Abstract
TATA-box-binding protein (TBP)-related factor 3, TRF3 (also called TBP2), is a vertebrate-specific member of the TBP family that has a conserved C-terminal region and DNA binding domain virtually identical to that of TBP1. TRF3 is highly expressed during embryonic development, and studies in zebrafish and Xenopus have shown that TRF3 is required for normal embryogenesis2,3. Here we show that Trf3-depleted zebrafish embryos exhibit multiple developmental defects and, in particular, fail to undergo hematopoiesis. Expression profiling for Trf3-dependent genes identified mespa, which encodes a transcription factor whose murine orthologue is required for mesoderm specification4, and chromatin immunoprecipitation verified that Trf3 binds to the mespa promoter. Depletion of Mespa resulted in developmental and hematopoietic defects strikingly similar to those induced by Trf3 depletion. Injection of mespa mRNA restored normal development to a Trf3-depleted embryo, indicating mespa is the single Trf3 target gene required for zebrafish embryogenesis. Zebrafish embryos depleted of Trf3 or Mespa also failed to express cdx4, a caudal-related gene required for hematopoiesis. Mespa binds to the cdx4 promoter, and epistasis analysis revealed an ordered trf3-mespa-cdx4 pathway. Thus, in zebrafish commitment of mesoderm to the hematopoietic lineage occurs through a transcription factor pathway initiated by a TBP-related factor.
To analyze the role of TRF3 during embryonic development, we used antisense morpholino oligonucleotides (MOs) to ablate Trf3, and as a control Tbp, function in zebrafish embryos. MOs were injected into wild-type one-cell stage fertilized embryos and depletion of Trf3 and Tbp was analyzed by immunoblotting at 6 hours post-fertilization (hpf), a time at which expression of both proteins was readily detectable (Supplementary Fig. 1a). Immunoblot analysis confirmed that injection of each MO efficiently and specifically depleted its target gene (Supplementary Fig. 1b). Consistent with previous studies5, Tbp-depleted embryos appeared to initiate gastrulation but failed to progress past 50% epiboly (Supplementary Fig. 2; n=122/150). By contrast, Trf3-depleted embryos appeared to develop normally until the tailbud stage, but by 14 hpf exhibited delayed development and necrosis compared to siblings injected with a randomized control MO (n=166/177). Inspection of Trf3-depleted embryos at 21 hpf revealed severe necrosis, although head, trunk and tail rudiments were apparent, suggesting that initial antero-posterior patterning was largely unaffected.
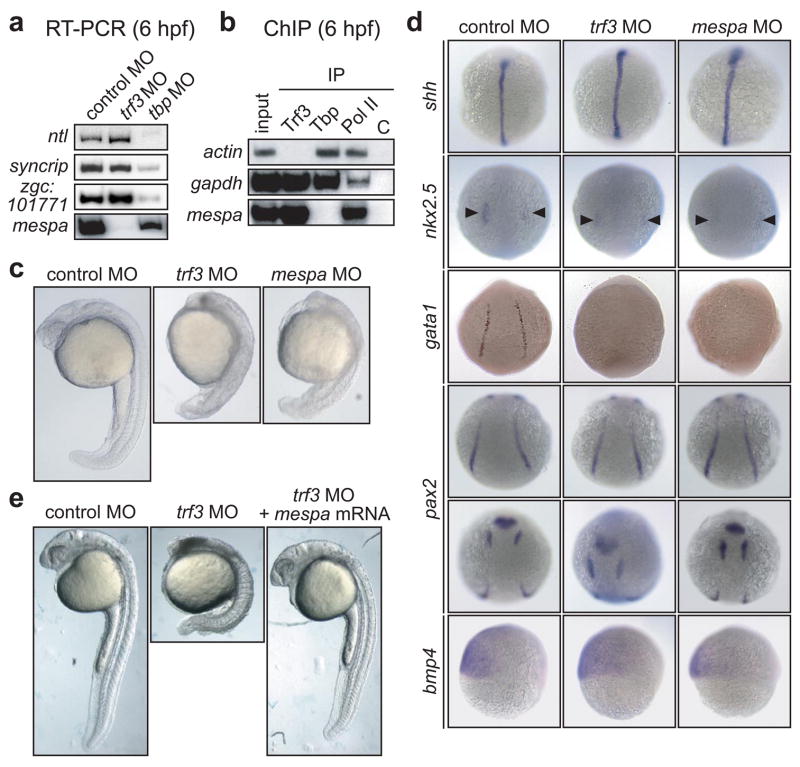
To identify Trf3 target genes, we performed expression profiling in trf3 MO-treated embryos (at 6 hpf) using a zebrafish oligonucleotide microarray representing ~12,800 genes. As expected, the vast majority of genes were unaffected by Trf3 depletion (Supplementary Data). Three such representative genes are shown as controls in the RT-PCR experiment of Fig. 1a, which confirms that their expression is dependent upon Tbp but not Trf3. Using a candidate-based approach, we selected genes whose expression were significantly decreased by Trf3 knockdown (Supplementary Table 1) and had been previously implicated in embryonic development. These candidates were further analyzed by RT-PCR for Trf3-dependent expression, chromatin immunoprecipitation (ChIP) for selective Trf3 occupancy, and finally for a role in zebrafish development (see below and data not shown). This combined analysis identified mespa, whose mouse orthologue, Mesp1, encodes a basic-helix-loop-helix (bHLH)-type transcription factor required for proper embryonic development4.
Figure 1.
mespa is the single Trf3 target gene required for proper embryonic development. a, RT-PCR analysis. b, ChIP analysis. C, negative control (yeast Gal4). c, Phenotypic analysis at 24 hpf. c, Whole-mount in situ hybridization with riboprobes to shh (14 hpf), nkx2.5 (10 hpf; arrowheads denote decreased expression in cardiac mesoderm in trf3 MO- and mespa MO-treated embryos), gata1 (12 hpf), pax2 (10 hpf; showing dorsal posterior (top) and anterior (bottom) views) and bmp4 (6 hpf). . d, Phenotypic analysis of embryos injected with a control MO or trf3 MO, or trf3 MO-treated embryos injected with mespa mRNA at 28 hpf.
The RT-PCR results of Fig. 1a show that Trf3 depletion eliminated mespa expression. To determine whether mespa was a direct Trf3 target, we analyzed promoter occupancy by ChIP assays using antibodies directed against Trf3, Tbp, RNA polymerase II (Pol II) or, as a negative control, an irrelevant protein (yeast Gal4). As controls, we analyzed two well-characterized housekeeping genes, actin and gapdh. Figure 1b shows that Pol II was bound to all three promoters, consistent with the transcriptional activity of these genes. Notably, Tbp but not Trf3 was bound to the actin promoter, whereas both Tbp and Trf3 were bound to the gapdh promoter. By contrast, the mespa promoter was selectively bound by Trf3 and not Tbp. Based upon the dependence of mespa expression on Trf3 (Fig. 1a) and the selective binding of Trf3 to the mespa promoter (Fig. 1b), we conclude that mespa is a direct Trf3 target gene.
Phenotypic analysis demonstrated that the mespa MO-injected embryo had a developmental defect that was strikingly similar to that of a Trf3-depleted embryo (n=61/89) (Fig. 1c and Supplementary Fig. 3). As a control, injection of a mespa mRNA bearing a silent mutation that prevented hybridization with the mespa MO restored normal development to the Mespa-depleted embryo (Supplementary Fig. 4). To compare the phenotypes of the Trf3- and Mespa-depleted embryos in greater detail, we performed whole mount in situ hybridization using several developmentally-regulated genes as markers. Figure 1d shows that depletion of either Trf3 or Mespa resulted in increased shh expression in axial mesoderm, decreased nkx2.5 expression in cardiac mesoderm, and decreased gata1 expression in lateral mesoderm. Surprisingly, expression of a second lateral mesoderm marker, pax2, was unaffected by loss of Trf3 or Mespa. Moreover, expression of the ventral marker bmp4 was normal in both Trf3- and Mespa-depleted embryos, indicating that the developmental defects were not due to loss of proper dorsal-ventral patterning. Thus, the developmental defect observed in Mespa-depleted embryos was very similar, if not identical, to that of Trf3-depleted embryos.
We next asked whether ectopic expression of mespa could restore normal development to a Trf3-depleted embryo. Figure 1e shows that injection of mespa mRNA (n=110/134), but not an unrelated control mRNA (n=0/63; data not shown), restored normal development to the trf3 MO-injected embryo. Completeness of rescue was verified by differential interference contrast microscopy (Supplementary Fig. 5) and in situ hybridization analysis (Supplementary Fig. 6). Collectively, these results indicate that in zebrafish, mespa is the single Trf3 target gene required for proper embryonic development.
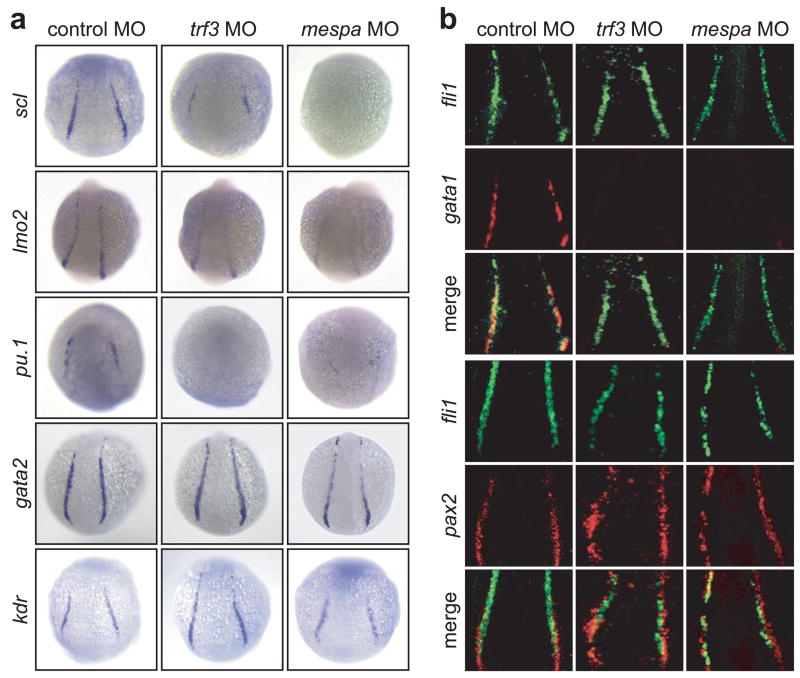
The results of Fig. 1d suggested a requirement for Trf3 and Mespa during development of cell types in the lateral mesoderm, which we elected to analyze in greater detail. We assayed the expression of several hematopoietic, vascular and pronephric markers in trf3- or mespa-MO injected embryos. The RT-PCR results of Supplementary Fig. 7 indicate that a number of blood cell-specific genes were significantly down-regulated in Trf3- and Mespa-depleted embryos, including hbae1 and hbae3, which are terminal markers of erythroid cell fate, as well as gata1, which is required for the expression of a variety of genes in the erythroid lineage. In addition, earlier markers of hematopoietic precursors, scl and lmo2, were similarly reduced although expression of gata2 was unaffected. Finally, expression of pu.1, a marker of myeloid cells that arise in the anterior lateral mesoderm, was also reduced in the absence of Trf3 or Mespa.
To confirm the RT-PCR results, we analyzed the expression of these marker genes by whole mount in situ hybridization. Figure 2a shows that in trf3- or mespa-MO injected embryos, scl expression was reduced in the posterior lateral mesoderm, although expression was maintained within more anterior cells whose position is consistent with that of endothelial cells6,7. Expression of lmo2 was moderately reduced in the posterior lateral mesoderm in Trf3- and Mespa-depleted embryos, whereas pu.1 expression was absent from anterior lateral mesoderm. Also consistent with the RT-PCR results, expression of the early hematopoietic marker gata2 was unaffected by loss of Trf3 or Mespa. Similarly, expression of the endothelial cell marker kdr was normal in embryos lacking Trf3 or Mespa. As expected, injection of mespa mRNA into Trf3-depleted embryos fully rescued expression of the hematopoietic markers gata1 (see below), hbae3 and scl (Supplementary Fig. 6), as well as the cardiac mesoderm marker nkx2.5 (Supplementary Fig. 6).
Figure 2.
Trf3- and Mespa-depleted embryos fail to undergo hematopoiesis. a, Whole-mount in situ hybridization with riboprobes to scl (12 hpf), lmo2 (14 hpf), pu.1 (12 hpf), gata2 (14 hpf) and kdr (12 hpf). b, Fluorescence microscopy monitoring expression of an fli1:EGFP transgene (top panels), and either gata1 or pax2 (middle panels). The merged signal is shown in the bottom panels.
Collectively, the results of Fig. 2a suggest a defect in the formation of hematopoietic cells in the posterior lateral mesoderm. To confirm this defect, we simultaneously assayed expression of fli1, a marker for both hematopoietic and endothelial cells, and either gata1 or pax2, markers of erythroid or pronephric lateral mesoderm, respectively. Figure 2b shows that expression of a fli1:EGFP transgene was maintained in the absence of Trf3 or Mespa, whereas gata1 expression was absent from the same embryo. By contrast, pax2 was unaffected by the loss of Trf3 or Mespa and continued to be co-expressed with fli1 in adjacent cells of the lateral mesoderm. Taken together, these results demonstrate a selective loss of hematopoietic cells within the posterior lateral mesoderm of zebrafish embryos lacking Trf3 or Mespa.
The defects in hematopoietic development described above are reminiscent of those observed in embryos lacking Cdx4, a caudal-related transcription factor that is required for hematopoiesis6 and functions by activating expression of homeobox genes involved in the commitment of mesoderm to the hematopoietic lineage6,8. RT-PCR and in situ hybridization analysis showed that cdx4 expression was substantially decreased in Trf3-depleted embryos, as well as in Mespa-depleted embryos (Fig. 3a). As expected, expression of the Cdx4-dependent genes hoxa9a, hoxb7a and hoxb5a8 was also substantially reduced in Trf3- and Mespa-depleted embryos (Supplementary Fig. 8). Depletion of Trf3 or Mespa did not affect expression of cdx1a (Supplementary Fig. 9), which has been reported to cooperate with cdx4 in hematopoietic development8.
Figure 3.
Trf3 initiates a transcription factor pathway required for cdx4 expression and hematopoiesis. a, (Top) RT-PCR analysis (9 hpf). (Bottom) In situ hybridization (6 hpf). b, In situ hybridization monitoring cdx4, trf3 and mespa expression in wild-type embryos (6 hpf). c, Epistasis analysis; gata1 expression was monitored by in situ hybridization at 14 hpf. d, In situ hybridization monitoring gata1 and scl induction (14 hpf). Arrowheads show a “third stripe” indicative of expanded scl expression. e, In situ hybridization monitoring cdx4 induction (14 hpf). Lateral (top) and dorsal (bottom) views are shown. Arrowheads indicate normal (left) and expanded (right) cdx4 expression.
In situ hybridization analysis revealed that at 6 hpf, cdx4, mespa and trf3 were co-expressed in the presumptive hematopoietic tissues based on the zebrafish fate map9,10 (Fig. 3b), suggesting that the three factors are components of a common pathway. To confirm this idea and determine the order of the pathway, we performed epistasis experiments using gata1 expression as a phenotypic read-out. Figure 3c shows that injection of trf3 mRNA restored gata1 expression in Trf3-depleted embryos, but not in Mespa- and Cdx4-depleted embryos, indicating that trf3 is upstream of both mespa and cdx4. Moreover, injection of mespa mRNA restored gata1 expression in Trf3- and Mespa-depleted embryos, but not in Cdx4-depleted embryos, indicating that mespa is upstream of cdx4 and downstream of trf3. Finally, injection of cdx4 mRNA restored gata1 expression in Trf3-, Mespa- and Cdx4-depleted embryos, indicating that cdx4 is downstream of both trf3 and mespa. ChIP analysis supports the possibility that cdx4 is a direct Mespa target gene (Supplementary Fig. 10a).
Previous studies have shown that ectopic expression of cdx4 in zebrafish results in increased number of blood cells, as evidenced by expanded expression of hematopoietic markers6. Ectopic expression of mespa also resulted in expanded expression of gata1 and scl (Fig. 3d) and cdx4 (Fig. 3e). The collective results of Figs. 2 and 3 reveal that trf3, mespa and cdx4 function in a common pathway, which is summarized in Supplementary Fig. 10b.
Collectively, our results indicate that binding of Trf3 to the mespa promoter is the earliest documented step in committing mesoderm to the hematopoietic lineage in the developing zebrafish embryo. A recent study has provided definitive evidence for the existence of hemangioblasts, bipotential progenitors that can give rise to both endothelial and hematopoietic cells, within the zebrafish embryo10. Significantly, in zebrafish embryos, hemangioblasts are present at the same time and place in which trf3, mespa and cdx4 are expressed. Thus, the timing and location of trf3, mespa and cdx4 co-expression suggests that this transcription factor pathway may function within hemangioblasts to specify hematopoietic progenitor cells (i.e., commitment of mesoderm to the hematopoietic lineage). Although the defects associated with loss of Trf3 or Mespa are restricted to hematopoietic cell types in the lateral mesoderm, the effects on other tissues appear to be more widespread. We predict that there will be additional Mespa target genes that function analogous to but independent of cdx4 in mediating other developmental pathways, such as specification of cardiac mesoderm, in the early embryo.
METHODS SUMMARY
Zebrafish maintenance, embryo production and microinjection
Zebrafish (Danio rerio) were maintained under standard conditions11, and embryos were produced and staged as described12. Antisense morpholino oligonucleotides (MOs) were designed against the start codon/5’UTR to block translation. MO sequences are as follows: control MO, 5’-CCTCTTACCTCAGTTACAATTTATA-3’; trf3 MO, 5'-GATGCCTC CTCATCCATGTTCA T-3'; mespa MO, 5'-GAAGAGAAAACGTGGAGGCGTCCAT -3'; and zfzb MO, 5'-GCCCAACTTCCCGTACCGCCATCAT-3'. The cdx4 MO (5'-CT CCAAAAGGTATCCAACGTACATG-3') was purchased from Open Biosystems, and the tbp MO has been previously described5. MOs were injected at a concentration of 5 mg ml−1, except the cdx4 MO, which was injected at 0.2 mg ml−1. Embryos were analyzed morphologically up to 24 h post injection.
To generate mRNAs for phenotypic rescue and epistasis experiments, full-length trf3, mespa and cdx4 genes were amplified from shield-stage cDNA by RT-PCR (primer sequences are listed in Supplementary Table 2), and subcloned into pCS2 (ref. 13) for mRNA synthesis. The mRNAs were then injected into MO-treated or wild-type embryos at the 1–2 cell stage at concentrations of 100 ng μl−1, 200 ng μl−1, and 30 ng μl−1 for trf3, mespa and cdx4, respectively.
Immunoblot analysis
Protein extracts were prepared from zebrafish embryos as described11. Blots were probed with either an α-zebrafish Trf3 polyclonal antibody raised to the unique N-terminal region of zebrafish Trf3 (CPQKESTQADIDTSNS; amino acids 90–105), or an α-human TRF3 polyclonal1, α-TBP monoclonal (3G3; Eurogentec), or α-RNA Pol II monoclonal (8WG16; Covance Research Products) antibody.
METHODS
Microarray analysis
Total RNA was prepared from 50 control MO- or trf3 MO-injected zebrafish embryos 6 h after MO treatment using Trizol reagent. The KCC 17K Zebrafish Microarray was constructed using a commercially available 17K Zebrafish OligoLibrary™ consisting of 16,399 oligos (XEBLIB96; Compugen/Sigma-Genosys), corresponding to ~12,800 genes, based on UniGene clusters. Microarray construction, hybridization, and scanning were performed at the Kimmel Cancer Center Microarray Core Facility at Thomas Jefferson University. Chips were scanned using a Perkin Elmer ScanArray XL5000 Scanner, software version 3.1 and images were quantified by PerkinElmer QuantArray Software 3.0. The raw data for all genes induced or repressed by trf3 MO treatment is provided in Supplementary Data. Background threshold values were determined for both microarrays and calculated as the mean plus two (for the trf3 MO microarray) or three (for the control MO microarray) standard deviations of the channel intensities for the five bacterial probes present on each array. The background threshold values were then subtracted from the channel intensity value of each probe set (representing a single gene). The resulting data from the two microarrays were compared to identify Trf3-affected genes, which were classified as those whose probe values satisfied the following condition: >0 in the control microarray (i.e., the gene is significantly expressed over background) and ≤0 in the trf3 MO microarray (i.e., the gene is significantly down-regulated, essentially “off”, upon Trf3 depletion); these genes are listed in the “Analyzed data” tab in Supplementary Data. To determine transcript identity and gene descriptions, we first annotated the gene list by identifying Unigene numbers (Unigene build #104) that corresponded with the Genbank accession number for each oligo on the array. We subsequently annotated gene names and descriptions through downloadable datasets obtained from the Zebrafish Information Network (ZFIN) and ENSEMBL (Zv7). In each case, we used FileMaker Pro to generate relational databases to annotate gene sets.
RT-PCR
RT-PCR analysis was performed according to standard protocols14. Total RNA was prepared using Trizol reagent (Invitrogen) and RT-PCR products were separated by agarose gel electrophoresis and visualized by autoradiography. Primer sequences are listed in Supplementary Table 2.
Chromatin immunoprecipitation
Embryos (~3000 at 6 hpf) were washed in 1X PBS three times, suspended in 1X PBS containing 1% formaldehyde (final concentration), transferred to a 10 ml dounce homogenizer and then dounced in one stroke. The reaction mixture was incubated at room temperature for 15 min and quenched by addition of glycine (0.125 M final concentration) for 10 min. The cells were centrifuged at 14K for 10 min at 4°C, and the pellets were washed twice with ice-cold 1X PBS containing protease inhibitors, and resuspended in 5 ml lysis buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 1% Triton-X 100, 0.1% SDS, 1 mM PMSF, and protease inhibitor cocktail). The suspension was then sonicated eight times on ice to generate ~500 bp fragments. The lysates were centrifuged, precleared with protein-A agarose beads (Upstate Biotech), and then divided into 1.5 ml aliquots per immunoprecipitation. Antibodies for immunoprecipitation were as follows: Trf3, Tbp and Pol II antibodies were described above; the Mespa antibody was raised to the C-terminal portion of the protein (CYQTQNPVQGDFHS; amino acids 191–204); yeast Gal4 (sc577; Santa Cruz). Following addition of the antibody, lysates were incubated for 12 h at 4°C, and then incubated with protein-A agarose beads (50 μl) for 2 h at 4°C (5% of the lysate was kept as “input” prior to the addition of the antibody). The beads were washed twice with lysis buffer without protease inhibitor, twice with the lysis buffer containing 1 M NaCl, and once with LiCl immune-complex wash buffer (50 mM Tris-HCl, pH 8.0, 0.25 M LiCl, 1 mM EDTA, 0.5% N-P40, 0.5% sodium deoxycholate). Finally the beads were washed thrice in TE and pelleted, and chromatin was eluted from the beads by adding 500 μl freshly prepared 0.1 M NaHCO3 and 1% SDS and incubated with rotation for 15 min at room temperature. Following addition of 2 μl proteinase K (18.6 ml ml−1), reverse cross-linking was performed for all samples for 6 h at 65°C. Chromatin was purified from the beads by phenol extraction followed by alcohol precipitation. The input sample was dissolved in 200 μl and the immunoprecipitated samples were dissolved in 40 μl, 2 μl of which was used in the PCR reaction. ChIP PCR primers are listed in Supplementary Table 2.
In situ hybridization
In situ hybridization was performed on whole-mount zebrafish embryos as described15, or on embryos flat-mounted in glycerol. Digoxygenin-labeled antisense probes were synthesized in vitro and obtained as follows: shh, pax2, cdx4, hoxa9a, hoxb5a and hoxb7a (Charles Sagerstrom, UMMS), bmp4 (Michael Tsang, NIH), gata1, ephrinb2a and kdr (Nathan Lawson, UMMS), nkx2.5 (The Zebrafish International Resource Center16), scl (NIH Zebrafish Gene Collection), lmo2 (IMAGE clone 7433557), pu.1 (IMAGE clone 6960940), gata2 (IMAGE clone 6789690). For the trf3 and mespa probes, N-terminal fragments of either trf3 and mespa were first subcloned into the vector pGEM-T (Promega), and riboprobes were synthesized from the corresponding constructs as T7 or SP6 transcripts using the DIG RNA labelling mix (Roche). Riboprobe primers are listed in Supplementary Table 2.
Differential interference contrast (DIC) microscopy
High-resolution DIC microscopy was carried out on a Zeiss Axiophot microscope equipped with a Zeiss AxioCam HRC digital camera.
Fluoresence microscopy
Embryos (14 hpf) expressing the fli1:EGFP transgene [TG(fli1:EGFP)y1; ref. 17] were hybridized with a riboprobe to either gata1 or pax2 (described above) for 16 h at 70°C. Samples were then washed twice in 2X SSC, 0.1% Tween 20 and 50% formamide for 30 min, once in 2X SSC and 0.1% Tween 20 for 15 min, and twice in 0.2X SSC and 0.1% Tween 20 for 30 min, all at 65°C, and blocked with Western Blocking Reagent (Roche) diluted in PBS for at least 1 h at room temperature. Primary antibody staining was performed in 100 μl of blocking buffer consisting of a 1:200 dilution of anti-GFP rabbit IgG (A11122; Molecular Probes) and a 1:200 dilution of pre-absorbed anti-DIG Fab fragments (Roche) for 2 h at room temperature with gentle shaking. Embryos were washed six times in blocking buffer for 20 min at room temperature. Secondary antibody staining was performed using goat anti-rabbit Alexa Fluor 488 (A11008; Molecular Probes) at a 1:200 dilution in blocking solution for 2 h at room temperature. Embryos were washed six times in blocking buffer for 20 min at room temperature. Fast Red tablets (Roche) were dissolved (1 tablet per 2 ml of buffer) in 0.1 M Tris-HCl, pH 8.3 and 0.1% Tween 20 by shaking, and embryos were stained in 1 ml per sample well for 4 h at room temperature. After staining was assessed, embryos were washed six times in PBST for 20 min and stored in 90% glycerol at 4°C. Embryos were flat-mounted in glycerol and fluorescence microscopy was carried out using a Leica TCS SP2 confocal microscope.
Supplementary Material
Supplementary Information accompanies the paper on Nature’s website (http://www.nature.com).
Acknowledgments
We thank C. Sagerstrom and M. Tsang for reagents, the Kimmel Cancer Center Microarray Core Facility at Thomas Jefferson University for performing the microarray analysis, members of the Lawson lab for technical support, and S. Evans for editorial assistance. The Zebrafish International Resource Center is supported by grant P40 RR12546 from the NIH-NCRR. This work was supported in part by a grant from the National Institutes of Health to M.R.G. M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing financial interests.
References
- 1.Persengiev SP, et al. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci USA. 2003;100:14887–14891. doi: 10.1073/pnas.2036440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartfai R, et al. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci USA. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 5.Muller F, Lakatos L, Dantonel J, Strahle U, Tora L. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol. 2001;11:282–287. doi: 10.1016/s0960-9822(01)00076-8. [DOI] [PubMed] [Google Scholar]
- 6.Davidson AJ, et al. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–306. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- 7.Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson AJ, Zon LI. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox gene expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Dev Biol. 2006;292:506–518. doi: 10.1016/j.ydbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Langeland J, Kimmel CB. In: Embryology: Constructing the Organism. Gilbert SF, Raunio AM, editors. Sinauer Associates; Sunderland, MA: 1997. pp. 383–407. [Google Scholar]
- 10.Vogeli KM, Jin SW, Martin GR, Stainier DY. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature. 2006;443:337–339. doi: 10.1038/nature05045. [DOI] [PubMed] [Google Scholar]
- 11.Westerfield M. A guide for the laboratory use of zebrafish (Danio rerio) University of Oregon Press; Eugene: 1993. The zebrafish book. [Google Scholar]
- 12.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 13.Turner DL, Weintraub H. Expression of achaetescute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 14.Ausubel FM, et al. John Wiley and Sons; New York, NY: 2001. [Google Scholar]
- 15.Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- 16.Thisse B, et al. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402) ZFIN Direct Data Submission. 2001 [Google Scholar]
- 17.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on Nature’s website (http://www.nature.com).