Abstract
The nuclear pore complexes (NPCs) are evolutionarily conserved assemblies that allow traffic between the cytoplasm and the nucleus. In this study, we have identified and characterized a novel human nuclear pore protein, hNup133, through its homology with the Saccharomyces cerevisiae nucleoporin scNup133. Two-hybrid screens and immunoprecipitation experiments revealed a direct and evolutionarily conserved interaction between Nup133 and Nup84/Nup107 and indicated that hNup133 and hNup107 are part of a NPC subcomplex that contains two other nucleoporins (the previously characterized hNup96 and a novel nucleoporin designated as hNup120) homologous to constituents of the scNup84 subcomplex. We further demonstrate that hNup133 and hNup107 are localized on both sides of the NPC to which they are stably associated at interphase, remain associated as part of a NPC subcomplex during mitosis, and are targeted at early stages to the reforming nuclear envelope. Throughout mitosis, a fraction of hNup133 and hNup107 localizes to the kinetochores, thus revealing an unexpected connection between structural NPCs constituents and kinetochores. Photobleaching experiments further showed that the mitotic cytoplasm contains kinetochore-binding competent hNup133 molecules and that in contrast to its stable association with the NPCs the interaction of this nucleoporin with kinetochores is dynamic.
Keywords: nucleoporin; nuclear pore; mitosis; kinetochores; GFP
Introduction
The nuclear pore complexes (NPCs)* are macromolecular assemblies that mediate molecular exchanges across the nuclear envelope. EM observations have revealed their highly conserved architecture characterized by an eightfold symmetric framework that encircles a central transporter and peripheral filamentous structures extending into both the cytoplasm and the nucleus (for review see Stoffler et al., 1999). The NPCs, which have an estimated molecular mass of 60 MD in the yeast Saccharomyces cerevisiae and 125 MD in vertebrates, are composed of 30–50 distinct proteins (termed nucleoporins) present in multiple copies. In recent years, rapid progress in the characterization of S. cerevisiae NPC components has led to an essentially complete list of NPC constituents in this organism (for review see Vasu and Forbes, 2001). In vertebrates, ∼20 nucleoporins have been characterized to date at the molecular level (for review see Vasu and Forbes, 2001). Sequence comparison of vertebrate and S. cerevisiae nucleoporins (subsequently referred to as scNupX) has led to the identification of homologous nucleoporins, and in few cases evolutionarily conserved NPC subcomplexes have also been characterized (for review see Belgareh and Doye, 1999; Stoffler et al., 1999; Vasu and Forbes, 2001).
An extensively characterized building block of the S. cerevisiae NPC is the scNup84 subcomplex, which consists of scNup84, scNup85, scNup120, scNup145-C (the in vivo–cleaved COOH-terminal half of scNup145), scSeh1 (Sec13 homologue 1), and a fraction of scSec13 (Siniossoglou et al., 1996; Teixeira et al., 1997). This complex has been shown recently to exhibit a Y-shaped structure with an average diameter of 25 nm (Siniossoglou et al., 2000). Mutations of nucleoporins that belong to the scNup84 complex lead to specific defects in the export of mRNA and to a constitutive clustering of the NPCs (for review see Doye and Hurt, 1997). Recently, the vertebrate homologue of scNup145-C, Nup96, was shown to comigrate on a sucrose gradient with Nup107 (the vertebrate homologue of scNup84) (Radu et al., 1994; Siniossoglou et al., 1996), mammalian sec13, and p37 (a sec13-related protein), suggesting that the scNup84 complex might have been conserved partly during evolution (Fontoura et al., 1999).
Although scNup133 shares very similar phenotypes and is genetically linked to the members of the scNup84 complex (Doye et al., 1994; Siniossoglou et al., 1996), biochemical approaches did not previously allow the characterization of any interaction between scNup133 and constituents of the scNup84 complex. To address this question in another organism and get access to the dynamics of these nucleoporins at the various stages of the cell cycle, we have characterized the human homologue of scNup133. Two hybrid screens and immunoprecipitation experiments indicated that hNup133 is part of a vertebrate NPC subcomplex that also contains hNup107, hNup96, and a novel mammalian nucleoporin homologous to scNup120. Immunoelectron microscopy revealed that like their S. cerevisiae counterparts hNup107 and hNup133 are localized on both sides of the NPC. Using immunofluorescence and in vivo analysis of GFP-tagged hNup133 and hNup107, we demonstrate that hNup133 and hNup107 are stable components of the NPC in interphase, remain associated with each other during mitosis, and are targeted at early stages to the reforming nuclear envelope. Unexpectedly, this study further revealed that a fraction of hNup133 and hNup107 associates with the kinetochores from prophase to late anaphase and that the interaction of hNup133 with kinetochores is dynamic.
Results
The human homologue of S. cerevisiae Nup133 displays an evolutionarily conserved interaction with Nup84/107
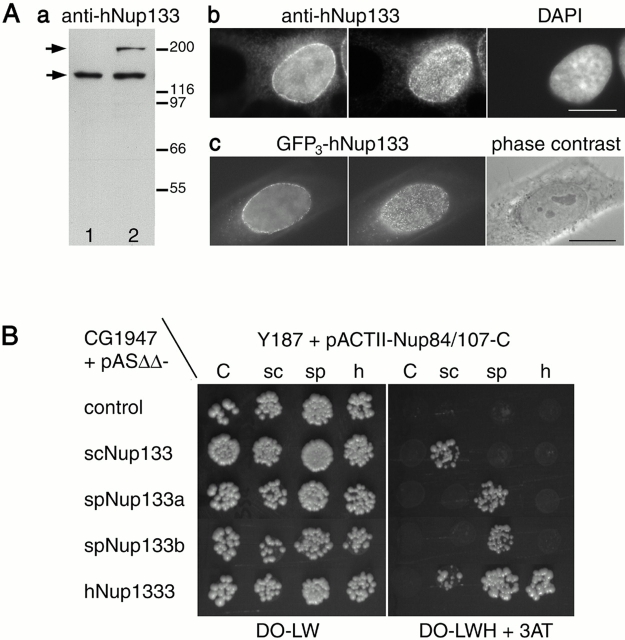
By screening Schizosaccharomyces pombe and higher eukaryote databases, we identified two distinct S. pombe ORFs (which we designated as spNup133a and spNup133b) and one human ORF (subsequently referred to as hNup133) that displayed 18–20% identity with scNup133 over their entire length (see Materials and methods; Fig. S1 available at http://www.jcb.org/content/vol154/issue6). Consistent with its homology with scNup133, indirect immunofluorescence analysis of hNup133 localization in HeLa cells revealed a punctuate nuclear rim staining typical for nuclear pores, a localization which was further confirmed in cells expressing GFP3-hNup133 (Fig. 1 A).
Figure 1.
hNup133 localizes to the NPC and interacts with hNup107 in the yeast two-hybrid assay. (A) hNup133 localizes to the NPC. (a) Whole cell extracts from HeLa cells either nontransfected (1) or transfected with the pEGFP3-hNup133 vector (2) were analyzed by Western blot using affinity purified anti-hNup133. Molecular mass markers are in kD. (b) Immunofluorescence analysis of methanol-fixed HeLa cells using affinity purified anti-hNup133 antibodies. Nuclear DNA was stained with DAPI. (c) Fluorescence analysis of living HeLa cells expressing the GFP3-hNup133 fusion. The shape of the nucleus is revealed on the corresponding phase–contrast image. In b and c, cells were photographed in two different focal planes that reveal the discontinuous nuclear rim (left) and the punctuate staining on the surface of the nuclear envelope (middle). (B) S. cerevisiae, S. pombe, and human Nup133 interact with Nup84/Nup107 in a yeast two-hybrid assay. Strain CG194 was transformed with pASΔΔ plasmids expressing the Gal4p-BD either alone (control) or in fusion with scNup133, spNup133a, spNup133b, or hNup133 as indicated. Strain Y187 was transformed with pACTII plasmids expressing the Gal4p-AD either alone (C) or in fusion with the COOH-terminal domain of scNup84 (sc), spNup107 (sp), or hNup107 (h). Following mating, diploids expressing both plasmids were spotted onto minimum medium either lacking leucine and tryptophan (DO-LW) or lacking leucine, tryptophan, and histidine but containing 5 mM 3-aminotriazole (DO-LWH + 3AT), and the plates were incubated at 30°C for 3 d. Bars, 10 μm.
To identify specific partners of this evolutionarily conserved nucleoporin, two-hybrid screens based either on scNup133 and an S. cerevisiae genomic library or on hNup133 and a human cDNA library were carried out (see Materials and methods). Strikingly, the most frequent candidates found in these screens were scNup84 and its human homologue, hNup107 (Radu et al., 1994; Siniossoglou et al., 1996). Comparison of the 5′ junctions and fragment sizes allowed the definition of the last 277 amino acids of scNup84 and the last 66 amino acids of hNup107 as the minimal domains sufficient for the interaction with scNup133 and hNup133, respectively. We further demonstrated that this interaction is evolutionarily conserved, since spNup133a and spNup133b also interact with spNup107, the S. pombe homologue of scNup84/hNup107 (Tran et al., 2001) (Fig. 1 B). Furthermore, the fact that scNup133 and both S. pombe Nup133 homologues interact almost exclusively with scNup84 and spNup107, respectively, in a species-specific manner (Fig. 1 B) strongly suggests that the two-hybrid interaction between Nup133 and Nup84/107 is direct (a putative bridging protein would indeed allow interaction between Nup133 and Nup84/107 from the various species).
hNup133 and hNup107 belong to an NPC subcomplex that contains hNup96 and a novel protein homologous to scNup120
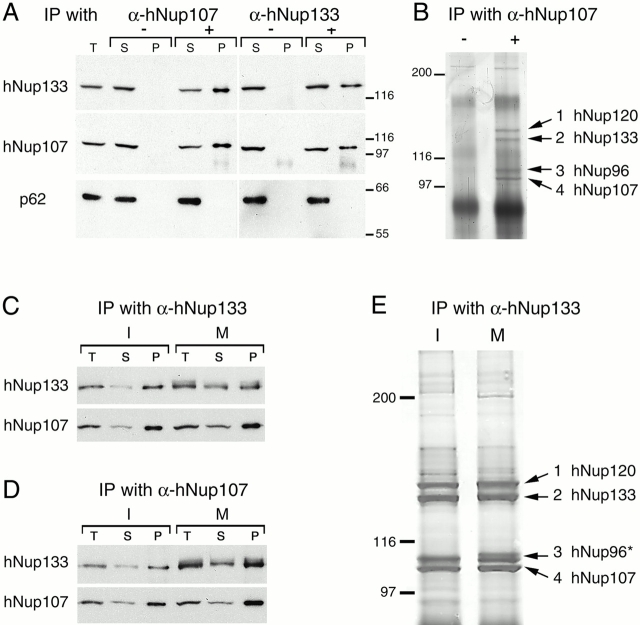
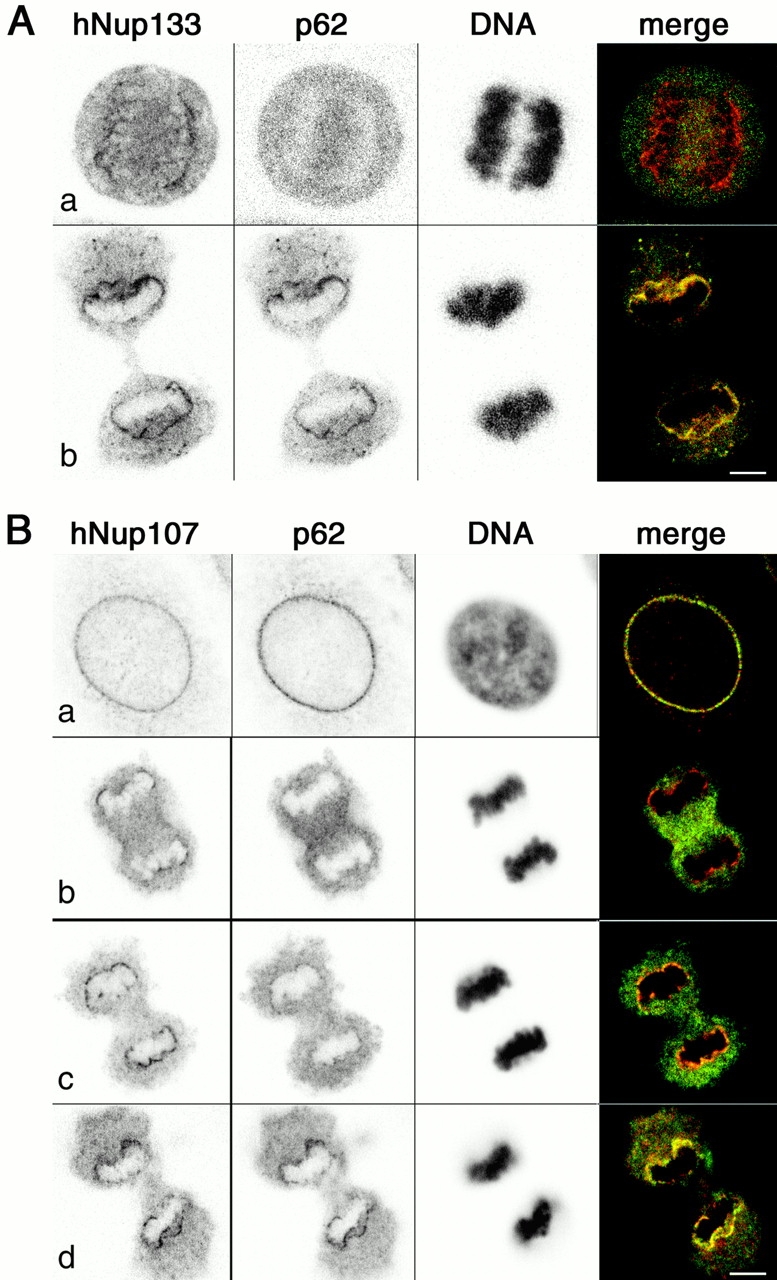
Although our data indicate that the two-hybrid interaction between Nup133 and Nup84/107 is highly specific and may also take place in wild-type yeast and human cells, scNup133 has never been reported to be present in the scNup84 complex (Siniossoglou et al., 1996, 2000; see Discussion). Therefore, we decided to characterize the interaction between hNup133 and hNup107 in human cells. Immunoprecipitation experiments performed on HeLa cell extracts using anti-hNup107 or anti-hNup133 antibodies revealed that these two nucleoporins coprecipitate efficiently with each other, whereas neither p62 nor any other nucleoporin recognized by the mAb414 antibody could be detected in the immune pellets (Fig. 2 A). Silver-staining analysis of the immunoprecipitates obtained with anti-hNup107 (Fig. 2 B) and anti-hNup133 (Fig. 2 E) reproducibly revealed four specific bands of similar intensity that migrated in the 100–150-kD range. Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) and quadrupole time-of-flight (Q-TOF) spectrometry analysis confirmed the identity of bands 2 and 4 as hNup133 and hNup107 and further identified band 3 as hNup96 (the human homologue of scNup145-C) and band 1 as an uncharacterized 149-kD protein referred to in the database as KIAA0197 (sequence data available from GenBank/EMBL/DDBJ under accession no. G02870). Blast searches revealed that the KIAA0197 protein showed significant homology to an S. pombe ORF coding for a 130-kD protein designated as SPBC3B9.16C (sequence data available from GenBank/EMBL/DDBJ under accession no. T40355) and to scNup120 (Fig. S2 available at http://www.jcb.org/content/vol154/issue6). Accordingly and irrespective of their apparent molecular weights, we have designated the human and the S. pombe proteins as hNup120 and spNup120, respectively.
Figure 2.
Immunoprecipitation of HeLa cell extracts using anti-hNup107 or anti-hNup133 antibodies. (A) Immunoprecipitation of HeLa interphasic cell extract using preimmune (−) or immune (+) sera directed against the NH2-terminal domain of hNup107 or against hNup133. Equivalent amounts of total extracts (T) and immune supernatants (S) and a fivefold equivalent of the immune pellets (P) were analyzed by immunoblot using anti-hNup133, hNup107, or the mAb414 antibody that mainly recognized p62. (B) Silver staining of immunoprecipitates from HeLa cell extracts obtained with preimmune (−) or immune (+) anti-hNup107 serum. (C and D) Same as A, using interphasic (I) or mitotic (M) extracts from HeLa cells. Note that hNup133 migrates as a more diffuse band in the mitotic extracts and that the slowly migrating forms of hNup33 are also precipitated efficiently by the anti-hNup107 antibody. (E) Interphasic (I) or mitotic (M) immune pellets obtained with affinity purified anti-hNup133 antibodies were analyzed by silver staining. In both samples, the same four prominent bands corresponding to hNup120, hNup133, hNup96, and hNup107 are present. In the immune pellet from mitotic extract, band 3 migrates as a doublet (*).
It was demonstrated previously that Nup107 sediments on sucrose gradients from enriched NPC fractions as a unique peak that also contains Nup96, mSec13, a novel sec13-related protein, and two additional proteins of ∼150 kD (Fontoura et al., 1999). Although the existence of distinct hNup107-containing complexes of similar properties cannot be ruled out formally, these sedimentation data together with our results, demonstrating the coprecipitation of the same subset of nucleoporins with both anti-hNup133 and anti-hNup107 antibodies, suggest strongly that hNup133 belongs to the hNup107 NPC subcomplex that also contains at least hNup120 and hNup96 (see Discussion). The two proteins of ∼150 kD observed by Fontura et al. (1999) thus likely correspond to hNup120 and hNup133.
During mitosis, the soluble components of mammalian NPCs are partially disassembled into subcomplexes and dispersed throughout the mitotic cytosol. To analyze the biological relevance of this biochemically identified NPC subcomplex in mitotic cells, we performed immunoprecipitation experiments from interphase and mitotic populations of HeLa cells. The four major constituents of the hNup133-hNup107–containing complex were found to coprecipitate in mitotic cell extracts with a stoichiometry similar in both interphasic and mitotic precipitates (Fig. 2, C–E), demonstrating that these nucleoporins remain as a stable subcomplex during mitosis.
hNup133 and hNup107 are localized on both sides of the nuclear pore complex
We determined the localization of these human nucleoporins within the NPC using at first a differential permeabilization protocol based on the fact that low concentrations of digitonin permeabilize the plasma membrane but leave the nuclear membrane intact (Adam et al., 1990). Although lamin B or Nup153, used as controls, were detected only in Triton X-100–permeabilized cells, anti-hNup133 and anti-hNup107 antibodies labeled the nuclear envelope in both digitonin- and Triton X-100–permeabilized cells, indicating that these nucleoporins are at least located at the cytoplasmic face of the NPC (unpublished data).
To determine whether these two nucleoporins are also present on the nuclear side of the NPC, ultrathin sections of HeLa cells fixed and processed for ultracryomicrotomy were immunogold-labeled using as primary antibodies an affinity purified anti-hNup133 antibody (Fig. 3 A) or two distinct anti-hNup107 antibodies (Fig. 3, B and C). Analysis of the distribution of the gold particles revealed that like their S. cerevisiae homologues hNup133 and hNup107 are localized to both the cytoplasmic and nucleoplasmic faces of the nuclear envelope (Fig. 3, right).
Figure 3.
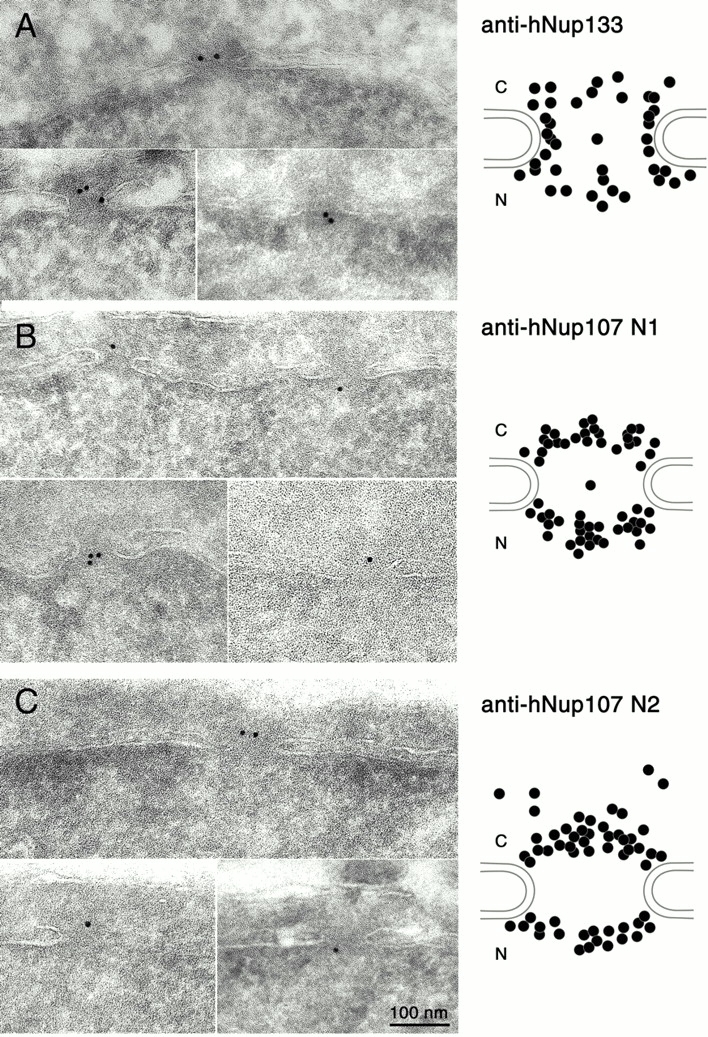
Immunolocalization of hNup133 and hNup107 on both sides of the NPC. Cryo-sections of paraformaldehyde-fixed HeLa cells were labeled with affinity purified anti-hNup133 antibody (A), anti-hNup107-N1 serum (B), or affinity purified anti-hNup107-N2 antibody (C). Typical patterns of nuclear pore labeling obtained with the various antibodies are shown. The cytoplasmic faces of the nuclear envelopes are oriented towards the top of each micrograph. Statistical analysis of the distribution of the gold particles over the NPC is presented on the right.
hNup133 and hNup107 are stably associated with the NPC during interphase
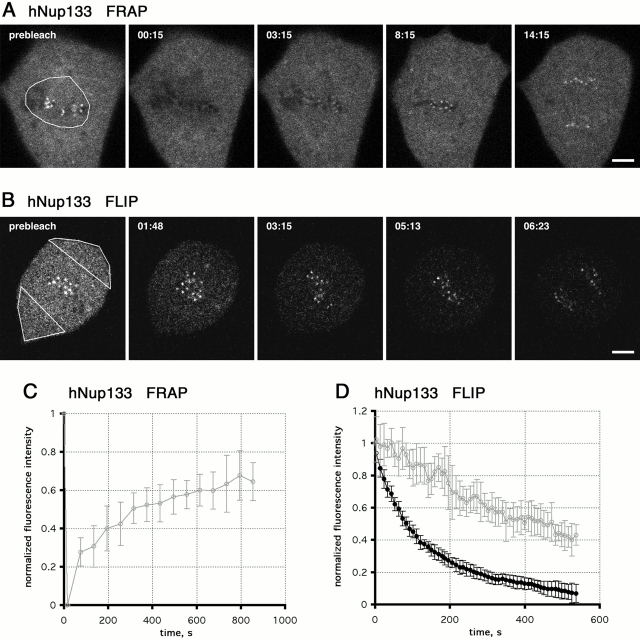
To gain insight into the dynamics of hNup133 and hNup107, we selected mammalian cell lines stably expressing triple GFP-tagged versions of both proteins. In these cells, FRAP was used to assay the turnover of both nucleoporins in the NPC from interphase cells. Strikingly, neither hNup133 nor hNup107 showed any detectable recovery up to 1 h after the photobleach (unpublished data). Thus, we performed FRAP experiments where we monitored the recovery in living cells for up to 30 h after the photobleach (Fig. 4 A). Although the kinetics of recovery tended to vary between individual cells depending on the length of interphase, we found that hNup107 and hNup133 did not recover >60% of their fluorescence during interphase even if cells were bleached in early G1 (4 h after anaphase onset) (Fig. 4, A–C; videos 1 and 2 available at http://www.jcb.org/content/vol154/issue6). Accordingly, both hNup133 and hNup107 remain very stably associated with the same NPC for the entire duration of interphase (∼16 h in the cell lined used). In addition, the bleached region did not intermix with the unbleached areas of the nucleus before the next mitosis (Fig. 4, A and B), indicating a low mobility of mammalian NPCs within the plane of the nuclear envelope (in agreement with our recent study, Daigle et al., 2001). Cells in which half the nuclear pores were bleached divided normally, and the daughter cells formed nuclei with a uniform distribution of the unbleached molecules in all nuclear pores (Fig. 4 D). Thus, in mitosis bleached and unbleached hNup133-hNup107–containing complexes are released from the whole NPC and randomly partitioned between the daughter cells where they assemble normally into new NPCs early after mitosis. The fact that the bleached cells divided normally and that both nucleoporins equilibrated with all new NPCs after mitosis also demonstrates that the low turnover was not due to photo damage.
Figure 4.
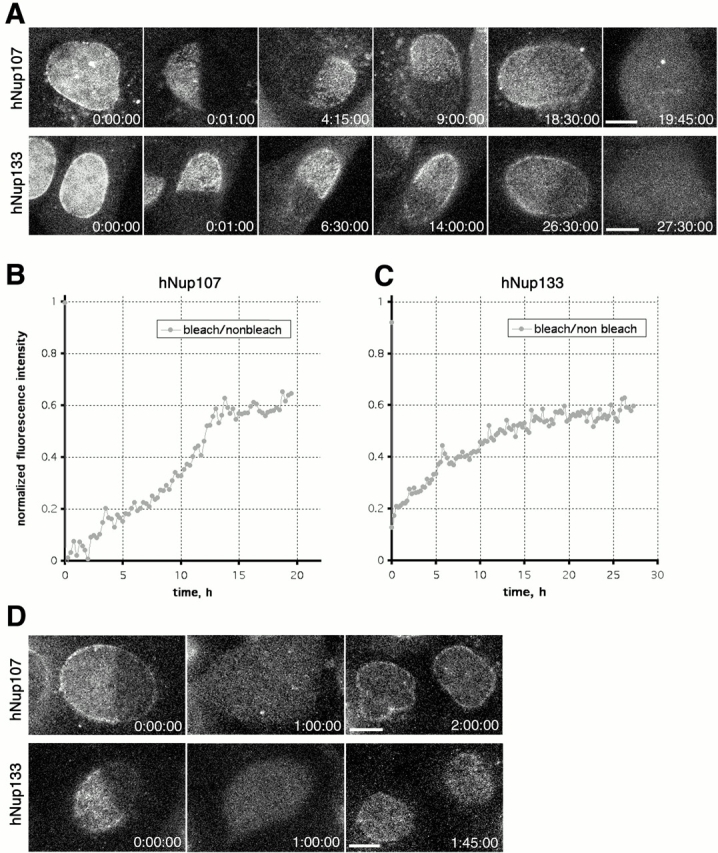
Turnover of GFP3-hNup107 and GFP3-hNup133 at the NPCs measured by FRAP. (A) Long-term FRAP experiment of NRK cells stably expressing GFP3-hNup107 or GFP3-hNup133. Half of the nucleus was bleached ∼5 h after mitosis in G1, and the fluorescence recovery was followed until the next mitosis. A confocal section focussed automatically on the lower part of the nucleus was acquired every 15 min. The images were contrast enhanced for display in a still figure. See also videos 1 and 2 available at http://www.jcb.org/content/vol154/issue6. (B and C) Ratios of background-subtracted average intensities of the bleached half versus the nonbleached half of the nuclei shown in A are plotted against time. Data points are 15-min intervals. (D) Confocal sections of an NRK cell stably expressing GFP3-hNup107 or GFP3-hNup133 before, during, and after mitosis. Bleaching and acquisition conditions were the same as in A. The images are contrast enhanced and smoothed with a 3 × 3 median filter. Time is indicated in hh:mm:ss. Bars, 5 μm.
hNup133 and hNup107 are recruited at early stages of nuclear envelope reassembly
We further analyzed the kinetics of recruitment of these two nucleoporins during nuclear envelope reassembly by comparing their behavior to that of two previously characterized nucleoporins, p62 and Nup153 (Bodoor et al., 1999) (Fig. 5). Double immunofluorescence analysis of paraformaldehyde-fixed HeLa cells revealed that hNup133 and hNup107 concentrated at the nuclear periphery during anaphase well in advance of p62 (Fig. 5, A and B) and with a timing roughly coincident with that of Nup153 (Fig. 5, C and D). As a complementary approach, we also followed the in vivo dynamics of GFP3-hNup133 and GFP3-hNup107 in stably transfected mitotic cells from metaphase to G1 of the cell cycle by fast high resolution four-dimensional confocal imaging (see Materials and methods). As a temporal reference, metaphase/anaphase transition identified in simultaneously acquired DIC image stacks (Fig. 6, A and B, right columns) was used as time zero. In agreement with data obtained in fixed cells, we found that both nucleoporins exhibited an early recruitment from the soluble cytoplasmic pool to the chromosome periphery, producing a well-defined rim in late anaphase (∼4 min after anaphase onset) (Fig. 6, A and B; videos 3 and 4 available at http://www.jcb.org/content/vol154/issue6). In telophase (∼8 min after anaphase onset), the majority of the soluble pool of both proteins had been recruited in a clear rim pattern enclosing the chromosomes. A diffuse recruitment of GFP3-hNup107 but not GFP3-hNup133 to the chromosome periphery was also observed from early anaphase (∼1 min after anaphase onset) until late anaphase when it started to accumulate in a clear rim pattern concomitant with hNup133's recruitment to the chromosome periphery (Fig. 6, A, 04:40, compared with B, 04:05).
Figure 5.
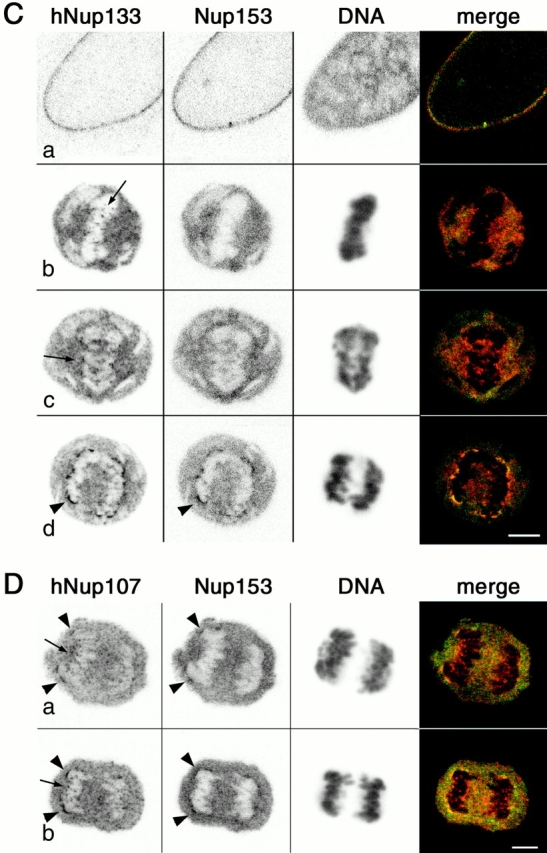
Fluorescence microscopy of HeLa cells labeled with anti-hNup133 and anti-p62 (A), anti-hNup107 and anti-p62 (B), anti-hNup133 and anti-Nup153 (C), and anti-hNup107 and anti-Nup153 (D) and stained with DAPI. Cells were fixed in formal dehyde and permeabilized with Triton X-100. Digital confocal micrographs of cells at various stages of mitosis are presented. (Merge) Superimposition of the hNup133 or hNup107 (red) and p62 or Nup153 (green) signals. hNup133 and hNup107 appear to be recruited to the chromatin periphery clearly ahead of p62 and coincident to or slightly ahead of Nup153 in anaphase (C and D, arrowheads). Arrows in C and D point to specific dot-like structures labeled with hNup133 or hNup107 but not Nup153. Bars, 5 μm.
Figure 6.
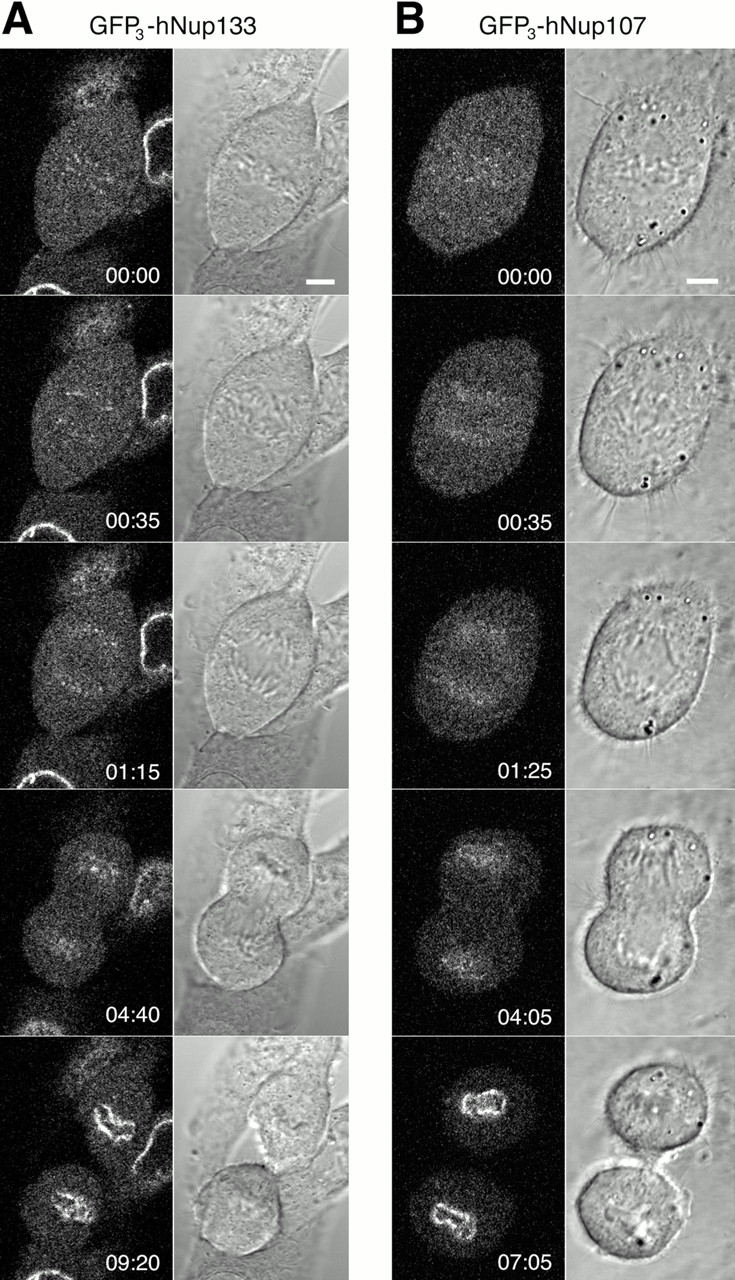
In vivo analysis of GFP3-hNup133 and GFP3-hNup107 dynamics in mitosis. (A) Four-dimensional confocal sequence of an NRK cell stably expressing GFP3-hNup133. The sequence extends from metaphase/anaphase transition to telophase. Confocal z-stacks with six slices every 1.5 μm were acquired in 5-s intervals. A single z-slice containing the best-focused GFP signal is displayed for representative time points. Left panels show GFP fluorescence to localize the nucleoporin; right panels show the simultaneously acquired DIC images to visualize the chromosomes. Due to the low signal to noise ratio in a single confocal section, fluorescence images were contrast enhanced and smoothed with a 3 × 3 median filter to remove detector shot noise. Time is indicated in mm:ss. See also video 3 available at http://www.jcb.org/content/vol154/issue6. (B) Same as in A but from a cell stably expressing GFP3-hNup107. Time interval between z-stacks is 10 s. Note the earlier diffuse recruitment of GFP3-hNup107 to the chromatin surface compared with GFP3-hNup133. See also video 4 available at http://www.jcb.org/content/vol154/issue6. Bars, 5 μm.
hNup133 and hNup107 localize to kinetochores in mitotic cells
Unexpectedly, immunofluorescence analysis of paraformaldehyde-fixed cells in metaphase or early stages of anaphase using anti-hNup133 or anti-hNup107 antibodies also revealed a faint albeit reproducible dot-like labeling at the center of the metaphase plate or adjacent to the spindle in anaphase cells (Fig. 5, C and D, arrows). A similar dot-like localization was also detectable over the surrounding cytoplasmic signal for GFP3-hNup133 and GFP3-hNup107 in stably transfected mitotic NRK cells (Figure 6, A and B). Confocal stacks and three-dimensional reconstructions from such live metaphase cells treated with low concentrations of the in vivo DNA dye Hoechst 33342 showed paired dots on both sides of each metaphase chromosome, a pattern characteristic of centromere/kinetochore localization (Fig. 7, A and B). Immunofluorescence analyses of methanol-fixed HeLa cells using an autoimmune CREST serum that labels constitutive constituents of the centromeres/kinetochores confirmed that the dot-like structures labeled by GFP3-hNup133 in stably transfected HeLa cells (Fig. 7 C) and by our various anti-hNup107 and anti-hNup133 antibodies in nontransfected HeLa cells (Fig. 8, A and B) correspond to kinetochores. Higher magnification of merged images from metaphase and early anaphase cells further revealed that the hNup107 or hNup133 signals were located at the outer region of centromeres adjacent to the CREST staining (Fig. 8 A; unpublished data).
Figure 7.
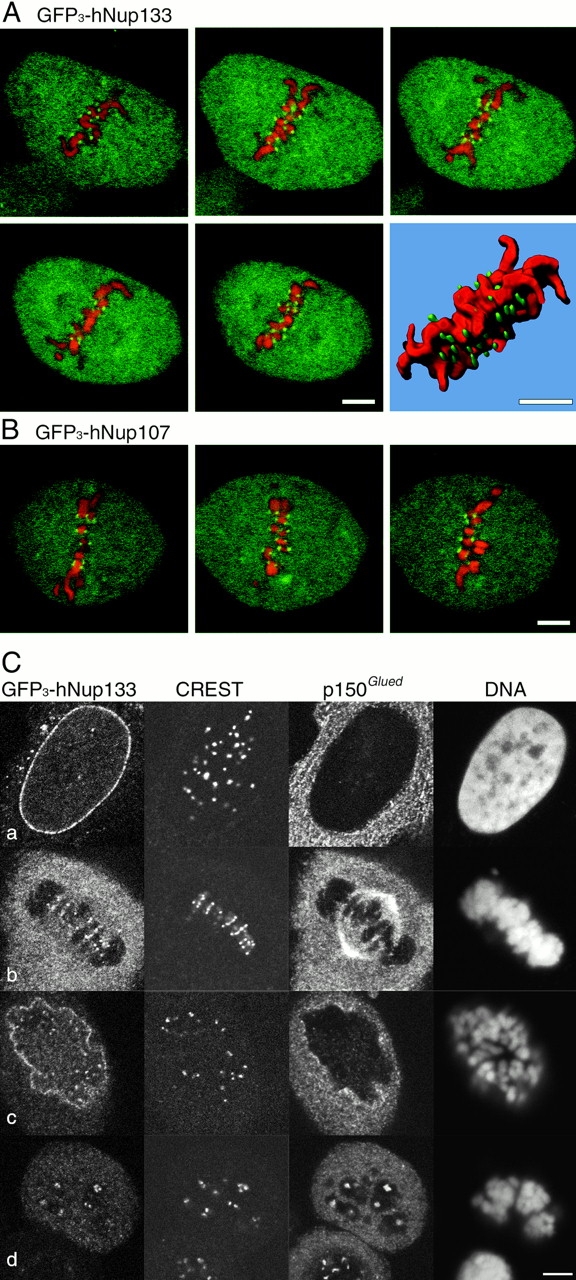
GFP3-hNup133 and GFP3-hNup107 localize to kinetochores of mitotic cells. (A and B) Live metaphase NRK cells stably expressing GFP3-hNup133 (A) or GFP3-hNup107 (B) (green) and stained with Hoechst 33342 (10 ng/mL; red) were imaged by confocal microscopy. (A) Five consecutive optical sections acquired every 1.5 μm show GFP3-hNup133 on kinetochores whose distribution on both sides of the metaphase plate is illustrated by the three- dimensional reconstruction of the entire z-stack. (B) The same localization pattern is observed in cells stably expressing GFP3-Nup107 shown in three representative optical sections. (C) HeLa cells expressing GFP3-hNup133 fixed with cold methanol and labeled with CREST serum, anti-p150Glued, and DAPI. Cells in c and d were treated for 14 h with nocodazole (5 × 10−7 M). Digital confocal micrographs of cells respectively in interphase (a), metaphase (b), prophase (c), or blocked in prometaphase (d) are presented. Bars, 5 μm.
Figure 8.
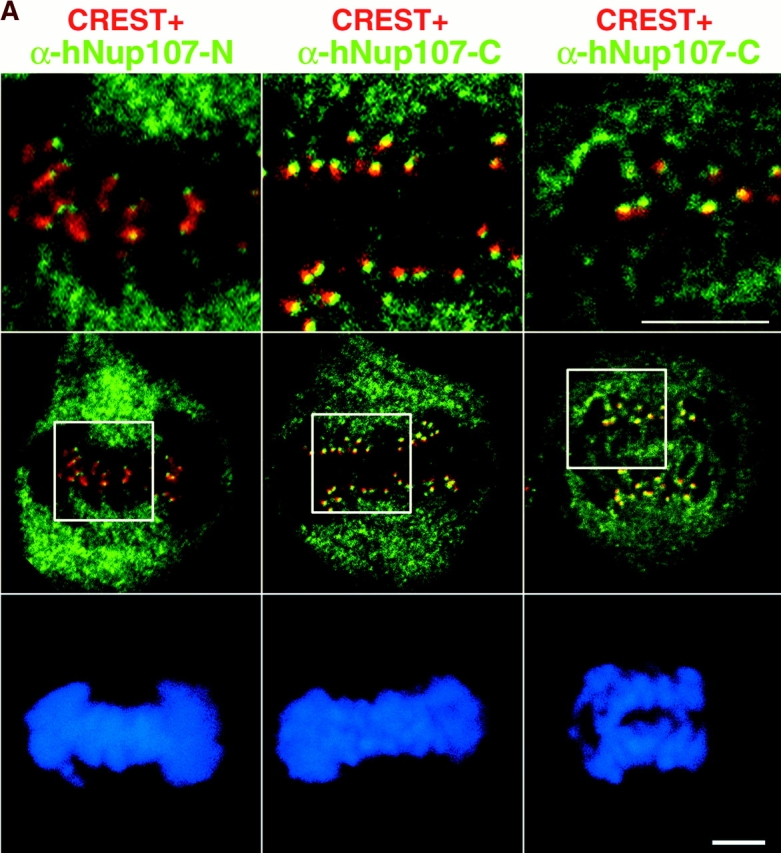
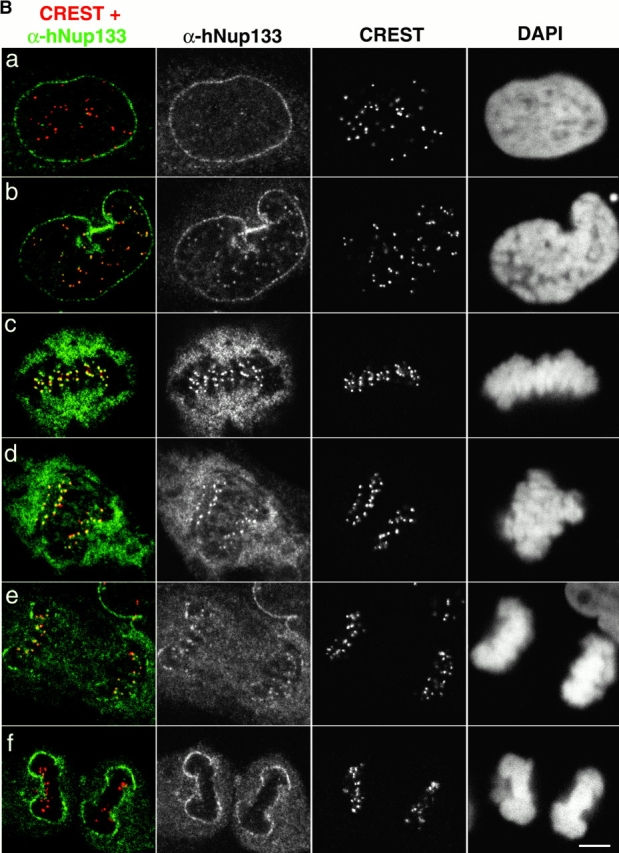
A fraction of endogenous hNup107 and hNup133 colocalizes with kinetochores in mitosis. (A) Mitotic HeLa cells were labeled with CREST serum, DAPI, and as indicated affinity purified antibodies against the NH2-terminal domain of hNup107 (α-hNup107-N) or its COOH-terminal domain (α-hNup107-C). Superimposition of the hNup107 (green) and CREST (red) signals are presented. Top, higher magnifications of the insets; bottom, DAPI. (B) HeLa cells at various stages of the cell cycle were labeled with anti-hNup133 (green), CREST serum (red), and DAPI. Digital confocal micrographs of cells respectively in interphase (a), prophase (b), metaphase (c), early anaphase (d), late anaphase (e), and telophase (f) are presented. Bars, 5 μm.
In contrast to the CREST serum that labels the centromeres throughout the cell cycle, the labeling observed with anti-hNup133 and anti-hNup107 was not observed in interphase cells (Fig. 8 B, a; unpublished data). Since assembly of the kinetochores is a hierarchical process (Chan et al., 1998), we further characterized the timing of appearance of hNup133 at the kinetochores. A faint yet specific staining was observed already in prophase, while hNup133 was still mainly localized to the nuclear envelope (Fig 7 C, c, and Fig. 8 B, b). At this stage p150Glued, a subunit of the dynactin complex that transiently associates with the nuclear envelope during prophase (Busson et al., 1998) and with kinetochores during prometaphase (Echeverri et al., 1996) was still excluded entirely from the nuclear interior (Fig. 7 C, c; unpublished data). Unlike several kinetochore-associated proteins, hNup133 staining persisted on the kinetochore of aligned chromosome and remained detectable on the kinetochores until late anaphase at a time point when it was associated already with the reforming nuclear envelope (Fig. 8 B, e). Finally, the kinetochore localization of a fraction of hNup133 was also observed in nocodazole-treated cells, indicating that it is independent of the continuous presence of microtubules (Fig. 7 C, d; unpublished data).
Dynamics of hNup133 at the kinetochore
The low turnover of hNup107/hNup133 in the nuclear envelope prompted us to investigate the dynamics of their interaction with kinetochores. To this end, NRK cells stably expressing GFP3-hNup133 that gave a slightly higher kinetochore signal than GFP3-hNup107 cells were used. From prometaphase to early anaphase, these cells contained only two pools of hNup133: diffuse cytoplasmic and kinetochore localized. Four-dimensional FRAP experiments (see Materials and methods) in which the entire kinetochore fraction of GFP3-hNup133 was photobleached were carried out to see if this pool could recover from the cytoplasmic reservoir. FRAPs were started in prometaphase and completed before anaphase onset when the imminent nuclear assembly would have complicated the measurement. All cells used for photobleaching initiated anaphase normally, serving as a control for photo damage (Fig. 9 A, fifth panel). Recovery of kinetochore hNup133 was observed after several minutes (Fig. 9 A) with a halftime of 132 ± 64 s (Fig. 9 C), meaning that it took over 2 min to exchange half of the GFP3-hNup133 molecules bound to kinetochores with freely diffusing molecules in the cytoplasm. An immobile fraction of 34 ± 16% that did not exchange at all over the course of ∼10 min was also characterized in these experiments (Fig. 9 C). Using a simple kinetic model for the exchange of the mobile fraction of GFP3-hNup133 with a single binding site on kinetochores (Fig. S3 and supplemental methods available at http://www.jcb.org/content/vol154/issue6), we could estimate its dissociation constant as k off = 0.0058−1 ± 0.0026−1 s. Thus 1/k off, the mean residence time, was 172 ± 77 s, meaning on average each GFP3-hNup133 molecule spent ∼3 min bound to kinetochores before it exited to the cytoplasm.
Figure 9.
Turnover of GFP3-hNup133 on kinetochores. (A and B) GFP3-hNup133 binding to kinetochores was analyzed by FRAP (A) or FLIP (B) experiments in stable NRK cells. Bleach regions are outlined in the prebleach images. Note the incomplete recovery in FRAP after 8 min (A, 8:15) and the significant fluorescence remaining on kinetochores after cytoplasmic fluorescence was depleted to background in the FLIP (B, 5:13). (C) Plot of mean kinetochore recovery over time of FRAP experiments similar to A. Error bars are standard deviations (n = 10). (D) Plot of mean kinetochore (gray) and cytoplasmic (black) intensity over time with standard deviation (n = 5) of FLIP experiments similar to B. See also Fig. S3 and supplemental methods available at http://www.jcb.org/content/vol154/issue6. Bars, 5 μm.
As an independent way of determining these parameters, we performed fluorescence loss in photobleaching (FLIP) experiments in which the kinetochore fraction would be depleted indirectly by repetitive bleaching of the cytoplasm if it was able to exchange from its binding site (Fig. 9 B). As expected, kinetochores lost ∼60% of their GFP3-hNup133 fluorescence over the course of ∼8 min, albeit with much slower kinetics than the directly bleached cytoplasm (Fig. 9, B and D). The different kinetics demonstrate that depletion of kinetochores is not limited by diffusion but by the slower dissociation of hNup133 from kinetochores. Using a simple two-compartment model of fluxes between kinetochores and cytoplasm (methods available at http://www.jcb.org/content/vol154/issue6), we estimate the dissociation from kinetochores to k off = 0.0054−1 ± 0.0013−1 s by fitting the experimental FLIP data (Fig. S3 and supplemental methods available at http://www.jcb.org/content/vol154/issue6). k off obtained by this independent approach is in excellent agreement with the FRAP result. In addition, it should be noted that we could not deplete >70% of the kinetochore fluorescence by repetitive bleaching and that the quality of the fit to the experimental data by the compartmental model was improved if ∼30% of the kinetochore-bound fraction was defined as immobile (Fig. S3 and supplemental methods available at http://www.jcb.org/content/vol154/issue6). This further validates the finding of a significant immobile fraction of hNup133 on kinetochores in our FRAP experiments.
Discussion
The S. cerevisiae Nup84- and the human Nup107-containing NPC subcomplexes: similarities and specificities
In this paper, we have identified hNup133, a human nucleoporin homologous to scNup133, and demonstrated its evolutionarily conserved interaction with Nup84/107. Immunoprecipitation experiments revealed that the same subset of nucleoporins (namely hNup120, hNup133, hNup96, and hNup107) can be pulled down with either anti-hNup133 or anti-hNup107 antibodies. Thus, these data strengthen and extend the previously reported interaction between Nup107 and Nup96, which was so far based on their cosedimentation as a unique peak on a sucrose gradient from solubilized NPCs (Fontoura et al., 1999). They also indicate the existence of a major hNup107 complex that includes, besides hNup133, other nucleoporins homologous to constituents of the scNup84 complex.
Although hNup133 thus appears as a constitutive subunit of the hNup107 complex, scNup133 had not been detected previously in the scNup84 complex (Siniossoglou et al., 1996, 2000). This could result from the high sensitivity of scNup133 to proteolysis. Alternatively, considering that scNup133 interacts directly with scNup84 (as demonstrated by the species-specific two-hybrid interaction) and that scNup84 resides peripherally within the scNup84 complex (Siniossoglou et al., 2000), scNup133 might be associated loosely to the periphery of the scNup84 complex. Conversely, although scNup85 is an essential constituent of the scNup84 complex (Siniossoglou et al., 2000), no major band could be detected in the 75-kD range (the theoretical molecular weight of the putative human homologue of scNup85; unpublished data) on silver-stained gels from hNup133 or hNup107 immune pellets. In addition, no specific protein migrating at ∼75 kD was recorded to cosediment specifically on the sucrose gradient with Nup96 and Nup107 (Fontoura et al., 1999). If present in the hNup107 complex, the putative mammalian homologue of scNup85 may, as hypothesized for scNup133, be either loosely associated with this complex or more sensitive to proteolysis. Alternatively, hNup85 may not migrate at its expected molecular weight. Finally, although mSec13 and a novel sec13-related protein were found also to cosediment with rat Nup107 and Nup96 (Fontoura et al., 1999), their presence in our immune pellets could not be assessed so far, since their putative signals could not be resolved from the immunoglobulin light chains.
Immunofluorescence experiments on semipermeabilized cells and immunoelectron microscopy revealed that hNup133 and hNup107 are localized on both sides of the NPC. However, previous studies performed with an antibody directed against a 200–amino acid domain of hNup96 revealed its specific localization on the nucleoplasmic side of the NPC (Fontoura et al., 1999). This could result from the specific enrichment on the nucleoplasmic side of the NPC of a fraction of Nup96, which would not associate with the hNup107 complex. Conversely, Nup96 may be present only in a subfraction of hNup107 complexes containing the other components and localized on the nucleoplasmic side of the NPC. However, the fact that Nup96 sediments as a unique peak on the sucrose gradient (Fontoura et al., 1999) together with the previously described localization of scNup133 and the various members of the scNup84 complex (Rout et al., 2000) rather suggests that the entire hNup107 NPC subcomplex displays a symmetric localization on both sides of the NPC (Fontoura et al., 1999). Thus, the observed discrepancy could be due to a biased accessibility of the epitope recognized by this anti-Nup96 antibody. Together, these data indicate that the scNup84- and hNup107-containing complexes are structurally and possibly functionally related.
The hNup107 complex behaves as a structural building block of the mammalian NPCs
The characterization in mammalian cells of this evolutionarily conserved NPC complex enabled us to address the dynamics of its subunits in both interphase and mitosis thereby providing a set of functional data that could not be obtained in yeast. Immunofluorescence studies and four-dimensional in vivo imaging during nuclear envelope reassembly revealed an early recruitment of both nucleoporins from the soluble cytoplasmic pool to the chromosome periphery, producing a well-defined rim in late anaphase. In vivo imaging also indicated that a fraction of GFP3-hNup107 was recruited diffusely to the chromosome surface ∼2–3 min earlier than hNup133, a rather unexpected difference, since our biochemical data indicate that hNup133 and hNup107 remain in a stable complex in metaphase-arrested cells. Because the in vivo approach relies on the use of GFP-tagged nucleoporins, this discrepancy could be due to the mild overexpression inherent to the use of stably transfected tissue culture cells. Alternatively, there might also be a pool of uncomplexed subunits for both proteins. The chromosome-recruited fraction of hNup107 might then provide a template for the formation of additional hNup107/hNup133 complexes during anaphase.
In either case, the early chromosome labeling, which precedes by several minutes the formation of the nuclear membrane (unpublished data) and the recruitment of many other nucleoporin characterized to date (Bodoor et al., 1999), makes the hNup107 complex a likely key player in an early step of NPC reassembly after mitosis. In addition, FRAP experiments revealed an extremely low turnover of both nucleoporins in single NPCs during interphase. The identical behavior of these two soluble nucleoporins strongly suggests that the entire hNup107 complex is bound tightly to the NPC and is exchanged only once per cell cycle, consistent with a structural function of this complex within the NPC. In comparison, Nup153 has a high turnover rate in the order of seconds, whereas such a stable association with the NPC was so far observed only for the membrane-anchored POM121 nucleoporin (Daigle et al., 2001).
Thus, in the future it will be of interest to correlate these data with the proposed structural intermediates in NPC assembly that have been imaged in Xenopus egg extracts using high resolution scanning EM (Goldberg et al., 1997). In particular, because of the Y shape of the scNup84 complex (Siniossoglou et al., 2000) it is tempting to speculate that this conserved NPC subcomplex might build up the star-ring region of the NPC, a structure consisting of eight triangular subunits that appears as an early intermediate in NPC assembly (Goldberg et al., 1997).
A novel link between NPC constituents and kinetochores
The most unexpected result arising from this study was the localization of a pool of both hNup133 and hNup107 to the mitotic kinetochores. This dual localization does not reflect the existence of specific splice variants recognized by the antibodies, since a fraction of transfected GFP-tagged hNup133 or hNup107 is recruited similarly to the kinetochores during mitosis. Our immunofluorescence data and in vivo labeling studies indicate that only a minor fraction of hNup133 and hNup107 is localized at the kinetochores, the remaining pool giving rise to a diffuse staining throughout the mitotic cytoplasm. The overall stability of the hNup107 complex in metaphase-arrested cells suggests that all of the constituents of the hNup107 complex may associate with kinetochores. However, since this kinetochore-associated fraction would not affect the overall yield of our immunoprecipitation experiments we cannot rule out that only hNup133 and hNup107 (which directly interact with each other) associate with the kinetochores.
Although we demonstrated that hNup133 is a stable constituent of NPC during interphase, both FRAP and FLIP experiments showed that the kinetochore fraction of GFP3-hNup133 exchanges with the diffuse cytoplasmic pool. 30% of the kinetochore-bound pool exchanged with the cytoplasm every minute, and we detected no change in the rate of this exchange from prometaphase to anaphase. The recovery during FRAP experiments shows clearly that the mitotic cytoplasm contains kinetochore-binding competent hNup133 molecules and that interaction of this nucleoporin with kinetochores is dynamic. In addition, these photobleaching experiments also pointed to a significant immobile fraction of ∼34% of hNup133 on kinetochores, suggesting the presence of a second higher affinity binding site for this nucleoporin on kinetochores.
Interestingly, while this work was in progress Mad1 and Mad2, two mitotic checkpoint proteins that transiently associate with the kinetochores during mitosis (for review see Shah et al., 2000), were described as being localized to the nucleocytoplasmic face of the NPC throughout interphase (Campbell et al., 2001). Conversely, the mRNA export factor hRae1 was shown to interact with the mitotic checkpoint protein mBUB1 and to be localized at kinetochores of prometaphase chromosome (Wang et al., 2001). In addition, a few other proteins including a fraction of the SUMO-1–modified form of RanGAP1 (Matunis et al., 1996), a fraction of tankyrase (Smith and de Lange, 1999), and the PCB68 antigen (Theodoropoulos et al., 1999), have been reported previously to relocalize from the NPCs to microtubule-associated structures during mitosis. Finally, it was demonstrated recently that a mutation in the S. cerevisiae nucleoporin gene NUP170 leads to defects in chromosome transmission fidelity and kinetochore integrity in this organism (Kerscher et al., 2001). Together with the involvement of the nuclear transport factor Ran in mitotic spindle formation (for review see Azuma and Dasso, 2000), these data support the notion that the nucleocytoplasmic transport and mitotic machineries contain multiple shared components. They further suggest that the shared localization of a subset of NPC/kinetochore constituents may be an important and so far neglected aspect of cell division or nuclear transport.
What could be the function(s) of these structural nuclear pore constituents at the kinetochores? One hypothesis is that this fraction of hNup133 and hNup107 may act as “kinetochore-associated passenger proteins” that use kinetochores as a means of conveyance to be positioned correctly during the subsequent mitotic events (for review see Adams et al., 2001). Unlike the currently described bona fide “passenger proteins,” this minor pool of nucleoporins does not redistribute to the spindle midzone in anaphase but persists as a distinctly localized pool during nuclear envelope assembly in late anaphase and would possibly be positioned properly to allow the assembly of a specific subset of NPCs in late anaphase or early telophase. Alternatively, hNup133 and hNup107 may play a specific role on kinetochores in mitotic cells. Although the dynamics of kinetochores constituents had been addressed so far only for Mad2 (Howell et al., 2000), our photobleaching experiments, demonstrating that the interaction of hNup133 with kinetochores is dynamic, argues against hNup133 as a structural constituent of kinetochores. Conversely, the dissociation rate of hNup133 from kinetochores is about fivefold slower than the one measured previously for GFP-Mad2 (Howell et al., 2000). This and the fact that hNup133 kinetochore localization is maintained even through the early stages of nuclear assembly in late anaphase actually argues against a direct involvement of hNup133 in the “spindle assembly” checkpoint. Although the function of the kinetochore fraction of hNup107/hNup133 still remains to be determined (a study that will probably be complicated by their dual NPC/kinetochore localization), it is intriguing to speculate that these nucleoporins could provide a link between cell cycle control and NPC disassembly/assembly, a question currently under investigation in our laboratories.
Materials and methods
Identification, cDNA cloning, and GFP-tagging of hNup133 and hNup107
Two distinct S. pombe homologues of scNup133, subsequently referred to as spNup133a (sequence data available from EMBL/GenBank/DDBJ under accession no. CAB87368.1; SPBP35G2.06c gene product) and spNup133b (accession no. CAB55845.1; SPAC1805.4 gene product) were initially identified by using the S. pombe BLAST server at the Sanger Center (Cambridge, UK). Further BLAST searches using these two sequences subsequently helped us to identify several human ESTs encoding a protein 100% identical to the Homo sapiens hypothetical protein FLJ10814 mRNA (NM_018230). A 3.9-kb I.M.A.G.E. Consortium (LLNL) human cDNA clone (ID 1069918) (Lennon et al., 1996), encoding most of hNup133 ORF (amino acids 12–1156), was obtained from the Resource Center of the German Human Genome Project at the Max-Planck-Institut for Molecular Genetics (http://www.rzpd.de). This cDNA was subcloned into the pEGFP3-C1 vector that contains a triple GFP concatamer constructed in pEGFP-C1 (Daigle et al., 2001).
A human Nup107 cDNA was obtained from total human RNA by reverse transcriptase-PCR amplification with PCR primers designed by using human EST sequences homologous to the rat NUP107 cDNA sequence (Radu et al., 1994). The primary sequence of hNup107 (which we found identical to NP_065134) revealed 91.6% identity to the published rat Nup107 peptide sequence (Radu et al., 1994). For in vivo studies, the hNup107 cDNA was subcloned into pEGFP3-C1.
Yeast two-hybrid screening
Two-hybrid screens using as bait either full-length scNup133 or scΔN-Nup133 (Doye et al., 1994) cloned into pASΔΔ or hNup133 cloned into pLex12 were performed by a mating strategy as described (Fromont-Racine et al., 1997, 2000) with the FRYL S. cerevisiae genomic library (Fromont-Racine et al., 1997) or a human Jurkat cell line cDNA library (Jullien-Flores et al., 1995). To investigate pairwise interactions between Nup133 and Nup84/Nup107 from various organisms, the spNup133a, spNup133b, and hNup133 cDNAs were also cloned into the pASΔΔ vector. Two-hybrid preys either recovered from the previous screens or constructed using pACTII were transformed into the Y187 strain (Fromont-Racine et al., 1997). Bait and prey strains were mated in rich medium, and diploids were tested on histidine-free medium containing 5 mM 3-aminotriazole.
Antibodies, immunofluorescence, and immunoelectron microscopy
Polyclonal antibodies against hNup133 and hNup107 were obtained by injecting recombinant GST-hNup133, 6His-hNup107-N, or 6His-hNup107-C into rabbits (Agrobio). The anti-hNup133 serum was depleted of GST antibodies and affinity purified using GST-hNup133 immobilized on an NHS-activated column. The anti–hNup107-N and anti–hNup107-C antibodies were affinity purified against recombinant GST-hNup107-N and GST-hNup107-C, respectively. Anti–lamin B (Guilly et al., 1987) and autoimmune CREST serum were obtained from J.C. Courvalin (Institut J. Monad, Paris, France), guinea pig anti-p62 (Cordes et al., 1991) was a gift from G. Krohne (Biocenter of the University, Würzburg, Germany); SA1 monoclonal antibody against Nup153 (Bodoor et al., 1999) was provided by Ricardo Bastos (University of Barcelona, Barcelona, Spain); monoclonal anti-p150Glued was from Transduction Laboratories. Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.
For immunofluorescence, cells were either fixed for 20 min in 3% fresh paraformaldehyde and permeabilized with 0.5% Triton X-100 or fixed for 5 min in methanol at −20°C. Ultracryomicrotomy and immunogold labeling of HeLa cells fixed with 4% paraformaldehyde was performed as described (Raposo et al., 1997).
Immunoprecipitation, MALDI-TOF, and Q-TOF spectrometry identification
Immunoprecipitation experiments from HeLa whole cell extracts (2 × 106 cells) were performed essentially as described (Grandi et al., 1997) using 25 μl of crude sera or 6 μg of affinity purified anti-hNup133 antibody bound to 25 μl Affi-prep protein A matrix (Bio-Rad Laboratories). For some experiments, HeLa cells synchronized using a double thymidine block were accumulated in prometaphase using 1μM nocodazole and collected by shake-off 4 h after nocodazole addition.
For MALDI-TOF or Q-TOF spectrometry analysis, protein spots were cut off from silver-stained polyacrylamide gels and digested in gel slices with trypsin as described (Shevchenko et al., 1996). Digests were resuspended in 20 μl formic acid 1%, desalted using Zip Tips C18 from Millipore, dried, and dissolved in 3 μl of formic acid 1%. The sample and the matrix (a saturated solution of 2,5-dihydroxybenzoic acid in TFA 0.1%) were loaded on the target using the dried droplet method. MALDI-TOF spectra of the peptides were obtained with a Voyager-DE STR Biospectrometry Workstation mass spectrometer (PE Corp.) and were calibrated externally using Des-Arg bradykin and ACTH peptides and internally using the trypsin autoproteolysis products. Data mining was performed using the ProFound software. A mass deviation of 0.1 D was allowed in the database searches. For Q-TOF spectrometry identification, the dried trypsin digests were dissolved in 3 μl of a mixture of water, formic acid, and methanol (49:1:50) and were analyzed by MS/MS on a nanoESI Q-TOF mass spectrometer (Micromass).
Photobleaching experiments and four-dimensional confocal microscopy
Photobleaching experiments and four-dimensional confocal microscopy were performed on a custom made ZEISS LSM510 system essentially as described (Zaal et al., 1999; Daigle et al., 2001). For three-dimensional reconstruction of confocal z-stacks, individual images were processed with an anisotropic diffusion filter to remove background noise while preserving edge information (Tvarusko et al., 1999). Then, chromosomes and kinetochores were segmented by thresholding and surface rendered using Amira (Template Graphics Software, Inc.).
FRAP experiments on kinetochores were started in prometaphase to allow maximum recovery time before anaphase onset. Bleaching was performed on the outlined regions in three optical sections (field width half maximum = 1.6 μm) distributed over the metaphase plate and completely bleached all kinetochores (unpublished data). Recovery was monitored every minute on three different focal planes spaced 1.5 μm apart in the center of the metaphase plate. In FLIP experiments, bleaching was repeated on the outlined cytoplasmic regions every 10 s from early metaphase to the onset of anaphase. One image was acquired after each bleach to monitor depletion of fluorescence from cytoplasm and kinetochores. Mean fluorescence intensity of the cytoplasm and the kinetochores was quantitated using the public domain software ImageJ (http://rsb.info.nih.gov/ij). Kinetochores were measured with constant circular regions centered on their peak intensity using an ImageJ macro. The values measured for all kinetochores at the same time point were averaged. Since optical slice thickness of 1.6 μm far exceeds the diameter of a kinetochore, we assumed that cytoplasmic GFP3-hNup133 contributes equally to kinetochore regions. Thus, to obtain GFP3-hNup133 bound to kinetochores mean cytoplasmic intensity was subtracted from mean kinetochore intensity. Bleaching due to the acquisition was corrected and was <10% in all experiments. For comparison, prebleach intensities were normalized to 1.
Online supplemental material
ClustalW alignment of scNup133 and scNup120 homologues (Figs. S1 and S2), Quicktime videos of the sequences shown in Fig. 4 A (videos 1 and 2) and Fig. 6 (videos 3 and 4), representative data set with theoretical fit from FRAP and FLIP experiment shown in Fig. 9 (Fig. S3), and the corresponding kinetic analysis that allowed us to determine the k off from FRAP and FLIP experiments are provided as supplemental material. Supplemental material is available at http://www.jcb.org/content/vol154/issue6.
Supplemental Material
Acknowledgments
We thank Ricardo Bastos, Jean-Claude Courvalin (Institut J. Monod, Paris, France), Dr. Georg Krohne (Biocenter of the University, Würzburg, Germany), and Jacques Camonis (Institut Curie, Paris, France) for generous gifts of antibodies and two-hybrid reagents, Danièle Tenza and Claude Antony for help with EM studies, Lucien Cabanier for antibody purification, Virginie Raedeker for assistance with mass spectrometry, the members of Michel Bornens' lab for helpful advice with cell culture and synchronization, and Michel Bornens and Anne Paoletti for critical reading of the article. We gratefully acknowledge Bob Phair for introducing us to the SAAM II software, and Thorsten Klee for help with kinetic modeling.
This work was supported by Centre National de la Recherche Scientifique, the Institut Curie, and the Association pour la Recherche contre le Cancer (grants to V. Doye and fellowship to N. Belgareh). J. Beaudouin was supported by a fellowship through the European Molecular Biology Laboratory International Ph.D. Program.
The online version of this article contains supplemental material.
O.V. Zatsepina's present address is A.N. Belozersky Institute of Physical and Chemical Biology, Moscow State University, Moscow 119899, Russia.
Footnotes
Abbreviations used in this paper: FLIP, fluorescence loss in photobleaching; MALDI-TOF, matrix-assisted laser desorption/ionization time of flight; NPC, nuclear pore complex; Q-TOF, quadrupole time-of-flight.
References
- Adam, S.A., R.S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, R.R., M. Carmena, and W.C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54. [DOI] [PubMed] [Google Scholar]
- Azuma, Y., and M. Dasso. 2000. The role of Ran in nuclear function. Curr. Opin. Cell Biol. 12:302–330. [DOI] [PubMed] [Google Scholar]
- Belgareh, N., and V. Doye. 1999. Yeast and vertebrate nuclear-pore complexes: evolutionary conserved, yet divergent macromolecular assemblies. Protoplasma. 209:133–143. [Google Scholar]
- Bodoor, K., S. Shaikh, D. Salina, W.H. Raharjo, R. Bastos, M. Lohka, and B. Burke. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264. [DOI] [PubMed] [Google Scholar]
- Busson, S., D. Dujardin, A. Moreau, J. Dompierre, and J.R. De Mey. 1998. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 8:541–544. [DOI] [PubMed] [Google Scholar]
- Campbell, M., G. Chan, and T. Yen. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114:953–963. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., B.T. Schaar, and T.J. Yen. 1998. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V., I. Waizenegger, and G. Krohne. 1991. Nuclear pore complex glycoprotein p62 of Xenopus laevis and mouse: cDNA cloning and identification of its glycosylated region. Eur. J. Cell Biol. 55:31–47. [PubMed] [Google Scholar]
- Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye, V., and E. Hurt. 1997. From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol. 9:401–411. [DOI] [PubMed] [Google Scholar]
- Doye, V., R. Wepf, and E.C. Hurt. 1994. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 13:6062–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri, C.J., B.M. Paschal, K.T. Vaughan, and R.B. Vallee. 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132:617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M., G. Blobel, and M.J. Matunis. 1999. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144:1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine, M., J.C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277–282. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., A.E. Mayes, A. Brunet-Simon, J.C. Rain, A. Colley, I. Dix, L. Decourty, N. Joly, F. Ricard, J.D. Beggs, et al. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 17:95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, M.W., C. Wiese, T.D. Allen, and K.L. Wilson. 1997. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J. Cell Sci. 110:409–420. [DOI] [PubMed] [Google Scholar]
- Grandi, P., T. Dang, N. Pane, A. Shevchenko, M. Mann, D. Forbes, and E. Hurt. 1997. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell. 8:2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilly, M.N., F. Danon, J.C. Brouet, M. Bornens, and J.C. Courvalin. 1987. Autoantibodies to nuclear lamin B in a patient with thrombopenia. Eur. J. Cell Biol. 43:266–272. [PubMed] [Google Scholar]
- Howell, B.J., D.B. Hoffman, G. Fang, A.W. Murray, and E.D. Salmon. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150:1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien-Flores, V., O. Dorseuil, F. Romero, F. Letourneur, S. Saragosti, R. Berger, A. Tavitian, G. Gacon, and J.H. Camonis. 1995. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem. 270:22473–22477. [DOI] [PubMed] [Google Scholar]
- Kerscher, O., P. Hieter, M. Winey, and M.A. Basrai. 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics. 157:1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon, G.G., C. Auffray, M. Polymeropoulos, and M.B. Soares. 1996. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 33:151–152. [DOI] [PubMed] [Google Scholar]
- Matunis, M.J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase–activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu, A., G. Blobel, and R.W. Wozniak. 1994. Nup107 is a novel nuclear pore complex protein that contains a leucine zipper. J. Biol. Chem. 269:17600–17605. [PubMed] [Google Scholar]
- Raposo, G., M.J. Kleijmeer, G. Posthuma, J.W. Slot, and H.J. Geuze. 1997. Immunoglod labeling of ultrathin cryosections: application in immunology. Handbook of Exp. Immunol. Vol. 4. L.A. Herzenberg, D. Weir, and C. Blackwell, editors. Blackwell Science, Inc., Cambridge, MA. 1–11.
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J.V., D.W. Cleveland, G.K. Chan, S.A. Jablonski, D.A. Starr, M.L. Goldberg, and T.J. Yen. 2000. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 103:997–1000. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. [DOI] [PubMed] [Google Scholar]
- Siniossoglou, S., C. Wimmer, M. Rieger, V. Doye, H. Tekotte, C. Weise, S. Emig, A. Segref, and E.C. Hurt. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 84:265–275. [DOI] [PubMed] [Google Scholar]
- Siniossoglou, S., M. Lutzmann, H. Santos-Rosa, K. Leonard, S. Mueller, U. Aebi, and E. Hurt. 2000. Structure and assembly of the Nup84p complex. J. Cell Biol. 149:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S., and T. de Lange. 1999. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112:3649–3656. [DOI] [PubMed] [Google Scholar]
- Stoffler, D., B. Fahrenkrog, and U. Aebi. 1999. The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 11:391–401. [DOI] [PubMed] [Google Scholar]
- Teixeira, M.T., S. Siniossoglou, S. Podtelejnikov, J.C. Benichou, M. Mann, B. Dujon, E. Hurt, and E. Fabre. 1997. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 16:5086–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoropoulos, P.A., H. Polioudaki, M. Koulentaki, E. Kouroumalis, and S.D. Georgatos. 1999. PBC68: a nuclear pore complex protein that associates reversibly with the mitotic spindle. J. Cell Sci. 112:3049–3059. [DOI] [PubMed] [Google Scholar]
- Tran, P.T., L. Marsh, V. Doye, S. Inoué, and F. Chang. 2001. Mechanism of nuclear positioning in fission yeast based upon microtubule pushing. J. Cell Biol. 153:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvarusko, W., M. Bentele, T. Misteli, R. Rudolf, C. Kaether, D.L. Spector, H.H. Gerdes, and R. Eils. 1999. Time-resolved analysis and visualization of dynamic processes in living cells. Proc. Natl. Acad. Sci. USA. 96:7950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu, S.K., and D.J. Forbes. 2001. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13:363–375. [DOI] [PubMed] [Google Scholar]
- Wang, X., J.R. Babu, J.M. Harden, S.A. Jablonski, M.H. Gazi, W.L. Lingle, P.C. de Groen, T.J. Yen, and J.M. van Deursen. 2001. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLEBS-containing proteins. J. Biol. Chem. 276:26559–26567. [DOI] [PubMed] [Google Scholar]
- Zaal, K.J., C.L. Smith, R.S. Polishchuk, N. Altan, N.B. Cole, J. Ellenberg, K. Hirschberg, J.F. Presley, T.H. Roberts, E. Siggia, et al. 1999. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 99:589–601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.