Abstract
Cdc25 phosphatases are required for eukaryotic cell cycle progression. To investigate mechanisms governing spatiotemporal dynamics of cell cycle progression during vertebrate development, we isolated two cdc25 genes from the zebrafish, Danio rerio, cdc25a and cdc25d. We propose that Zebrafish cdc25a is the zebrafish orthologue of the tetrapod Cdc25A genes, while cdc25d is of indeterminate origin. We show that both genes have proliferation promoting activity, but that only cdc25d can complement a Schizosaccharomyces pombe loss of function cdc25 mutation. We present expression data demonstrating that cdc25d expression is very limited during early development, while cdc25a is widely expressed and consistent with the mitotic activity in previously identified mitotic domains of the post-blastoderm zebrafish embryo. Finally, we show that cdc25a can accelerate the entry of post-blastoderm cells into mitosis, suggesting that levels of cdc25a are rate limiting for cell cycle progression during gastrulation.
Keywords: zebrafish, cell cycle, Cdc25, mitotic domain, mitosis
Introduction
Over the last quarter-century, our understanding of the mechanisms that govern the cell division cycle has advanced rapidly. Studies in yeast and cultured cells have identified critical regulators of cell cycle progression and have provided detailed mechanisms underlying cell cycle control. However, the manner in which these factors interface with the genetic program of vertebrate development, although critical to a complete understanding of developmental processes is less clear, and requires analysis in intact embryos.
Early work in the yeast Schizosaccharomyces pombe identified many of the factors involved in eukaryotic cell cycle regulation (Nurse et al., 1976; Nurse and Thuriaux, 1980; Nasmyth and Nurse, 1981). In S. pombe, cell cycle progression is regulated in G2 by the Cdk1 kinase, the prototypic member of the cyclin-dependent kinase class of proteins (Simanis and Nurse, 1986). Cdks are regulated by the periodic availability of their obligate cyclin cofactors and by reversible inhibitory phosphorylation at adjacent threonine and tyrosine residues near the N-terminus (Gould and Nurse, 1989). This inhibitory phosphorylation is mediated by the Wee and Myt kinases (Gould and Nurse, 1989) and reversed by the dual-specificity phosphatase Cdc25 (Russell and Nurse, 1986; Gautier et al., 1991).
Studies in Drosophila have established the importance of Cdc25 in coordinating cell proliferation with developmental events. Early syncitial cell cycles are driven by maternally provided mRNA and protein from the Drosophila cdc25 homolog, string (Edgar and Datar, 1996). After the maternal-zygotic transition (MZT, cycle 10), maternal string mRNA is degraded (Edgar and Datar, 1996) and spatio-temporally patterned zygotic string expression begins (Edgar and O’Farrell, 1990). Expression of zygotic string mRNA anticipates mitoses in the early post-blastoderm cell cycles, and the highly stereotyped mitotic domains of the early Drosophila embryo correlate tightly with domains of string expression (Edgar and O’Farrell, 1990). Strong hypomorphic mutations in the Drosophila string gene cause an arrest in G2 of cycle 14 (Edgar and O’Farrell, 1990), suggesting a requirement for this protein in continued mitotic divisions during development. Further studies have shown that expression of string mRNA is directly controlled by patterning genes and that correct embryonic development requires precise control and integration of string expression and cell proliferation with patterning events (Edgar, 1994). Expression of a second Drosophila cdc25 homologue, twine, is restricted to the germline, and loss-of-function mutations in this gene cause both male and female sterility (Edgar and Datar, 1996).
Tetrapod vertebrates posses three Cdc25 genes, designated Cdc25A, Cdc25B and Cdc25C. The developmental role of these genes, however, remains unclear. Cdc25B knockout mice are largely phenotypically normal, although females are sterile, suggesting that this gene may play a role in female germ cell formation (Lincoln et al., 2002). Both Cdc25C (Chen et al., 2001) knockout and Cdc25B/C double knockout (Ferguson et al., 2005) mice develop normally, suggesting that either these phosphatases are dispensable for embryonic development or that Cdc25A can functionally compensate for loss of Cdc25B and C. The inaccessibility of the early mouse embryo and the difficulty of experimental manipulation in this model has made elucidation of the developmental roles of Cdc25 genes problematic.
The role of Cdk phosphoregulation in vertebrate development has been most extensively investigated in the amphibian Xenopus laevis. As in Drosophila, maternal Cdc25A appears to be important in early cleavage phases and is degraded rapidly at the midblastula transition (MBT) (Kim et al., 1999; Shimuta et al., 2002). Later spatio-temporal regulation of expression of the inhibitory kinase Wee1 is critical for development (Leise and Mueller, 2002); disruption of Wee 1-mediated inhibition of cell proliferation by morpholino knockdown results in gastrulation defects (Murakami et al., 2004; Leise and Mueller, 2004). However, the extent to which the multiple Xenopus Cdc25 isoforms contribute to cell cycle progression and their potential role in coupling of cell proliferation and patterning during Xenopus development remains unclear.
In recent years, the zebrafish has emerged as a model for studies of vertebrate development. Despite the range of genetic tools available in this organism, including reverse and forward genetic techniques and a wide array of mutations in developmentally important genes, very little is known about the presence, expression and function of cell cycle regulatory genes and the placement of these cell cycle controls in the genetic hierarchy of development. Here we identify two zebrafish cdc25 genes, cdc25a and cdc25d, and show that they are differentially expressed during early development, but neither transcript shows a dramatic redistribution at or shortly after the MBT within the embryonic cells. We conclude that cdc25a is a paralogue of the tetrapod Cdc25a gene, while the relationship between cdc25d and vertebrate Cdc25 genes is unclear. Furthermore, we demonstrate that ectopic cdc25a mRNA expression can induce deep cells of mid-gastrula zebrafish embryos to rapidly enter mitosis, suggesting that Cdc25 activity is rate-limiting for cell cycle progression in these cells.
Materials and Methods
Fish husbandry
Adult zebrafish (Danio rerio) were maintained at 28°C as described in Westerfield (2000). Embryos were collected from natural mating and staged according to (Kimmel et al., 1995). All experiments were carried out in the DZ strain.
cDNA library screening and phylogenetic reconstruction
A mid-gastrula cDNA library (Sagerstrom et al., 2001) was plated to a density of 5000 colonies/200mm plate. These colonies were transferred to nitrocellulose filters, lysed using NaOH and crosslinked to the filter by UV irradiation. 32P radiolabelled probes for cdc25a and cdc25d were synthesized by PCR followed by nick-end labelling (Takara Biosciences). Filters were incubated overnight with appropriate probes, washed and then exposed to film. Positive colonies were selected and plasmid DMA was isolated and sequenced using standard techniques. RACE was performed using the SMARTRACE kit (Clontech) following the manufacturer’s directions. Protein sequence alignments were performed using ClustalX (Higgins and Sharp, 1988) and phylogenetic analysis of full-length protein sequences was performed in MEGA version 3.1 (Kumar etal., 2004).
In Situ Hybridization
Whole-mount in situ hybridization of wild-type embryos was carried out as described previously (Sagerstrom et al., 1996). Templates for zebrafish cdc25a and cdc25d in situ probe transcription were prepared by cloning the complete open reading frames into pBluescript KS+ followed by linearization with either Xho1 (antisense) or Xba1 (sense) (New England Biolabs). Transcription was carried out using either T3 (Promega) or T7 (New England Biolabs) RNA polymerase in the presence of Digoxigenin labelling mix (Boehringer-Mannheim).
RT-PCR
Total RNA was extracted from collections of five embryos each at 256 cells, 512 cells, 1K cells, high and sphere stages using Trizol reagent (Ambion). cDNA synthesis was performed with the superscript III RT (Invitrogen) using random hexamer primers. Standard PCR reactions were performed for each stage, including a RT-minus control cDNA reaction using NovaTaq (Novagen) and an Eppendorf Mastercycler PCR machine. Aliquots were removed at 18, 21, 24, 27 and 30 cycles and analyzed by agarose gel electrophoresis. The following primers were used to amplify cdc25a: AACACGCCTGTCCGAGTGAAGAGG and CTCTAACGTAGCGACACATCCGTGG, amplifying a fragment of approximately 500bp, as well as previously described primers specific for Ornithine Decarboxylase (ODC) (Draper et al., 2001).
Yeast complementation assay
For expression in S. pombe, wild type zebrafish cdc25a and cdc25d genes were amplified by PCR with primers ATGGATATTGATATGGTTCCAGGCG and TCAGAGTTTTTTGAGACGGCTGTAC (cdc25a) and ATGGCCGGTGATGCGCTAGAGG and TTATCGTTTTGTTCTGTGATGTC (for cdc25d) and subcloned into the S. pombe pREP3x expression vector (Forsburg, 1993) in which transcription is regulated by the thiamine repressible nmt1 gene promoter (Maundrell, 1990). Phosphatase-dead mutant versions cdc25aC470S and cdc25dC295S were constructed using the Quickchange site directed PCR mutagenesis kit (Promega). The S. pombe cdc25 construct was previously described (Wolfe and Gould, 2004). The S. pombe cdc25-22 leu1-32 h temperature sensitive strain (Nurse et al., 1976; Russell and Nurse, 1976) was grown to mid-log phase and transformed using Merlin Core Services™ EZ-Yeast Transformation kit.
Cells were grown on Edinburgh Minimal Medium (EMM) (Moreno et al., 1991). Colony growth and cell viability were monitored using the vital dye Phloxine B (PB) (Sigma), which accumulates in dead cells resulting in a dark pink color.
Cells were fixed in formaldehyde (Moreno et al., 1991), mounted on slides with Vectashield Mounting Medium with DAPI (Vector Laboratories) to visualize the DNA, examined using a Zeiss Axioskop fluorescence microscope and photographed with a DVC 1300 Black and White CCD camera using QED software (Media Cybernetics).
Heat-shock overexpression and immunostaining
A 1.5 kb Spel-EcoRV fragment containing the zebrafish hsp70 promoter was taken from pzHSP70/4prom, described in Halloran et al. (2000). This was subcloned into the Nhe1-EcoRV sites of Tol2MCS.2, a gift of Maria Dorsett and Steve Johnson (Washington University, St. Louis), and is derived from the Tol2 transposon described in Kawakami et al. (2004). cdc25a and cdc25d and their phosphatase-inactive mutant forms were cloned into this vector fused to a N-terminal 6-myc tag. Comparing the activity of tagged and untagged protein expressed from mRNA injected during cleavage stages suggested that the Myc-tag does not impair function of either Cdc25 protein (Dalle Nogare et al., manuscript in preparation). DNA was purified before injection by phenol-extraction/ethanol precipitation followed by purification on a QIAQuick PCR-purification column (Qiagen) and diluted to 20ng/μL in ddH2O/phenol red. This solution was mixed 1:1 with 50ng/μL transposase mRNA to yield a final injection solution of 10ng/μL plasmid with 25ng/μL transposase mRNA. 1nL of this solution was injected into a single blastomere at the 8-cell stage using a Harvard Apparatus PLI-90 microinjector. Embryos were kept at 28°C in E3 media until shield stage, at which point they were moved to a 37°C air incubator for 30 minutes, followed by a 20-minute recovery period at 28°C and fixation in 4% paraformaldehyde in PBS. Immunostaining was carried out using the 9E10 anti-myc monoclonal antibody (Roche, 1:500) and polyclonal anti-phosphohistone H3 antibody (upstate biotech, 1:200). Embryos were incubated with primary antibodies overnight at 4°C followed by extensive washes in PBT. Embryos were then incubated in Alexafluor secondary antibodies (goat anti-mouse 546 and goat anti-rabbit 488, Invitrogen) at a1:500 dilution for 2 hours at room temperature. Embryos were then washed in PBT containing 1:1000 Hoechst 33342 (Invitrogen) and mounted in Vectashield (Vector laboratories).
Mitotic index calculation
Confocal stacks (10 planes at 1μm spacing) were acquired on a Zeiss LSM510 confocal microscope using a 40X objective lens. Images were imported into Imaged (Wayne Rasband, NIH) and quantification of total nuclei in the field was performed using the 3D Object Counter Plugin (Fabrice Cordelières, Institut Curie). A typical field was 200–300 cells, of which 15% were Myc-positive. Phosphohistone and Myc positive cells were manually assigned and counted. The mitotic index of transgene expressing cells was calculated by dividing the number of myc-positive/PH3-positive nuclei by the total number of myc-positive nuclei. The mitotic index of non-expressing cells was calculated by dividing the number of myc-negative/PH3-positive nuclei by the total number of Myc-negative nuclei. To ensure consistency, only PH3-positive nuclei were counted; in the rare cases where mitotic figures were observed that were not positive for the phosphohistone antigen (late telophase), these cells were not considered in mitosis.
Results
Cloning and phylogenetic characterization of two zebrafish cdc25 homologues
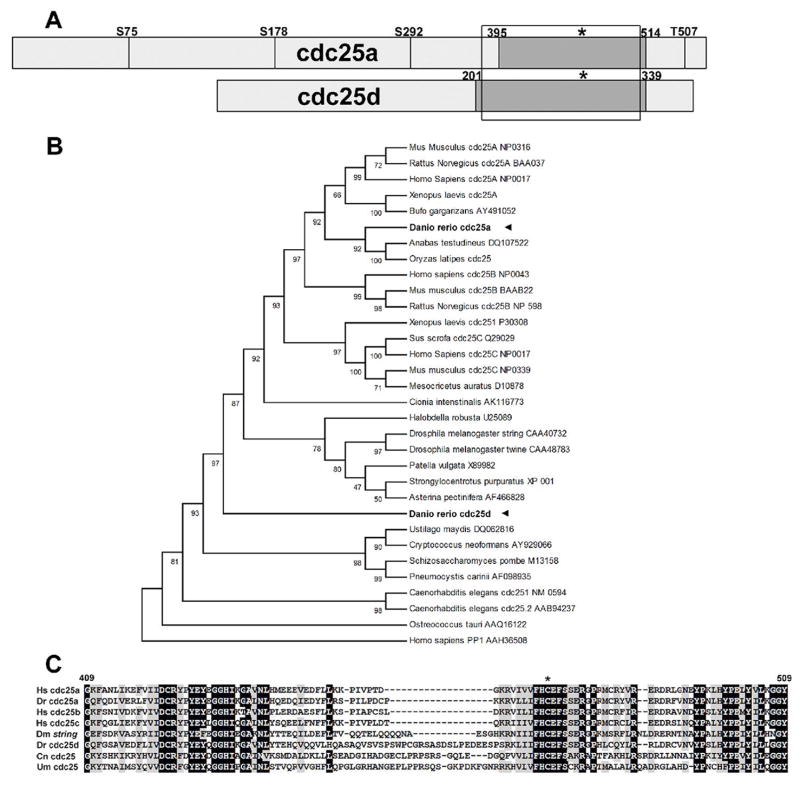
In order to clone cdc25 homologues from Danio rerio, a BLAST search of the NCBI NR database, as well as the partially completed zebrafish genome was performed using various vertebrate Cdc25 genes. These searches revealed the presence of two distinct cdc25 genes in the zebrafish. To determine the full cDNA sequences of these genes, we conducted a hybridization screen of a mid-gastrula zebrafish cDNA library using probes designed against the cdc25 fragments found in the database. From this screen we recovered 10 cDNAs representing a first isoform of cdc25 and three cDNA’s representing a second isoform. To confirm that the cDNA sequences recovered were full length, we performed 3′ and 5′ RACE on mRNA from mid-gastrula embryos. The more abundant cdc25 isoform cDNA maps to chromosome 13 and is 3042bp in length containing a 1692bp (563 amino acid) open reading frame, with a 290bp 5′ and a 1060bp 3′ UTR. A minority of these cDNAs (2/10) contained a truncated 3′ UTR (157bp), however the biological significance of this remains unknown. The second, smaller cDNA maps to chromosome 1 and is 1830bp in length, containing an 1160bp (386 amino acid) open reading frame (Fig. 1a).
Figure 1.
a: Schematic of cdc25a and cdc25d from zebrafish, to scale. Conserved Chk1 phosphorylation target sites from human Cdc25A are shown as vertical lines, labeled according to the residue in human Cdc25A. Boxed region shows alignment in figure 1c and dark box delineates cdc25 phosphatase domain as defined by NCBI conserved domain search. Asterisk indicates position of HCXXXXXR catalytic motif. b: UPGMA phylogenetic reconstruction of cdc25 genes from various species. Accession numbers used are noted in the figure. c: Alignment of the phosphatase domain of Cdc25 proteins: Dr Danio rerio Dm Drosophila melanogaster Hs Homo sapiens Cn Cryptococcus neoformans Um Ustilago maydis. Accession numbers are as in panel a.
To determine the evolutionary relationship of these two zebrafish cdc25 genes to other known members of this gene family, we conducted phylogenetic reconstruction using the full-length protein sequences of both zebrafish cdc25 genes along with a variety of cdc25 genes from both vertebrate and invertebrate lineages (Fig. 1b). The results of this reconstruction suggested that the larger 563 amino acid isoform falls within the cdc25a group when compared with mouse, human, rat and Xenopus genes and thus we have designated this isoform cdc25a. This placement is supported by the presence of conserved serine residues known to be targets of the Chk1 kinase (S178 and T507 in human cdc25a, S213 and T546 in zebrafish cdc25a, Chen et al 2003). T546 is unique to mammalian A-type Cdc25 proteins, while S178 is unique to A- and B-type Cdc25 proteins. The conservation of these and other Chk1 phoshorylation sites (Goulodina et al., 2003; Sorensen et al., 2003; see Fig. 1a) suggest that zebrafish Cdc25a may be subject to similar regulatory mechanisms as the human Cdc25a.
The second, less abundant, and more divergent cdc25 isoform is placed basal to the mammalian and Xenopus Cdc25ABC group and the invertebrate Drosophila/echoniderm/mollusc group. Clear orthologues of this gene cannot be found by BLAST in other fish species, including within the completely sequenced genomes of the pufferfish Takifugu rubripes and Tetraodon nigraviridis or within the partially sequenced medaka (Oryzias latipes) genome, all of which only appear to contain a single gene orthologous to zebrafish cdc25a. To reflect this divergence from any known vertebrate cdc25 isoforms, we have designated this gene cdc25d.
Interestingly, cdc25d contains a 19-residue insertion near the active site that is not seen in other vertebrate cdc25 genes (see alignment in Fig. 1c). Insertions at this position are present in two described cdc25 genes from the Basidiomycete fungi Cryptococcus neoformans and Ustilago maydis and, although shorter in length, string from Drosophila melanogaster. We have no evidence to suggest that these insertions are homologous.
Cdc25a and Cdc25d have proliferation-promoting activity in S. pombe
In order to confirm that both genes are functional cdc25 homologues, we assayed the ability of each gene to rescue a conditional mutation in the fission yeast Shizosaccharomyces pombe. The S. pombe cdc25-22 mutant is viable and essentially wild-type in growth rate and cell morphology at 25°C, but fails to grow at 34°C (Russell and Nurse, 1986; compare Fig. 2 panels f and I with r). The cdc25-22 mutant will grow at 34°C when housing a functional cdc25 gene on a plasmid, and for a positive control cells were transformed with fission yeast cdc25+Sp (Russell and Nurse, 1986) in plasmid pREP1 (Maundrell, 1993; Fig 2q). We used the pREPSx plasmid, containing the nmtl promoter to expresses zebrafish cdc25 genes at low levels in the presence of thiamine and at much higher levels in the absence of thiamine (Forsburg, 1993). Strains carrying the cdc25d gene grew at both the restrictive and permissive temperature (Figs. 2i, o). This rescue is dependent on the phosphatase activity of cdc25d, as strains carrying phosphatase-dead cdc25dC295S were unable to grow at the restrictive temperature (Fig. 2p). However, neither wild type cdc25a (Fig. 2m) nor the phosphatase dead mutant cdc25aC470S (Fig. 2n) were able to rescue cdc25-22. Interestingly, cells transformed with cdc25a plasmid grown at the permissive temperature in the absence of thiamine (high pREP3X promoter activity) also grew poorly (compare Figs. 2a and g), suggesting that expression of cdc25a is toxic.
Figure 2.
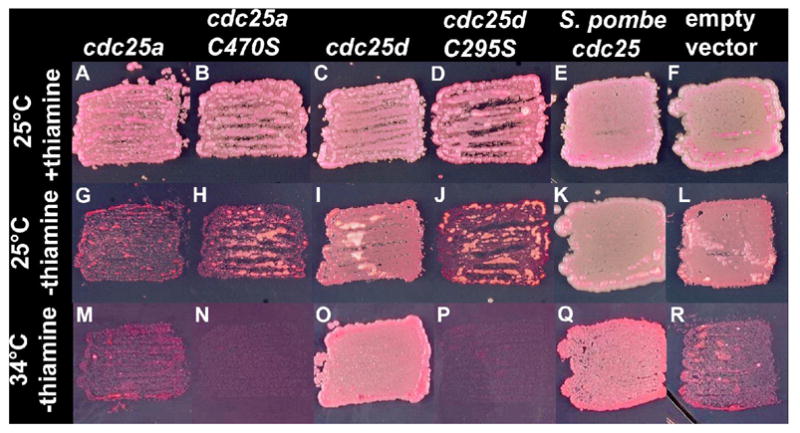
Rescue of the cdc25-22 mutation in S. pombe by various cdc25 constructs. All experiments were performed in the cdc25-22 background. Growth conditions are indicated at left and all genes expressed from the nmt1 promoter are noted above.
To determine the nature of cdc25a toxicity, we examined the morphology of cells at the permissive temperature in the presence and absence of thiamine. Cells expressing cdc25a (Fig. 3b) at low levels were shorter than control cells (Fig. 3a). The short size indicates rapid progression through the cell cycle, and is characteristic of wee mutants (Nurse, 1975), which are deficient in phosphoinhibition of Cdk1 activity. The phenotype is more severe in the absence of thiamine (compare Figs. 3c and d). Specifically, the presence of small binucleated cells indicates that this strain was entering mitosis prematurely, suggesting that the failure to rescue results from excessive activity of cdc25a. Multiple attempts achieve cdc25a activity levels sufficient for rescue, but without toxicity were unsuccessful. However, these data are consistent with the conclusion that both zebrafish cdc25 genes have proliferation-promoting activity in S. pombe cells.
Figure 3.
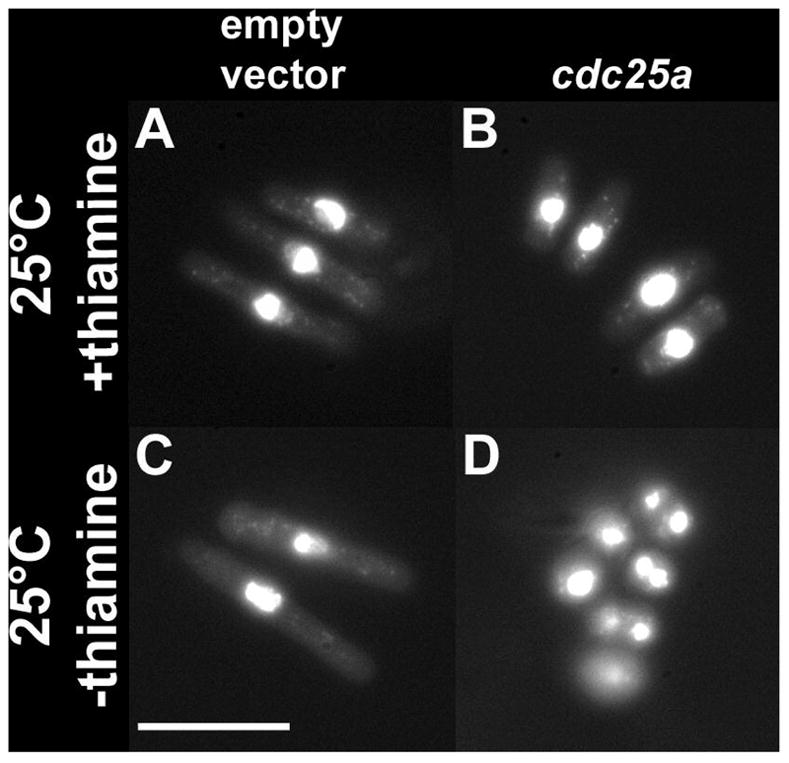
Morphology of fixed DAPI stained S. pombe cdc25-22 cells transformed with cdc25a or empty vector. Growth conditions are as indicated at left; All panels are to same scale. Scale bars are 10μm.
cdc25a and cdc25dare expressed during early development
To determine the developmental expression of the zebrafish cdc25 homologues, we performed in situ hybridization on fixed embryos. The results of this analysis are shown in Figures 4 and 5. cdc25a is present maternally (Fig. 4a) and very little change in expression is observed at the MBT. Shortly after the MBT, three mitotic domains emerge. The extraembryonic yolk syncitial layer (YSL) and enveloping layer (EVL) cease or delay mitotic proliferation. The deep layer gives rise to all of the embryonic tissue and continues to proliferate rapidly (Kane et al., 1992). cdc25a transcripts are observed in all deep cells (Fig. 4b), but not in the YSL (Fig. 4c) or EVL (Fig. 4d), which represent separate mitotic domains. Expression is maintained throughout early epiboly (Fig. 4e, f), decreasing toward the end of epiboly (Fig. 4g). To further test whether maternal cdc25a mRNA is degraded at the MBT, we performed semi-quantitative RT-PCR on mRNA extracted from staged pools of embryos around the MBT. The results of this analysis (Fig. 4j and k) suggested that cdc25a mRNA is not globally degraded at the MBT as in Drosophila, although we cannot preclude destruction followed by extremely rapid zygotic resynthesis.
Figure 4.
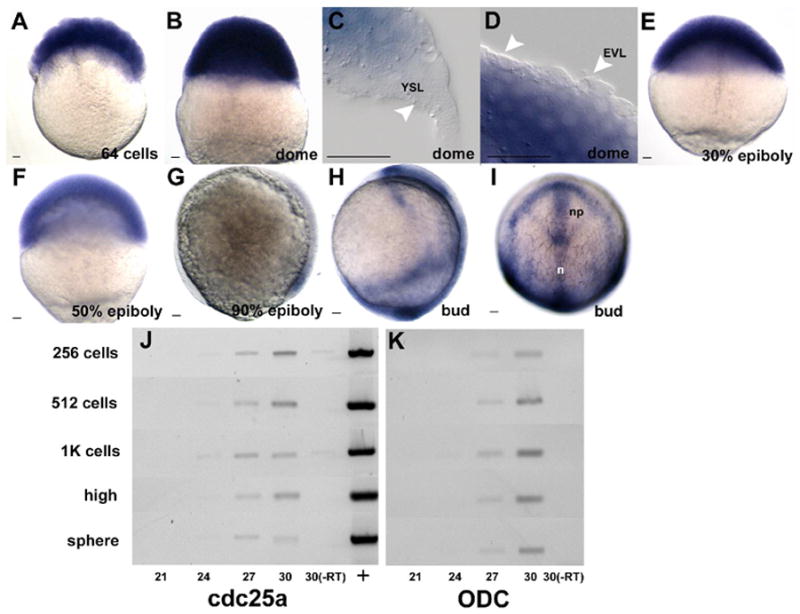
In situ hybridization using a cdc25a antisense probe during epiboly (a–i). Panels represent staged semi-quantiative RT-PCR results using either cdc25a (j) or ODC (k) specific primers. Cycle numbers are as indicated below panels, with stages at left. (+) plasmid positive control; (-RT) no RT control reactions. All embryos are oriented animal pole up. g and h are lateral views with the dorsal side to the right and i is a dorsal view. YSL is yolk syncitial layer, EVL is enveloping layer, n is notochord and np is neural plate. Scale bar is 20μm.
Figure 5.
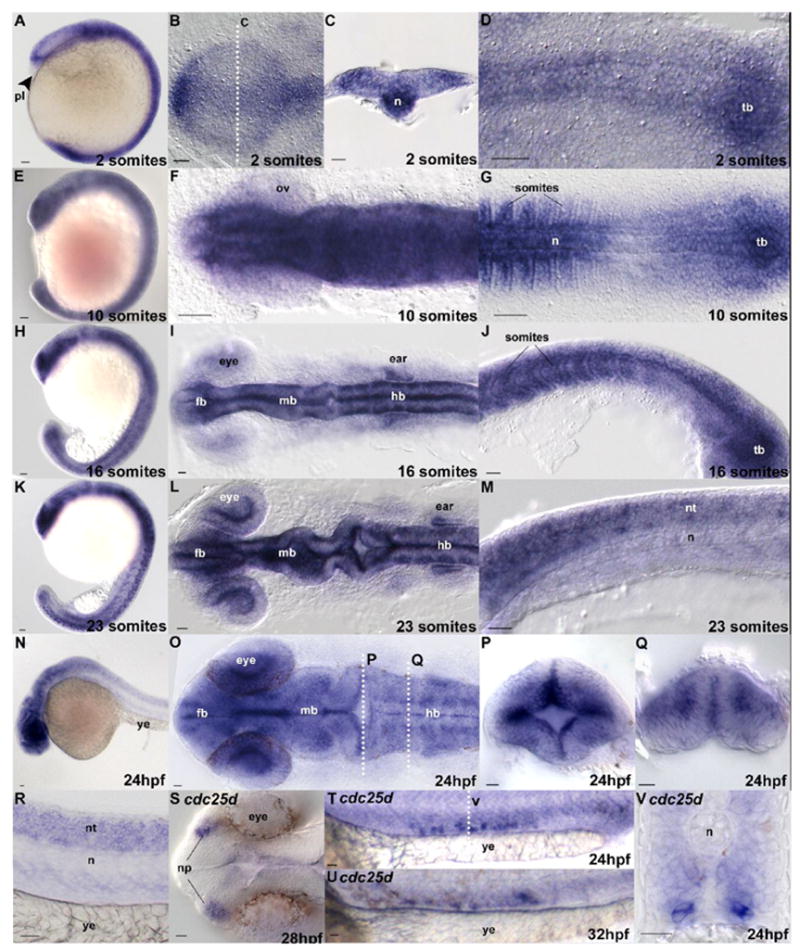
In situ hybridization using cdc25a (a–r) and cdc25d (s–v) antisense probes. Stages are indicated in the lower-left of each panel. Panels p and q are cross sections through a 24hpf embryo at the positions labeled in panel o. Panel v is a section as indicated in panel t. Anatomical structures, where appropriate for orientation, are labeled. Embryo orientations are as follows: a, e, h, and k are lateral views, b, f, I, I, o and s are dorsal views of the anterior regions of the embryo, anteriormost to the left. d, g are similarly oriented but focused on the posterior region of the embryo. J, m, n, r, t, and u are lateral views. Cross sections c, p, q and v are dorsal up. tb is tailbud; n is notochord; pl is polster; fb is forebrain; mb is midbrain; hb is hindbrain; ov is optic vesicle; ye is yolk extension; nt is neural tube; np is nasal placodes. Scale bar is 20μm.
Patterned expression is first observed at bud stage in the midline and anterior neural plate (Fig. 4h and i) and expression in the midline and trunk of the embryo continues throughout early development (Fig. 5a–d). We have not observed obvious restriction of mitotic cells, detected by anti-Phosphohistone-H3 immunostaining, to regions expressing cdc25a after gastrulation (not shown). We note, however, that expression is absent in the polster (Fig. 5a, arrow), one of the first tissues to exit the cell cycle in Xenopus (Saka and Smith, 2001) and Zebrafish (D.E. Dalle Nogare and M.E. Lane, unpublished observation). Widespread expression of cdc25a is maintained throughout early segmentation (Fig. 5e–m), and as development proceeds expression appears to become gradually restricted to the anterior portion of the embryo (Fig. 5n), although transcripts can be detected in the neural tube through 24hpf (Fig. 5r). At 24hpf, patterned anterior expression of cdc25a is observed in the rhombomeres and proliferative zones of the hindbrain (Fig. 5o–q), correlating with increased cell proliferation in the latter tissue. Little cdc25a expression is observed after 24hpf, concentrated mostly in the developing CNS (data not shown).
Expression of cdc25d appears to be more restricted than cdc25a. While RT-PCR data suggest that this gene is present throughout early development (data not shown), the first detectable patterned expression by in situ is observed between 24hpf and 32hpf in a small cluster of bilaterally distributed cells in the ventral mesoderm above the yolk extension (Fig. 5t–v) and in the nasal placodes (Fig. 5s). The number of labeled cells in the ventral mesoderm appears to be variable, although the position of these cells does not change with time (compare Fig. 5t with u). No other patterned expression of cdc25d is observed, and the significance of these expression domains remains unclear.
Cdc25a, but not Cdc25d, can rapidly induce cells in gastrulating embryos to enter mitosis
Direct observation of cell cycle lengths demonstrated that cycle length increases progressively during epiboly (Kimmel et al., 1994). Our finding that cdc25a expression levels appear to decrease during this same interval suggested the possibility that availability of cdc25a is directly regulating cell cycle progression. To test this possibility, we asked whether ecoptic expression of cdc25a can promote proliferation in these cells. We first injected mRNA for either cdc25a or cdc25d into embryos at the one-cell stage, but this resulted in morphologically abnormal embryos (DDN and MEL, unpublished observations). To overcome this early effect of widespread overexpression, we mosaically expressed either cdc25a, cdc25d or their phosphatase-inactive mutant forms by injecting single blastomeres at the 8-cell stage with DMA constructs in the ToL2 transposon (Kawakami et al., 2004) containing Myc-tagged versions of these genes under the control of the hsp70 promoter (Halloran et al., 2000). In Drosophila, such ectopic overexpression of string mRNA by a heatshock-inducible transgene during cycle 14, when approximately half of the embryonic cells are in G2, induces these cells to enter a precocious mitotic division. If zebrafish cdc25a is the rate-limiting regulator of cell cycle, we expected to see an increase in the number of mitotic cells following overexpression of cdc25a.
Injected shield stage embryos were subjected to 37°C heatshock for 30 minutes, allowed to recover at 28°C for 20 minutes then fixed and stained with antibodies against the Myc epitope to mark cells expressing the transgene, and phosphohistone H3 to visualize cells in mitosis. Following heatshock and recovery, cells expressing the transgene (blue cells in Fig. 6) were intermixed with non-expressing cells as a result of cell movement and intercalation and mosaic inheritance of the DMA construct. This allowed us to directly compare the mitotic index (defined at number of mitotic cells/total number of cells) of cells expressing cdc25 transgenes with non-expressing cells within the same embryo.
Figure 6.
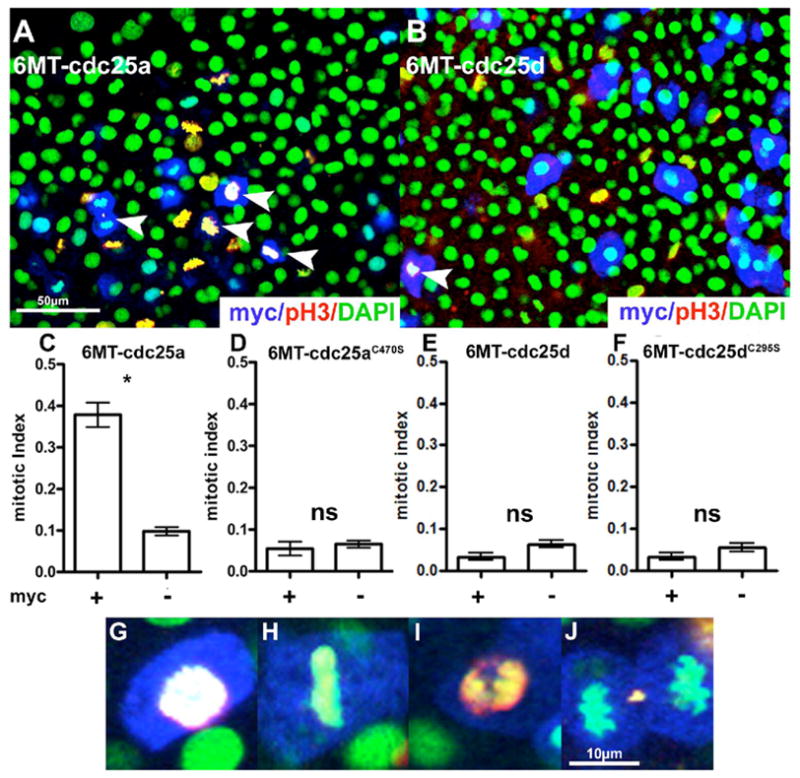
Overexpression of cdc25a and cdc25d in gastrulating zebrafish embryos. Panels a and b are example fields from either cdc25a or cdc25d expressing embryos following heatshock and staining. Mitotic index quantification is shown in panels c–f. (*) p <0.01 (ns) Not Significant. For cdc25a, n=14 fields, and for cdc25d, n=8 fields. See Materials and Methods for details. Panels g–j are enlarged images of cdc25a expressing (myc+) cells in various phases of mitosis. Stains are as indicated in the lower-right of panels a and b. White arrowheads in a and b indicate Myc-positive (therefore cdc25-positive) mitotic figures.
The mitotic index of cdc25a expressing cells is increased three-fold over non-expressing control cells (Fig. 6a and c, p < 0.01, 99% confidence interval), suggesting that expression of cdc25a in these cells is sufficient to induce cells to enter mitosis. The increase in the mitotic index of cells expressing cdc25a was dependent on phosphatase activity, as the phosphatase-inactive mutant cdc25aC470S had no significant effect on the mitotic index of expressing cells (Fig. 6d, p > 0.1). Analysis of mitotic figures in cdc25a-expressing cells (Fig. 6g–j) shows that all stages of mitosis are represented, suggesting that these cells are undergoing complete mitotic divisions. We do not see any evidence of mitotic catastrophe in these cells. In contrast, expression of either cdc25d (Fig. 6b and e p > 0.05) or the catalytically inactive cdc25dC295S (Fig. 6f, p > 0.1) was not sufficient to increase the mitotic index of expressing cells. We note, however, that cdc25d-expressing cells do undergo mitosis (white arrows in Fig. 6f). These data suggest that availability of cdc25a, but not cdc25d, is rate-limiting for cell cycle progression during gastrulation.
Discussion
We have identified two non-orthologous genes encoding Cdc25 mitotic activators in zebrafish. Our phylogenetic analysis clearly indicates that the cdc25a gene is related to the tetrapod Cdc25A genes, while the cdc25d gene is not related to tetrapod genes and is of indeterminate origin. We have demonstrated distinct phenotypic effects of overexpression of the cdc25 genes in yeast and in zebrafish embryonic cells, and differences in relative expression levels and patterns in developing embryos.
We found a single member of the tetrapod Cdc25 gene family, and the most recent zebrafish genome assembly (Zv7) does not show additional members of this family. The absence of other family members, coupled with the observations of higher expression and apparently stronger activity in both yeast and zebrafish assays suggests that in fish, most or all of the Cdc25 activity may come from the cdc25a gene. We also find a single gene in other teleosts where genome data is available, suggesting that the ancestral vertebrate condition is a single Cdc25A-like gene, and that the other tetrapod family members arose after the divergence of teleosts and tetrapods. These observations enhance the prospect for future genetic analysis in zebrafish and Medaka of developmental roles for vertebrate Cdc25 function.
Comparing the dynamics of zebrafish cdc25a expression with that of the Drosophila string gene reveals both similarities and important differences that are correlated with distinct cell behaviors during and immediately after the MBT/MZT. Whereas cdc25a and string mRNAs are abundant in early embryos as a result of maternal transcription, string mRNA is abruptly degraded at the MZT by factors dependent on zygotic transcription, and transcribed dynamically during gastrulation. We see no evidence for a dramatic alteration in cdc25a or mRNA levels at the MBT in the deep cells. In contrast, our observations are consistent with a gradual decay of maternally provided cdc25a message throughout gastrulation followed by dynamic spatially restricted expression upon completion of gastrulation. The drop in Cdc25 activity at the Drosophila MZT is required for a long G2 phase during cellularization of the embryo (Edgar and Datar, 1996), and is followed by pulses of string transcription that drive mitosis in domains corresponding to cell fate assignments. In zebrafish, only three mitotic domains have been described (Kane et al., 1992), and we see correlation between cdc25a expression and these domains. The domains comprise two extraembryonic layers, the YSL and the EVL, and the deep layer, which gives rise to all of the embryonic cells. The YSL and EVL are not actively proliferating during epiboly and gastrulation, and we do not observe cdc25a expression in these cells. While cells of the deep layer are actively proliferating, no mitotic subdomains linked to cell fate have been observed within the deep layer (Kane et al., 1992; Kimmel et al., 1994), in stark contrast to what is seen in the Drosophila embryo. Instead, deep cells proceed through up to three cell divisions throughout gastrulation, with no apparent spatial patterning. However cell cycles do progressively increase in length, and this correlates well with our observations that cdc25a transcripts are not spatially restricted but appear to decrease in abundance as gastrulation progresses. Our demonstration that overexpression of cdc25a during gastrulation forces cells into mitosis supports the proposal that cdc25a is limiting for cell cycle progression during gastrulation. Thus, while the spatiotemporal dynamics of cell cycle progression following the MBT/MZT are very different between Drosophila and zebrafish, in both cases, mechanisms that limit abundance and distribution of cdc25 transcripts may control the timing of entry into mitosis.
Acknowledgments
We thank Geraldine Mercedes Barrutia for assistance with cdc25 cloning. We thank Steve Johnson and Maria Dorsett for providing the modified ToL2 vector. We thank Michael Gustin, Michael Stern, and members of the Lane and Wagner fish labs for helpful discussion during the course of the work, and we thank Daniel Wagner for critical reading of the manuscript. We are grateful to Patrick Connor and the late Ronald W. Brannon for excellent fish care and Ravi Munjal for technical assistance. We thank Mary Dickinson and members of the Dickinson laboratory for use of their confocal microscope. We acknowledge helpful assistance from the Zebrafish Nomenclature Committee on guidelines for gene names. The work was supported by a March of Dimes Basil O’Connor Starter Scholar Award to MEL and NIH grant R01 EY015305 to MEL. This material is based in part on work supported by the National Science Foundation under Grant No. 0344471 to SS. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
References
- Chen MS, Hurov J, White LS, Woodford-Thomas T, Piwnica-Worms H. Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Mol Cell Biol. 2001;21(12):3853–61. doi: 10.1128/MCB.21.12.3853-3861.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Ryan CE, Piwnica-Worms H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol Cell Biol. 2003;23(21):7488–97. doi: 10.1128/MCB.23.21.7488-7497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30(3):154–6. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev. 1996;10(15):1966–77. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Lehman DA, O’Farrell PH. Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development. 1994;120(11):3131–43. doi: 10.1242/dev.120.11.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O’Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62(3):469–80. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AM, White LS, Donovan PJ, Piwnica-Worms H. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol Cell Biol. 2005;25(7):2853–60. doi: 10.1128/MCB.25.7.2853-2860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21 (12):2955–6. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J, Solomon MJ, Booher RN, Bazan JF, Kirschner MW. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67(1):197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342(6245):39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Goloudina A, Yamaguchi H, Chervyakova DB, Appella E, Fornace AJ, Jr, Bulavin DV. Regulation of human Cdc25A stability by Serine 75 phosphorylation is not sufficient to activate a S phase checkpoint. Cell Cycle. 2(5):473–478. [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127(9):1953–60. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73(1):237–44. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Kane DA, Warga RM, Kimmel CB. Mitotic domains in the early embryo of the zebrafish. Nature. 1992;360(6406):735–7. doi: 10.1038/360735a0. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7(1):133–44. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kim SH, Li C, Maller JL. A maternal form of the phosphatase Cdc25A regulates early embryonic cell cycles in Xenopus laevis. Dev Biol. 1999;212(2):381–91. doi: 10.1006/dbio.1999.9361. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Kane DA. Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development. 1994;120(2):265–76. doi: 10.1242/dev.120.2.265. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Leise W, 3rd, Mueller PR. Multiple Cdk1 inhibitory kinases regulate the cell cycle during development. Dev Biol. 2002;249(1):156–73. doi: 10.1006/dbio.2002.0743. [DOI] [PubMed] [Google Scholar]
- Leise W, 3rd, Mueller PR. Inhibition of the cell cycle is required for convergent extension of the paraxial mesoderm during Xenopus neurulation. Development. 2004;131(8):1703–15. doi: 10.1242/dev.01054. [DOI] [PubMed] [Google Scholar]
- Lincoln AJ, Wickramasinghe D, Stein P, Schultz RM, Palko ME, De Miguel MP, Tessarollo L, Donovan PJ. Cdc25b phosphatase is required for resumption of meiosis during oocyte maturation. Nat Genet. 2002;30(4):446–9. doi: 10.1038/ng856. [DOI] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth Enzymol. 1991:194795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami MS, Moody SA, Daar IO, Morrison DK. Morphogenesis during Xenopus gastrulation requires Wee1-mediated inhibition of cell proliferation. Development. 2004;131(3):571–80. doi: 10.1242/dev.00971. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182(1):119–24. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256(5518):547–51. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics. 1980;96(3):627–37. doi: 10.1093/genetics/96.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146(2):167–78. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45(1):145–53. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Sagerstrom CG, Grinbalt Y, Sive H. Anteroposterior patterning in the zebrafish, Danio rerio: an explant assay reveals inductive and suppressive cell interactions. Development. 1996;122(6):1873–83. doi: 10.1242/dev.122.6.1873. [DOI] [PubMed] [Google Scholar]
- Sagerstrom CG, Kao BA, Lane ME, Sive H. Isolation and characterization of posteriorly restricted genes in the zebrafish gastrula. Dev Dyn. 2001;220(4):402–8. doi: 10.1002/dvdy.1119. [DOI] [PubMed] [Google Scholar]
- Saka Y, Smith JC. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol. 2001;229(2):307–18. doi: 10.1006/dbio.2000.0101. [DOI] [PubMed] [Google Scholar]
- Shimuta K, Nakajo N, Uto K, Hayano Y, Okazaki K, Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. Embo J. 2002;21(14):3694–703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V, Nurse P. The cell cycle control gene cdc2+ of fission yeast encodes a protein kinase potentially regulated by phosphorylation. Cell. 1986;45(2):261–8. doi: 10.1016/0092-8674(86)90390-9. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3(3):247–58. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A guide for the laboratory use of zebrafish (Danio rerio) 4. Univ. of Oregon Press; Eugene: 2000. The zebrafish book. [Google Scholar]
- Wolfe BA, Gould KL. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 2004;23(4):919–929. doi: 10.1038/sj.emboj.7600103. [DOI] [PMC free article] [PubMed] [Google Scholar]