a tap42 ts mutant (tap42-11; Di Como and Arndt 1996) to induce autophagy at a nonpermissive temperature. This mutation was previously found to confer rapamycin resistance at permissive temperatures. Accumulation of vacuolar autophagic bodies in tap42 cells was found to be comparable to that in wild-type cells, confirming that induction of autophagy is not controlled by Tap42 (Fig. 5). Furthermore, deletion of NPR1, whose product is a protein kinase negatively regulated by Tap42 (Schmidt et al. 1998), did not affect the induction of autophagy (data not shown). Therefore, we concluded that Tap42 does not transmit a signal from Tor in the autophagy induction pathway.
Figure 5.
Tap42 does not mediate the induction of autophagy. Autophagy is normal in tap42 ts cells. Wild-type (CY5754) and tap42-11 ts (CY5755) cells were incubated for 4 h at the indicated temperature with or without rapamycin (0.2 μg/ml) in the presence of 1 mM PMSF to visualize autophagic bodies. For incubation at the nonpermissive temperature, 1 h preincubation was carried out. DIC images were shown. Bar, 2 μm.
Discussion
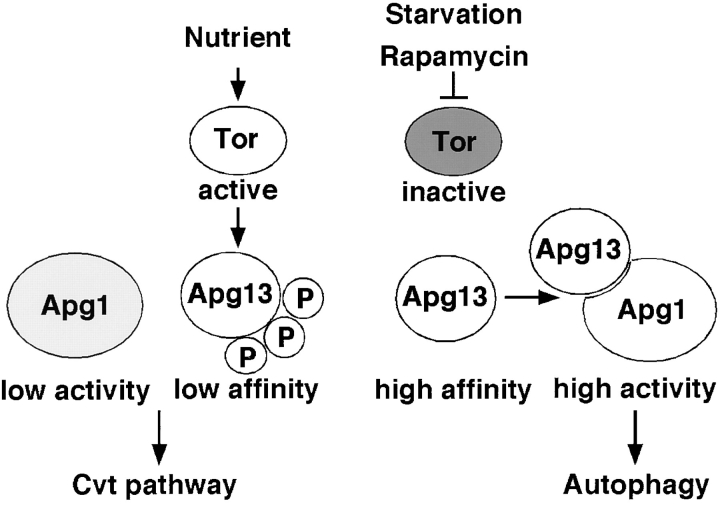
We have shown that Apg1 forms complexes with Apg13, Apg17, and Cvt9. The association between Apg1 and Apg13 is negatively regulated by Tor signaling. In nutrient-rich conditions, active Tor causes hyperphosphorylation of Apg13, which prevents its association with Apg1. Starvation or rapamycin treatment represses Tor activity, and Apg13 is immediately dephosphorylated. Dephosphorylated Apg13 possesses a high affinity for Apg1, which is activated upon Apg13 binding, leading to induction of autophagy. This tight Apg1–Apg13 association is required for autophagy, but not for the Cvt pathway, an observation that supports a model in which the Apg1–Apg13 complex plays an important role in switching from the Cvt pathway to autophagy in response to nutrient conditions (Fig. 6). The phenotypes of apg17Δ and cvt9Δ cells lend further support to this idea. The function of Apg17 is still unclear, but our preliminary results suggest that it may be involved in the Apg1–Apg13 interaction. When both Apg1 and Apg13 were overexpressed, Apg1–Apg13 binding was observed, even in cells grown in YEPD (Fig. 3 D), resulting in a small amount of Apg1 activation, insufficient to induce autophagy (e.g., see Fig. 4 C, lanes 1 and 10). Presumably, additional factors such as Apg17 are needed for full activation of Apg1.
Figure 6.
Model of Tor-mediated regulation between the Cvt pathway and autophagy. See text for details.
Protein sorting to the plasma membrane and vacuolar degradation of amino acid permeases, Gap1 and Tat2, are also regulated in response to nutrient conditions, and are dependent upon Tor, Npr1, and Tap42 (but not on Apg1; Roberg et al. 1997; Beck et al. 1999). Tap42 plays a key role in the rapamycin-sensitive Tor pathway, suppressing Npr1 activity (Schmidt et al. 1998). Our observation that Tor regulation of Apg1 activity and autophagy induction is rapamycin-sensitive, but Tap42-independent, implies that the Apg1–Apg13 interplay comprises a novel Tor signaling pathway regulating autophagy induction. It is possible that Tor directly phosphorylates Apg13, but this remains to be investigated. Recently, nuclear transport of transcription factor Gln3 has been shown to be under the control of Tor signaling, but the involvement of Tap42 in this system is still controversial (Beck and Hall 1999; Cardenas et al. 1999). Deletion of GLN3 did not affected autophagy (data not shown). In mammalian cells, two targets of mammalian Tor (mTOR) have been identified: 4E-BP1 and p70S6K (Thomas and Hall 1997). Another recent study reported a relationship between mammalian Tor and autophagy (Shigemitsu et al. 1999). APG1 homologues of Caenorhabditis elegans (Ogura et al. 1994) and mammals (Yan et al. 1998) have also been reported, suggesting that the Tor–Apg1 pathway might be conserved in eukaryotic cells.
Acknowledgments
We thank K.T. Arndt, M.N. Hall, and P. James for strains and plasmids, D.J. Klionsky for anti-API antibody, communicating results before publication, and critical reading of this manuscript, K. Fujimura-Kamada, K. Matsumoto, and members of Ohsumi's laboratory for helpful discussions, and NIBB Center for Analytical Instruments for technical assistance.
This work is partly supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Culture and Sports of Japan.
Footnotes
Yoshiaki Kamada and Tomoko Funakoshi contributed equally to this work.
Tomoko Funakoshi's present address is Department of Biochemistry, College of Pharmacy, Nihon University, Funabashi, Chiba 274-8555, Japan.
Abbreviations used in this paper: API, aminopeptidase I; Cvt, cytoplasm-to-vacuole targeting; HA, hemagglutinin.
References
- Abeliovich H., Darsow T., Emr S.D. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE–Sec1p complex composed of Tlg2p and Vps45p. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Ohsumi M., Scott S.V., Klionsky D.J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Hall M.N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Beck T., Schmidt A., Hall M.N. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M.E., Cutler N.S., Lorenz M.C., Di Como C.J., Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como C.J., Arndt K.T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Funakoshi T., Matsuura A., Noda T., Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae . Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Harding T.M., Hefner-Gravink A., Thumm M., Klionsky D.J. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Hardwick J., Kuruvilla F.G., Tong J.K., Shamji A.F., Schreiber S.L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. Genomic libraries and a host strain designated for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Broach J. Tor proteins and protein phosphatase 2A reciprocally regulated Tap42 in controlling cell growth in yeast. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- Kamada Y., U.S. Jung, J. Piotrowski, D.E. Levin The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N.R., Hall M.N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y., Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae . Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae . Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Ogura K., Wicky C., Magnenat L., Tobler H., Mori I., Muller F., Ohshima Y. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- Roberg K.J., Rowley N., Kaiser C.A. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae . J. Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Beck T., Koller A., Kunz J., Hall M.N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Hefner-Gravink A., Morano K.A., Noda T., Ohsumi Y., Klionsky D.J. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemitsu K., Tsujishita Y., Hara K., Nanahoshi M., Avruch J., Yonezawa K. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J. Biol. Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- Thomas G., Hall M.N. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae . FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Yan J., Kuroyanagi H., Kuroiwa A., Matsuda Y., Tokumitsu H., Tomoda T., Shirasawa T., Muramatsu M. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem. Biophys. Res. Commun. 1998;246:222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]