Abstract
By screening a yeast two-hybrid library with COOH-terminal fragments of vinculin/metavinculin as the bait, we identified a new protein termed raver1. Raver1 is an 80-kD multidomain protein and widely expressed but to varying amounts in different cell lines. In situ and in vitro, raver1 forms complexes with the microfilament-associated proteins vinculin, metavinculin, and α-actinin and colocalizes with vinculin/metavinculin and α-actinin at microfilament attachment sites, such as cell–cell and cell matrix contacts of epithelial cells and fibroblasts, respectively, and in costameres of skeletal muscle. The NH2-terminal part of raver1 contains three RNA recognition motifs with homology to members of the heterogeneous nuclear RNP (hnRNP) family. Raver1 colocalizes with polypyrimidine tract binding protein (PTB)/hnRNPI, a protein involved in RNA splicing of microfilament proteins, in the perinucleolar compartment and forms complexes with PTB/hnRNPI. Hence, raver1 is a dual compartment protein, which is consistent with the presence of nuclear location signal and nuclear export sequence motifs in its sequence. During muscle differentiation, raver1 migrates from the nucleus to the costamere. We propose that raver1 may coordinate RNA processing and targeting as required for microfilament anchoring in specific adhesion sites.
Keywords: α-actinin; hnRNP; microfilament attachment; PTB/hnRNPI; vinculin/metavinculin
Introduction
Adhesive junctions are essential for cell–cell and cell matrix adherence and for cellular signaling (Barth et al., 1997; Weisberg et al., 1997; Ben Ze'ev and Geiger, 1998; Critchley, 2000). All types of adhesive junctions contain cytoskeletal proteins and connect cytoskeletal elements of the cell with the plasma membrane, but as they differ in their tasks they may differ in protein composition. For example, whereas all cell matrix and cell–cell contact sites contain vinculin as a general and major component, metavinculin, the larger splice isoform of vinculin is confined to adhesive junctions of muscle cells (Gimona et al., 1987; Belkin et al., 1988). α-Catenin, a relative of vinculin, is found only in the adhesive junctions of cell–cell contacts of polarized cells (Ozawa et al., 1989) where it participates in the actin-anchoring complex with β-catenin and plakoglobin (Nathke et al., 1994). A defined subset of tropomyosin isoforms is found in the adhesive belt, which connects to these epithelial junctions (Temm-Grove et al., 1998).
Generation of this architectural and functional diversity is a complex process comprising cell type–specific transcription and/or splicing of the components combined with selective transport of mRNAs and even site-specific translation. These events must be interconnected and regulated tightly, and several proteins are known that are involved in either one or several of these steps. A cross-talk between cytoskeletal components of adhesive junctions and gene transcription has been demonstrated for β-catenin (Ben Ze'ev and Geiger, 1998), actin (Scheer et al., 1984; Gonsior et al., 1999; Rando et al., 2000), zyxin (Nix and Beckerle, 1997), and the desmosomal plaque proteins plakophilin-1a and -1b (Schmidt et al., 1997), plakophilin-2 (Mertens et al., 2001), and plakophilin-3 (Bonne et al., 1999; Schmidt et al., 1999). Differential RNA splicing resulting in junctional diversity may be executed by protein components of the heterogeneous nuclear RNPs (hnRNPs)* like the polypyrimidine tract binding protein (PTB)/hnRNPI that is engaged in the tissue- and/or differentiation stage–specific splicing of RNAs coding for signaling and cytoskeletal proteins such as c-src, α-actinin, and tropomyosin (Valcarcel and Gebauer, 1997). mRNA export from the nucleus, directed cytoplasmic transport, and anchoring at the site of translation are additional steps required. Export might also involve cytoskeletal proteins such as nuclear actin (Hofmann et al., 2001), and directed transport may require specific proteins that recognize and bind to selected nucleotide sequences. The well-characterized zip code binding protein (ZBP)1 that binds to a motif within the 3′ untranslated region of the β-actin mRNA (Ross et al., 1997) is an example of such a function. Large ribonucleoprotein particles containing various mRNAs, “zip code” proteins, adaptors, and possibly motor proteins may then move along cytoplasmic microtubules and/or microfilaments (Oleynikov and Singer, 1998; Jansen, 1999, 2001), and this transport mechanism can discriminate between different destinations: for example, β-actin and γ-actin mRNAs are found segregated in myoblasts and concentrate at the cell periphery or the perinuclear region, respectively (Hill and Gunning, 1993), and vinculin mRNA is localized at microfilament sarcolemma attachment sites (costameres) in skeletal muscle (Morris and Fulton, 1994). For tropomyosins, it was shown that mRNAs coding for different splice forms target to discrete subcellular sites during the development of neuronal polarity (Hannan et al., 1995).
Although several of the components involved in the different steps towards the generation of adhesive junctions as described above are identified, our knowledge is far from being complete. In this study, we present a new protein, termed raver1, that comprises RNA-binding motifs with homology to PTB/hnRNPI. Raver1 is a dual compartment protein, localized at microfilament plasma membrane attachment sites in fibroblasts, epithelial cells, and muscle, and like PTB/hnRNPI (Ghetti et al., 1992) within perinucleolar structures that are engaged in RNA metabolism (Huang, 2000; Spector, 2001). Raver1 forms complexes with PTB/hnRNPI and the junction proteins vinculin/metavinculin and α-actinin, and it can shuttle between the nucleus and the cytoplasm. In skeletal muscle, a translocation of raver1 from the nucleus to the cytoplasm is correlated with the differentiation of specific microfilament attachment sites.
Results
Identification and expression of raver1
In a search for new vinculin/metavinculin ligands, the COOH-terminal tail domain of metavinculin was used as the bait to screen a mouse embryonic (E17.5) yeast two-hybrid library. In two independent screens, a total of 45 blue colonies was obtained, and the isolated cDNAs were probed for the specificity of their interaction with vinculin/metavinculin by comparison with appropriate controls (see Materials and methods).
One particular cDNA coding for a 625 amino acid (1875 nt) protein fragment was identified five times. Subsequently, a cDNA comprising the entire ORF and the 3′ untranslated region was isolated from an embryonic (E12.5) cDNA library. The deduced amino acid sequence predicted a 748 amino acid protein (M w 80 kD) containing three RNA recognition motifs (RRMs), two nuclear localization signals (NLSs), and one Rev-like nuclear export sequence (NES; Fig. 1 A) identified by computational analysis. The RRMs (each ∼90 amino acids long) were each found to comprise conserved motifs of six to eight residues called RNP-2 and RNP-1 (Shamoo et al., 1995; Siomi and Dreyfuss, 1997). As a modification of the family consensus, aromatic residues at position two in RNP-2 and several residues at varying positions in RNP-1 were found substituted by nonaromatic amino acids (Fig. 1 B). Similar substitutions have been identified in PTB/hnRNPI (Conte et al., 2000).
Figure 1.
Raver1 is an 80-kD multidomain protein. (A, top) The ORF of raver1 is shown as a grey box. The cDNA fragment identified in the yeast two-hybrid screen (raver1 prey) and two deletion fragments (raver1NΔC and raver1CΔN) are indicated (double arrows). Three bona fide RRMs are presented as striped blue/yellow boxes, and the bona fide NLS and NES sequences are indicated in black and red, respectively. (A, bottom) The deduced amino acid sequence of raver1. The bipartite RRMs are shown in yellow (RNP-1 motifs) and blue (RNP-2 motifs), two NLS motifs (comprising a RKR-X10-RRK and KRKR motif, respectively) are in black, and the Rev-like NES motif is in red. (B) Sequence comparison of the putative RNP-2 and RNP-1 motifs of raver1 with RRM3 and RRM4 of PTB/hnRNPI and the RRM family consensus motifs. Note that raver1 and PTB/hnRNPI contain similar amino acid substitutions relative to the RRM family consensus (indicated as white boxes or lanes). (C) Western blot analysis of raver1 expression. Extracts from C2C12 (1), untransfected (2), and raver1-transfected HeLa cells (3) were resolved by SDS-PAGE. Immunoblotting was performed using a monoclonal antibody raised against a recombinant raver1 fragment. Note the absence of an endogenous raver1 signal in HeLa cells.
Apart from the RNP-1 and RNP-2 motifs, no significant homology was found with other proteins. For further analysis of raver1 expression and localization, we generated a monoclonal antibody against a recombinant fusion protein consisting of the same raver1 fragment identified as metavinculin prey (Fig. 1 A) and maltose-binding protein (MBP). Immunoblot analysis of cell extracts from untreated and raver1-transfected cells revealed only one band of ∼80 kD (Fig. 1 C), supporting the conclusion that the isolated cDNA comprises the entire ORF.
We then analyzed the expression and tissue-specific distribution of raver1 mRNA. Using two cDNA fragments, encoding the entire ORF as probes for Northern blotting, we detected a single mRNA transcript with an estimated size of 3.4 kb in all of the probed adult mouse tissues (Fig. 2 A). When compared with the amounts of β-actin mRNA, raver1 transcripts exhibited tissue-specific expression levels. High levels of mRNA were found in striated muscle and in liver, kidney, and testis, whereas low amounts of transcript were detected in brain, lung, and spleen (Fig. 2 A). Raver1 mRNA was detected already at early embryonic (E7) stages (Fig. 2 B).
Figure 2.
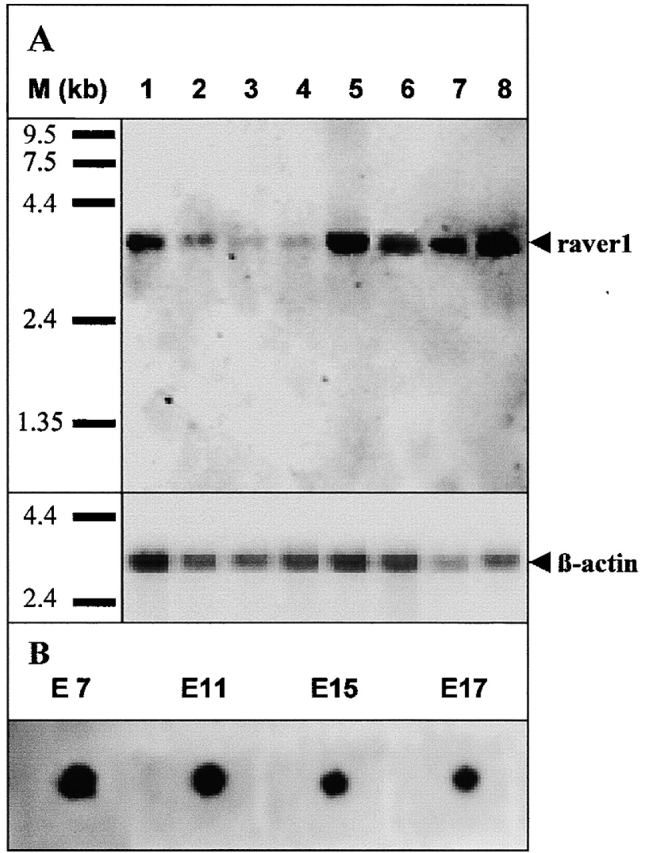
Raver1 is widely expressed as revealed by Northern blot analysis. Two cDNA fragments (bp 1–1326 and 1327–2257) comprising the whole ORF were amplified by PCR using DIG-labeled nucleotides and hybridized with total RNA on a mouse multiple tissue Northern (A) and a mouse RNA master (B) blot. Note that raver1 mRNA is detected in all adult mouse tissues tested, although in varying intensities (A, 1, heart; 2, brain; 3, lung; 4, spleen; 5, liver; 6, kidney; 7, skeletal muscle; 8, testis) and embryonic stages 7–17 (B, indicated above samples). Equal RNA load is demonstrated by probing for β-actin mRNA.
Raver1 binds to the COOH-terminal domain of vinculin and metavinculin
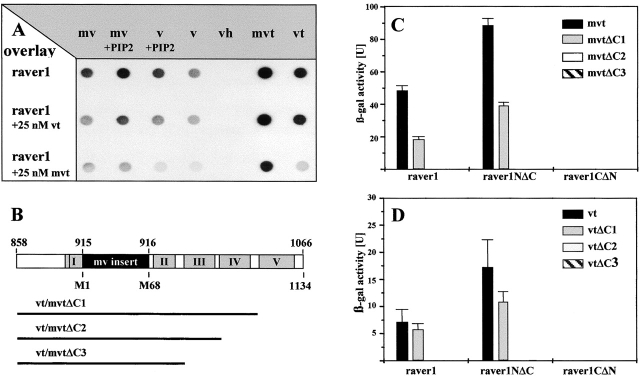
To confirm the interaction of raver1 with vinculin/metavinculin seen in the two-hybrid assay, in vitro–translated [35S]methionine-labeled raver1 was probed for binding to immobilized full-length proteins and isolated vinculin head (vh) and vinculin tail (vt) domains in an overlay assay (Fig. 3 A). vh and vt domains were either generated by proteolytic cleavage of genuine turkey gizzard vinculin with the endoprotease Glu-C (Kroemker et al., 1994; Hüttelmaier et al., 1998) or purified as recombinant proteins bearing an NH2-terminal His tag. In accordance with the yeast two-hybrid data, raver1 bound exclusively to the tail not to the head domain of vinculin (Fig. 3 A), although the vh domain, when adsorbed to the membrane, was still active in ligand binding (Fig. 4 B). Complex formation was also analyzed in the presence of phosphatidylinositol 4,5-bisphosphate (PIP2), an acidic phospholipid which activates ligand binding of vinculin (Gilmore and Burridge, 1996; Weekes et al., 1996; Hüttelmaier et al., 1998). PIP2 significantly enhanced raver1 binding to the full-length proteins. The specificity of this assay was challenged by the addition of soluble recombinant vt or metavinculin tail (mvt). Binding of radiolabeled raver1 was reduced significantly for all immobilized ligand proteins, with mvt being a more efficient competitor than vt.
Figure 3.
Raver1 binds to the tail domain of vinculin/metavinculin. (A) Overlay spots obtained with [35S]methionine-labeled raver1 on immobilized chicken metavinculin (mv), PIP2-activated metavinculin (mv + PIP2), activated vinculin (v + PIP2), vinculin (v), vinculin head domain (vh), recombinant murine mvt (mvt), and vt. Binding is enhanced by activation of v/mv with PIP2 and is reduced in the presence of soluble vt (raver1 + 25 nM vt) or mvt (raver1 + 25 nM mvt). Some binding was also noted for nonactivated v/mv, which may be due to partial conformational changes upon immobilization to the membrane. (B) A schematic representation of vt/mvt and deletion fragments used for two-hybrid analysis shown in C and D. Highlighted areas comprise the metavinculin insert (black box) and the five α-helices within the vt domain (grey boxes numbered according to Bakolitsa et al., 1999). (C and D) Raver1–vt/mvt interaction assayed by β-galactosidase activity in a yeast two-hybrid liquid assay. Samples were analyzed in three independent experiments, and means and SD are shown. Note that binding of mvt and vt to the NH2-terminal portion of raver1 (raver1NΔC, amino acids 1–441) involves α-helices IV and V of vt and the NH2-terminal portion of raver1 (raver1NΔC, amino acids 442–748).
Figure 4.
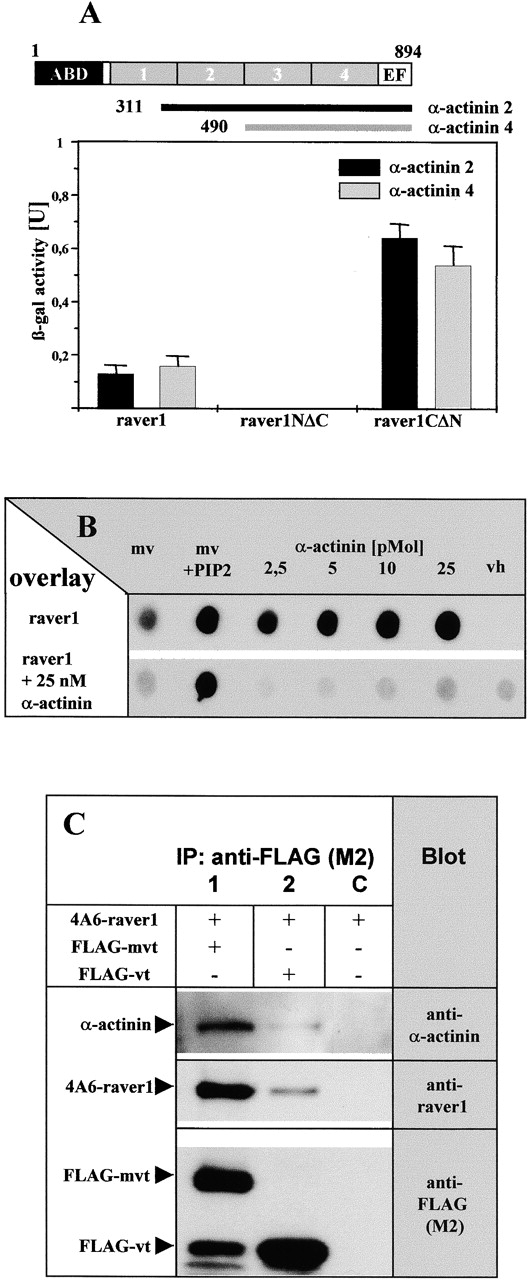
Binding to α-actinin is mediated by the COOH terminus of raver1. (A) Interaction of raver1 and its deletion fragments with COOH-terminal fragments of α-actinin 2 and 4 assayed by β-galactosidase activity in the yeast two-hybrid system. Each experiment was repeated three times, and means and SD are indicated. Organization of the α-actinin polypeptide and the fragments used are shown on top (ABD, actin binding domain; boxes 1–4, the four spectrin-like repeats; EF, EF-hand motif). (B) [35S]methionine- labeled raver1 overlay on immobilized metavinculin (mv), PIP2- activated metavinculin (mv + PIP2), α-actinin, and the vh in the presence or absence of increasing amounts of soluble α-actinin. Binding of raver1 to immobilized α-actinin is reduced significantly in the presence of soluble α-actinin. Note that binding to the vh requires the presence of α-actinin. (C) Coimmunoprecipitation of raver1 and α-actinin with vt/mvt. Bipro-tagged raver1 (4A6-raver), FLAG-mvt, or FLAG-vt were coexpressed in HeLa cells as indicated. After in situ cross-linking with DSP, immunoprecipitation (IP) with anti-FLAG (M2) was performed. Precipitates were analyzed by SDS-PAGE and immunoblotted with the antibodies indicated. A proteolytic FLAG-tagged mvt fragment with an apparent molecular mass similar to FLAG-vt is seen in lane 1. Note that raver1 and α-actinin are coprecipitated with mvt and vt.
Raver1 interaction with vt and mvt was further characterized using three COOH terminally truncated deletion constructs of vt and mvt containing two to four of the five COOH-terminal α-helices of the vt (Bakolitsa et al., 1999; Fig. 3 B). These fragments were tested for their binding to full-length raver1 and two raver1 fragments, comprising the NH2-terminal (raver1NΔC) or COOH-terminal (raver1-CΔN) half of the protein (Fig. 1 A). In a liquid yeast two-hybrid assay (see Materials and methods), β-galactosidase activity was determined and revealed that binding to the vt/mvt domain is mediated by the NH2-terminal portion of raver1 (Fig. 3, C and D). The presence of helices IV and V in the tail domain is crucial for the interaction with raver1, since no binding was observed for a vt fragment comprising helices I–III. Remarkably, the interaction with mvt was much stronger than with vt as seen by a 10-fold increase in β-galactosidase activity (Fig. 3, C compared with D), although the expression levels of both proteins in the yeast cells were comparable (data not shown). Qualitatively similar results had been obtained already in the overlay assay (Fig. 3 A). Hence, the metavinculin insert significantly enhances vinculin binding to raver1 even though it is not involved directly in this interaction.
Raver1 forms ternary complexes with vinculin and α-actinin
To identify further raver1 ligands, the full-length protein was used as the bait to screen a mouse embryonic (E17.5) and a human heart muscle cDNA library in the yeast two-hybrid system. From both screens, with a total of ∼106 yeast transformants, we isolated 18 cDNA clones that were considered to encode candidate raver1-specific ligands (Table I). A metavinculin clone comprising the entire tail domain was isolated, confirming the results obtained with the reciprocal yeast two-hybrid screen (see above) and the overlay assays (Fig. 3 A). In addition, several COOH-terminal fragments of the α-actinin isoforms 2 and 4 were identified (Fig. 4 A, top). Their interaction with raver1 was confirmed in a liquid yeast two-hybrid assay (Fig. 4 A, bottom). Binding was observed with the full-length raver1 protein and its COOH-terminal portion, comprising amino acids 442–748 (raver1CΔN). In contrast, the NH2-terminal fragment of raver1, which bound to vinculin/metavinculin (Fig. 3), failed to interact with α-actinin (Fig. 4 A).
Table I. Raver1 ligands identified by the two-hybrid system.
| Bait | Prey | Accession no. of prey | Number of isolated clones |
|---|---|---|---|
| raver1 | α-actinin 2 (COOH-terminal fragment) | NM_001103.1 | 5 |
| raver1 | α-actinin 4 (COOH-terminal fragment) | NM_001103.1 | 2 |
| raver1 | metavinculin (COOH-terminal fragment) | NM_014000 | 1 |
| raver1 | PTB (full-length) | NM_008956 | 8 |
| raver1 | murine Rod1 (COOH-terminal fragment) | AB023966 * | 2 |
| mvt | raver1 prey fragment | - | 5 |
Accession no. of rat Rod1.
Again, the yeast two-hybrid data were confirmed in an overlay assay where in vitro–translated 35S-radiolabeled raver1 was probed for binding to immobilized turkey gizzard α-actinin (Fig. 4 B). Raver1 bound to immobilized α-actinin in a dose-dependent manner, and binding could be competed for by the soluble ligand. Again (Fig. 3 A), raver1 did not interact with vh, but when preincubated with α-actinin it was bound to this vinculin fragment (Fig. 4 B). This result is explained by our previous data, showing that vh interacts with α-actinin (Kroemker et al., 1994). Thus, α-actinin can act as a mediator in this assay. Remarkably, preincubation of raver1 with α-actinin did not prevent binding to immobilized PIP2-activated metavinculin, suggesting either different affinities of raver1 for metavinculin and α-actinin or the formation of ternary complexes.
To probe for the existence of such ternary complexes under physiological conditions, we double transfected HeLa cells with raver1 bearing a sequence tag derived from birch profilin (4A6-raver1) (Rüdiger et al., 1997) and either a FLAG-tagged mvt or vt fragment. After stabilizing protein complexes in situ with a membrane permeant crosslinker (see Materials and methods), cell lysates were subjected to immunoprecipitation with FLAG antibody. As shown in Fig. 4 C, α-actinin and the epitope-tagged raver1 both coimmunoprecipitated with mvt and vt, respectively. These results confirm the overlay data and suggest that these proteins can form a ternary complex in cells.
Raver1 is a dual compartment protein
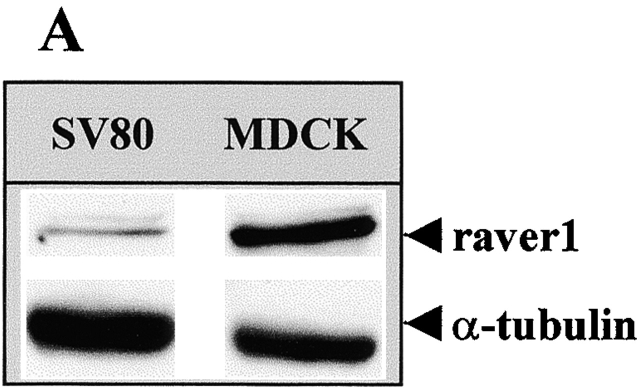
The complex formation of raver1 with microfilament anchoring proteins like α-actinin and vinculin suggested raver1 as a component of cell matrix and cell–cell contacts. Hence, we examined expression and localization of raver1 in a fibroblastic line displaying focal contacts (SV80) and in MDCK epithelial cells demonstrating adhesion belts and cell–cell contacts in culture. By immunoblotting, both cell types were found to contain raver1, but expression in the well-organized epithelial cells was ∼10-fold higher than in the fibroblasts (Fig. 5 A).
Figure 5.
Raver1 is a dual compartment protein. (A) Western blot analysis of raver1 and α-tubulin. Normalized protein samples of MDCK and SV80 cells were resolved by SDS-PAGE and immunoblotted with either raver1 or α-tubulin antibodies. Note that MDCK epithelial cells express raver1 to an ∼10-fold higher level than SV80 fibroblasts. (B and C) Immunofluorescence analysis of SV80 fibroblasts (B) and MDCK epithelial cells (C). Cells were fixed and subsequently stained with the monoclonal raver1 antibody. Note the strong nuclear signal for raver1 (B and C, arrowheads) in addition to the location at large, probably mature (B, arrow) and small peripheral focal contacts (B, inset) of the fibroblasts and at cell–cell contacts of epithelial MDCK cells (C, arrow). The exposure time of the picture in B was optimized for revealing focal contact staining, resulting in an overexposed nuclear image in the SV80 cell. Bars, 10 μm.
Indirect immunofluorescence revealed raver1 in focal contacts (Fig. 5 B) and in cell–cell contacts (Fig. 5 C) as expected by the immunoprecipitation data, showing that the contact proteins vinculin and alpha actinin are raver1 ligands. However, a prominent raver1-specific signal was also seen in the nucleus of both cell types (Fig. 5, B and C). Hence, raver1 is a dual compartment protein. Its association with microfilament attachment sites is probably due to complex formation with vinculin/metavinculin and α-actinin and may require the activity of the NES (Fig. 1 A). The presence in the nucleus probably involves the nuclear import sequences (NLS) (Fig. 1 A) and additional nuclear components (see below).
Raver1 binds to PTB/hnRNPI
Two members of the hnRNP family, PTB/hnRNPI (Valcarcel and Gebauer, 1997) and its homologue ROD1 (Yamamoto et al., 1999), were identified as additional raver1 ligands in the yeast two-hybrid system (Table I). The specificity of these results was challenged in localization and immunoprecipitation studies with murine C2C12 myoblasts. Immunofluorescence showed both endogenous raver1 and endogenous PTB/hnRNPI in the nucleus and concentration and colocalization of both proteins in perinucleolar bodies (Fig. 6 A). Immunoprecipitation of the endogenous proteins from C2C12 cells revealed that antibodies against PTB/hnRNPI coprecipitated raver1. In contrast, analogous experiments with an antibody against a control protein, ZBP1, did not result in raver1 precipitation (Fig. 6 B). Conversely, PTB/hnRNPI was coprecipitated with raver1 by a FLAG antibody from HeLa cells transfected with FLAG-raver1 but not from cells transfected with FLAG-ZBP1 (Fig. 6 C). In both cases, the PTB/hnRNPI-raver1 complex was not sensitive to RNAse treatment, indicating that it was neither generated nor stabilized by RNA (Fig. 6, B and C).
Figure 6.
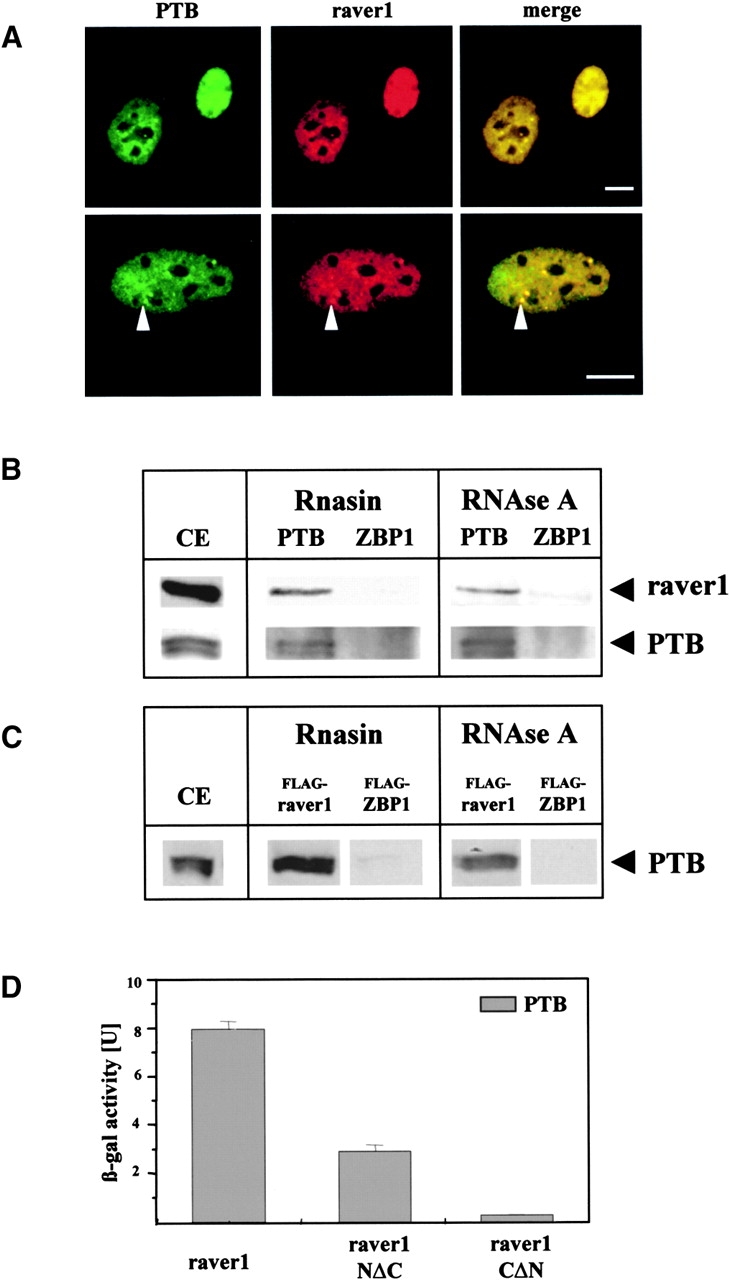
Raver1 interacts with the PTB/hnRNPI protein. (A) Colocalization of PTB/hnRNPI and raver1 in C2C12 myoblasts as seen by double immunofluorescence with the respective monoclonal antibodies. Both proteins are localized in the nucleus and highly concentrated in the perinucleolar bodies (arrowheads). (B and C) Coprecipitation of endogenous raver1 with PTB/hnRNPI from nontransfected C2C12 myoblasts (B) and HeLa cells transfected with FLAG-equipped raver1 or FLAG-ZBP1 (C), and analysis of the precipitates by SDS-PAGE and immunoblotting. Antibodies used for immunoprecipitation (against PTB/hnRNPI, raver1, ZBP1, and FLAG) are indicated on the top; those used for immunoblots are shown on the right. Cell extracts were treated with RNAsin or with RNAse A to either protect or remove putatively bound RNA. Note that PTB/hnRNPI and raver1 coprecipitate independently of RNA, whereas ZBP1 does not form complexes with raver1 under either experimental condition. CE, immunoblots of the total extracts, to demonstrate the expression of the relevant proteins. (D) Liquid β-galactosidase assay of the yeast two-hybrid system to identify the raver1 domain interacting with murine PTB/hnRNPI. β-Galactosidase activity was found highest with intact raver1, and some activity remained with the COOH-terminal deletion fragment, whereas the NH2-terminal deletion fragment was inactive. Means and calculated standard deviation of three independent experiments are indicated. Bars, 10 μm.
Binding sites for PTB/hnRNPI on raver1 were characterized with full-length raver1 and two deletion fragments in the liquid yeast two-hybrid assay (Fig. 6 D). Highest β-galactosidase activity was measured with the full-length protein. Although the NH2-terminal portion of raver1 (raver1NΔC) still showed some binding activity, the COOH-terminal fragment (raver1CΔN) was completely inactive.
Raver1 shuttles between the nuclear and cytoplasmic compartment
The observed localization of raver1 in both the nuclear and cytoplasmic compartment raised the question of whether raver1 can translocate from one compartment to the other. This was investigated in heterokaryons obtained as fusion products between HeLa (human) and C2C12 (murine) cells (Michael et al., 1995). HeLa cells were transfected with FLAG-tagged raver1 and fused with C2C12 myoblasts to yield heterokaryons by polyethyleneglycol treatment (Fig. 7, A and D) in the presence of cycloheximide to prevent de novo protein synthesis. Human and murine nuclei can unambiguously be discriminated by DAPI staining that reveals small dots in the mouse nuclei (Fig. 7, B, E, and insets). As shown in Fig. 7, C and F, 4 h after fusion and in the absence of protein synthesis FLAG-raver1 appeared in both HeLa and C2C12 cell nuclei of heterokaryons. FLAG-tagged protein hnRNP C2β, which is a nonmobile nuclear resident protein (Williamson et al., 2000), was used as a negative control in analogous experiments (data not shown). These data indicate that raver1 molecules expressed in HeLa cells and residing in the nucleus can leave this compartment and translocate through the cytoplasm into myoblast nuclei in a shuttle mechanism.
Figure 7.
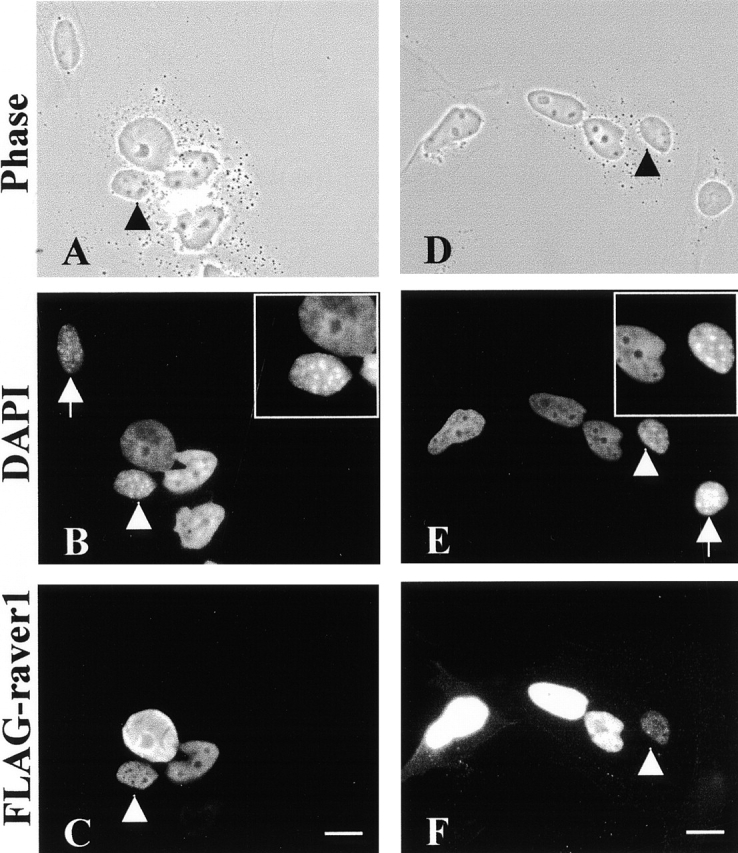
Raver1 shuttles between nuclear and cytoplasmic compartments in heterokaryons. HeLa cells were transiently transfected with FLAG-raver1, mixed and subsequently fused with mouse C2C12 cells, and incubated for another 4 h in the presence of cycloheximide. The coculture was fixed and stained using FLAG antibody. (A and D) Phase–contrast images of two examples, showing larger and smaller (arrowheads) nuclei within one common cytoplasm. (B, E, and insets) Identification by DAPI staining of the smaller nuclei (arrowheads) as of murine origin containing multiple DNA dots and the larger nuclei as human without prominent dots. (C and F) FLAG-raver1 is found within murine nuclei in fusion products with the raver1-transfected HeLa cells (arrowheads) but not in the corresponding nuclei in the untransfected unfused C2C12 cells (B and E, arrows). Bars, 10 μm.
During myotube formation and differentiation, raver1 redistributes from the nuclear to the cytoplasmic compartment and colocalizes with microfilament attachment sites in skeletal muscle
In an attempt to reveal the biological significance of raver1 shuttling, we investigated the expression and localization of raver1 protein during cell differentiation. Since raver1 expression in muscle was high (Fig. 2), we studied raver1 localization in undifferentiated myoblasts and during myotube formation (Fig. 8). Immunofluorescence analysis of C2C12 myoblasts showed that endogenous raver1 was localized mainly in the nucleus (Fig. 8 A) as also seen in the corresponding nuclei of heterokaryons (Fig. 7, C and F). The raver1-specific fluorescence signal was comparable for all nuclei (Fig. 8 A). To follow raver1-specific translocation, we performed counterstaining for the transcription factor MEF-2 that is resident in myogenic nuclei and engaged in myogenesis (Ornatsky et al., 1997). During myotube formation, the raver1-specific nuclear signal decreased, and concomitantly there was a significant increase in cytoplasmic signal of C2C12 myotubes (Fig. 8 B), although no changes in subcellular localization were observed with MEF-2 (Fig. 8 B). When cross-striated myotubes were formed at later stages of differentiation, raver1 was seen associated with these cross-striations, whereas myotube nuclei were negative (Fig. 8 C). These results indicate that raver1 as a dual compartment protein can specifically migrate from the nucleus to the cytoplasm during muscle differentiation.
Figure 8.
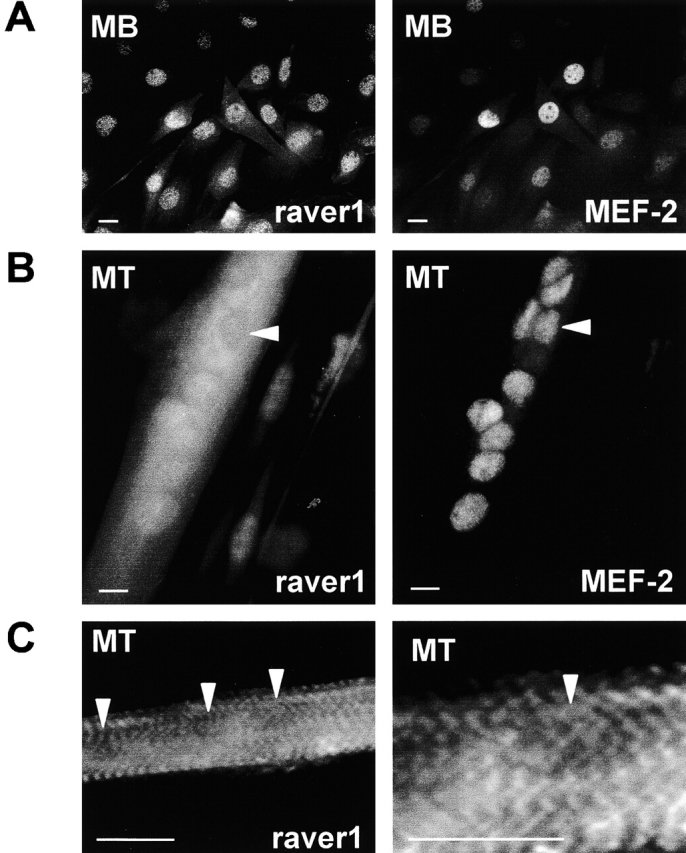
During myotube formation, raver1 translocates to the cytoplasmic compartment. (A) Double immunofluorescence of C2C12 myoblasts (MB) to reveal raver1 and the transcription factor MEF-2 in myoblast nuclei. (B) Distribution of raver1 and MEF-2 in myotubes (MT) as seen at 4 d after induction of differentiation. In the multinucleated myotubes, raver1 is present in both compartments, the nucleus and the sarcoplasma, but at that stage some nuclei exhibit already a very weak signal (arrowhead), whereas MEF-2 is a prominent component confined to the nuclear compartment. (C) Part of a myotube 6 d after induction of differentiation. Both images show staining for raver1 but at different magnifications. Differentiation has proceeded to cross-striated myotubes, and raver1 is now localized in sarcoplasmic cross-striations, whereas the nuclei appear negative (arrowheads). Bars, 10 μm.
In immunofluorescence studies on sections of mature murine skeletal muscle (musculus soleus), raver1 was detected in equivalent cross-striations (Fig. 9, A and D) that were identified as Z-disks by costaining for vinculin (Fig. 9 B) and α-actinin (Fig. 9 E). Raver1 colocalized with both proteins in these structures (Fig. 9, C and F). The peripheral part of the Z-disks, which connects the sarcomeres to the sarcolemma, has been termed costamere (Pardo et al., 1983). Vinculin is restricted to the costamere (Terracio et al., 1990), whereas α-actinin is a component of both costameres and the more central region of the Z-disk (McKenna et al., 1986; Danowski et al., 1992). Based on our findings that raver1 forms complexes with vinculin, we propose that it is also a component of the costamere in striated muscle where it may participate in the processes required for precise microfilament attachment at the sarcolemma. However, this does not preclude the possibility that raver1 is also located in the central part of the Z-disk together with α-actinin.
Figure 9.
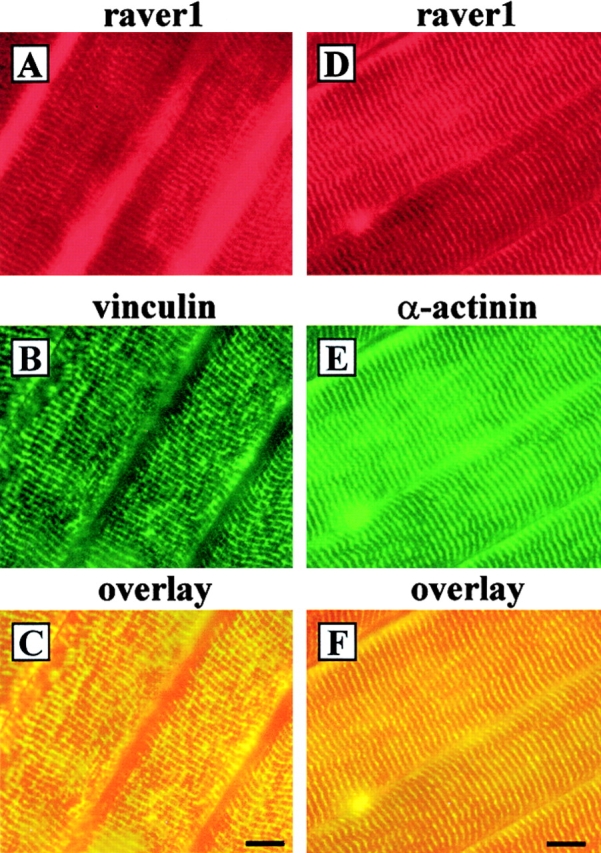
Raver1 colocalizes with actin attachment sites in striated muscle. Frozen tissue sections (6–8 μm) of adult murine skeletal muscle (m. soleus) were double stained with the monoclonal raver1 antibody (A and D) and a polyclonal vinculin antibody (B) or a monoclonal antibody against skeletal muscle α-actinin (E). Note that raver1 colocalizes partially with vinculin and α-actinin in costameres (C and F, overlays). Bars, 10 μm.
Discussion
We identified and characterized a hitherto unknown protein, which we termed raver1. Raver1 is a multidomain hnRNP-like protein and forms complexes with other members of the hnRNP family and actin-binding proteins involved in microfilament membrane attachment.
In accordance with these properties, raver1 was seen resident in the nucleus and in various types of microfilament attachment sites. This dual compartment localization may be due to both bona fide NLS and NES motifs present in the raver1 sequence. Both motifs seem functional, since raver1 can leave the nucleus, traverse the cytoplasm, and even target to a second nucleus as seen in the heterokaryon assay.
For both the nuclear compartment and the cytoplasmic membrane attachment sites, we identified partner proteins for raver1. Vinculin/metavinculin and α-actinin, essential components of microfilament attachment sites, form complexes with raver1 in in vitro assays and under physiological conditions. As different parts of raver1 bind to these ligands, raver1 can bind simultaneously to both these proteins and may thus participate in the formation of adhesive complexes.
Immunofluorescence of various cell lines and transfection of HeLa cells with GFP raver1 (data not shown) displayed a prominent signal of raver1 in the nuclei of different cell types, arguing for an additional nuclear function. Raver1 colocalizes with PTB/hnRNPI in perinucleolar bodies that harbor several RNA-binding proteins, including hnRNPs, and are actively engaged in RNA metabolism (Huang, 2000; Spector, 2001). In accordance with this location, we found that raver1 contains three RRMs with homology to those of members of the hnRNP family (Shamoo et al., 1995; Siomi and Dreyfuss, 1997) such as PTB/hnRNPI (Valcarcel and Gebauer, 1997), its homologue ROD1 (Yamamoto et al., 1999), and the PTB/hnRNPI-associated splicing factor PSF (Patton et al., 1993). All hnRNP proteins contain RNA-binding motifs (RRMs or KH domains), form heterooligomers with other members of the family, and some shuttle between the nucleus and the cytoplasm (Krecic and Swanson, 1999). In all these criteria, raver1 is hnRNP like. However, raver1 is also unique in providing a direct link to microfilament proteins.
hnRNP proteins are thought to be involved in RNA splicing, trafficking, and translational control (Krecic and Swanson, 1999; Shyu and Wilkinson, 2000). For the prototype PTB/hnRNPI that participates in the RNA splicing and generation of tissue-specific isoforms of α-actinin (Southby et al., 1999) and tropomyosin (Gooding et al., 1998; Mulligan et al., 1992) but is also involved in RNA splicing of nonmicrofilament proteins, it has been proposed that its differential activities require different protein ligands (Valcarcel and Gebauer, 1997). With raver1, we identified a PTB/hnRNPI partner, which may be an important regulator of these processes. On the other hand, we have preliminary evidence that raver1, without PTB/hnRNPI, is able to direct RNA splicing of α-actinin (data not shown). Hence, again in analogy to PTB/hnRNPI and its relatives the RRMs in raver1 seem functional and bind to RNA, but the complex between raver1 and PTB/hnRNPI does not require the presence of RNA. RNA-independent complexes of PTB/hnRNPI-like and microfilament-associated proteins in the same hnRNP particle have also been observed recently for PSF and the focal adhesion adaptor protein paxillin (J.C. Norman and D.R. Critchley, personal communication).
The proteins PTB/hnRNPI, hnRNPA2, and ZBP-1 have also been implicated in mRNA trafficking within the cytoplasmic compartment (Ross et al., 1997; Cote et al., 1999; Munro et al., 1999; Zhang et al., 2001). Bearing this in mind, one could speculate that raver1 might bind to mature mRNA of α-actinin or vinculin, mediating their transport and anchorage to microfilament attachment sites as a prerequisite for site-specific translation and junction assembly. We tested a putative association of raver1 with α-actinin and vinculin mRNA in several experiments (data not shown) but failed to obtain positive results. However, even if raver1 does not bind directly to these or other mature mRNA species it may still contribute to mRNA trafficking as a member of a PTB/hnRNPI–mRNA complex.
Raver1 mRNA and protein were found widely expressed in different tissues and cultured cell lines but to different levels. High levels of raver1 mRNA were found in striated muscle and in organs consisting largely of epithelial tissue (kidney and liver), and raver1 protein was strongly expressed in epithelial cells (MDCK), whereas transformed fibroblasts (SV80) contained little, and HeLa cells no detectable raver1. In addition, although focal contact or cell–cell contact staining was observed in SV80 or MDCK cells, raver1 was found almost exclusively in the nuclear compartment of undifferentiated C2C12 and the smooth muscle–derived PAC-1 cells (data not shown). These variations in mRNA and protein levels indicate that raver1 expression and/or subcellular localization depend on the degree of polarization or differentiation of a particular cell. For murine-striated muscle, we could show that raver1 localization is linked to differentiation. As seen by immunofluorescence, the raver1 signal moves from an exclusively nuclear (C2C12 myoblasts) to a cytoplasmic (early myotubes) position and finally to Z-lines (cross-striated myotubes) where it colocalizes with α-actinin and vinculin in costameres (mature muscle). It will be interesting to investigate raver1 mobility during smooth or heart muscle differentiation, since in these muscle types the membrane attachment sites (costameres, intercalated discs, and dense plaques, respectively) contain metavinculin in addition to vinculin, which we identified as a raver1 ligand with an apparently higher affinity.
In conclusion, we suggest that raver1 communicates between a PTB/hnRNPI-guided splicing machinery and sites of cytoskeletal assembly. In concert with its nuclear and cytoskeletal ligands, raver1 may operate in a multifunctional complex essentially involved in tissue- and differentiation-specific gene expression and/or mRNA trafficking. Additional functions can of course not be excluded. At present, we cannot predict whether raver1 will exert all of these functions simultaneously or how its proposed activities may be regulated. Further studies are required to solve these questions.
Materials and methods
Two-hybrid analysis
To identify new ligands for vinculin/metavinculin, a plasmid (pGBT-9; CLONTECH) containing the murine mvt (amino acids 851–1134) was used to screen an embryonic (E17.5) mouse two-hybrid library in the yeast strain HF7c, essentially according to the manufactures's protocol (matchmaker I–III; CLONTECH). After identification of the raver1 fragment and the full-length sequence (see below), the complete raver1 cDNA was cloned into the bait plasmid pGBKT7 (CLONTECH) and used to screen the mouse embryonic (E17.5) and a human heart two-hybrid library, both fused to the GAL4 transcription-activating domain (CLONTECH). Potential prey plasmids were recovered and reintroduced into yeast with the above indicated bait plasmids or a control pGBKT7-lamC (CLONTECH) plasmid or the empty bait vectors to exclude false positives. Two-hybrid liquid assays were performed according to the manufacturer's protocol (matchmaker III; CLONTECH) using the yeast strain Y187.
Sequence analysis
To isolate the raver1 cDNA (ICRF p523J246), a mouse embryonic (12E) library (library no. 523; RZPD) was screened. A DIG-11-UTP–labeled PCR fragment according to the raver1 prey fragment was used as a probe. The probe was labeled using a DIG-PCR nucleotide mix purchased from Roche.
Three cDNA clones obtained were sequenced completely. RRMs and the two NLS sequence motifs were predicted by use of the PSORT II (http://psort.nibb.ac.jp) program. The Rev-like NES sequence was identified by direct sequence comparison (Moore, 1996).
Cloning and vectors
Constructs used in this study were amplified according to standard PCR protocols and verified by DNA sequencing. Vinculin/metavinculin cDNA fragments encoding for the tail domain (amino acids 858–1066 for vt and amino acids 858–1134 for mvt) were equipped with an NH2-terminal FLAG tag and cloned into pQE-30 (QIAGEN) and pcDNA3 (Invitrogen). Fragments used in two-hybrid analysis were cloned into pGBT-9, pGBKT7, or pGADT7 (CLONTECH).
The raver1 full-length cDNA equipped either with an NH2-terminal FLAG tag or a birch profilin epitope tag (4A6; Rüdiger et al., 1997) was cloned into pcDNA3. For protein expression, the raver1 prey fragment (Fig. 1 A) was cloned into pMalc2 (New England Biolabs, Inc.). The raver1 full-length cDNA and the deletion fragments raver1NΔC (amino acids 1–441) and raver1CΔN (amino acids 442–748) used in yeast two-hybrid analysis were cloned into pGBKT7. Murine and human PTB/hnRNPI (provided by C.W. Smith, Cambridge University, Cambridge, UK) and cloned into pGADT7 (CLONTECH). Chicken ZBP1 was equipped with an NH2-terminal FLAG tag and cloned into pcDNA3 (Invitrogen). All constructs generated for this study are listed in Table II.
Table II. Constructs generated for this study.
| Construct | Insert | Vector |
|---|---|---|
| His-FLAG-VT | His/FLAG-tagged murine vinculin tail (vt), amino acids 858–1066 | pQE-30 |
| His-FLAG-MVT | His/FLAG-tagged murine metavinculin tail (mvt), amino acids 858–1134 | pQE-30 |
| MBP-raver1/prey | MBP-tagged murine raver1 fragment, amino acids 1–625 | pMalc2 |
| FLAG-VT | FLAG-tagged vt, amino acids 858–1066 | pcDNA3 |
| FLAG-MVT | FLAG-tagged mvt, amino acids 858–1134 | pcDNA3 |
| FLAG-raver1 | FLAG-tagged raver1, amino acids 1–748 | pcDNA3 |
| FLAG-ZBP1 | FLAG-tagged ZBP1, amino acids 1–576 | pcDNA3 |
| 4A6-raver1 | Birch profilin epitope-tagged raver1, amino acids 1–748 | pcDNA3 |
| Gal4BD-VT | Gal4(BD-domain)-tagged vt, amino acids 858–1066 | pGBT-9 |
| Gal4BD-MVT | Gal4(BD-domain)-tagged mvt, amino acids 858–1134 | pGBT-9 |
| Gal4AD-VT | Gal4(AD-domain)-tagged vt, amino acids 858–1066 | pGADT7 |
| Gal4AD-VTΔC1 | Gal4(AD-domain)-tagged vt, amino acids 858–1012 | pGADT7 |
| Gal4AD-VTΔC2 | Gal4(AD-domain)-tagged vt, amino acids 858–975 | pGADT7 |
| Gal4AD-VTΔC3 | Gal4(AD-domain)-tagged vt, amino acids 858–942 | pGADT7 |
| Gal4AD-MVT | Gal4(AD-domain)-tagged mvt, amino acids 858–1134 | pGADT7 |
| Gal4AD-MVTΔC1 | Gal4(AD-domain)-tagged vt, amino acids 858–1080 | pGADT7 |
| Gal4AD-MVTΔC2 | Gal4(AD-domain)-tagged vt, amino acids 858–1043 | pGADT7 |
| Gal4AD-MVTΔC3 | Gal4(AD-domain)-tagged vt, amino acids 858–1010 | pGADT7 |
| Gal4BD-raver1 | Gal4(BD-domain)-tagged raver1, complete | pGADT7 |
| Gal4BD-raver1NΔC | Gal4(BD-domain)-tagged raver1, amino acids 1–441 | pGADT7 |
| Gal4BD-raver1CΔN | Gal4(BD-domain)-tagged raver1, amino acids 442–748 | pGADT7 |
| Gal4AD-PTB1 | Gal4(AD-domain)-tagged PTB1, amino acids 1–557 | pGADT7 |
Northern blotting
Two PCR fragments comprising the entire ORF of raver1 were DIG labeled as described above and used to probe a mouse multiple tissue Northern blot and a mouse master blot, both purchased from CLONTECH.
Expression and purification of recombinant proteins
For recombinant expression of His-tagged proteins, Escherichia coli (M15) were transformed with the vectors pQE-30-FLAG-vt (amino acids 858–1066) or pQE-30-FLAG-mvt (amino acids 858–1134) and grown in 2 × YT medium at 37°C. Protein expression was induced, and proteins were purified according to the manufacturer's instructions (QIAGEN). Additional purification was achieved by FPLC (Amersham Pharmacia Biotech) on a Mono S column in 20 mM Tris-HCl, pH 6.2, 0.25 mM NaCl, 20 mM β-mercaptoethanol, 100 μm pefabloc SC, 10 μm pepstatin A, and 0.46 μm aprotinin using an NaCl gradient for protein elution. Purity of all proteins was checked by SDS-PAGE and immunoblots using a FLAG antibody (M2; Sigma-Aldrich). Protein concentration was determined by standard methods. Expression and purification of MBP-tagged proteins were performed essentially as described in Hüttelmaier et al. (1997).
Purification of gizzard proteins and PIP2 activation
Turkey gizzard α-actinin, metavinculin, and vinculin were purified according to Feramisco and Burridge (1980) using an FPLC Q-Sepharose (Amersham Pharmacia Biotech) instead of a DEAE column. Vinculin tail and head fragments were obtained by Glu-C protease (V8) digestion and purified essentially as described (Hüttelmaier et al., 1998). For activation of vinculin/metavinculin by PIP2, micelles were prepared as described in Hüttelmaier et al. (1998), and protein solutions were supplemented with a 50-fold molar excess of PIP2.
In vitro transcription/translation and dot overlay assays
The isolated raver1 cDNA was cloned into the vector pcDNA3 (Invitrogen), and [35S]methionine-labeled protein was synthesized by in vitro transcription/translation using the TNT-coupled reticulocyte lysate system (Promega).
Purified and PIP2-activated proteins were immobilized on nitrocellulose membrane (2.5–25 pmol) using a dot blotter with circular slots (Biometra). The transfer was controlled by Ponceau staining, and unspecific binding sites were blocked with 5% nonfat milk powder in TBST (25 mM Tris-HCl, 150 mM NaCl, 0.1% vol/vol, Tween-20, pH 7.6). In vitro–translated proteins were diluted (1:40–1:80) in TBST containing 20 mM β-mercaptoethanol. The membrane was incubated in this solution overnight at 4°C, and after intensive washing in TBST bound proteins were detected by autoradiography.
Antibodies
The monoclonal raver1 antibody 5G6 was raised against a recombinant MBP-tagged raver1 prey fragment. The antibody-producing hybridoma cells were grown according to standard protocols. The secreted antibody was classified as an IgG1 type. A polyclonal rabbit antibody against turkey gizzard vinculin was generated according to standard protocols. The serum was purified by FPLC (Amersham Pharmacia Biotech) on a protein G–Sepharose column according to the manufacturer's recommendations. The specificity of the antibodies was checked by immunoblot and immunofluorescence analyses. Substitution of the pre-immune sera for the purified antivinculin antibody was used as negative control.
Monoclonal antibodies against vinculin, α-actinin, and FLAG tag were purchased from Sigma-Aldrich, and a polyclonal antibody against MEF-2 was purchased from Santa Cruz Biotechnology, Inc. Polyclonal antibodies against PTB/hnRNPI and ZBP1 were provided by C.W. Smith (Cambridge University) and K. Farina (Albert Einstein College of Medicine), respectively. Secondary antibodies for immunoblots were HRP-coupled goat anti–mouse or HRP-coupled anti–rabbit (Dianova). For immunofluorescence, goat anti–mouse conjugated with FITC, TRITC, or Cy3 (Sigma-Aldrich) or goat anti–mouse Fab conjugated with FITC (Dianova) and goat anti–rabbit conjugated with FITC or Cy2 (Sigma-Aldrich) were used.
Cell lines
MDCK, HeLa, and C2C12 cell lines were purchased originally from the American Type Culture Collection or European Collection of Cell Cultures. For reference to the SV80 cell line see Flint et al. (1985). All cells were maintained in DME supplemented with 10% FBS. Fusion of C2C12 myoblasts was induced by replacing 10% FBS with 10% fetal horse serum. For immunofluorescence, cells were replated on collagen-coated glass coverslips and maintained at least 48 h before fixation.
Immunoprecipitation, immunoblotting, and immunofluorescence procedures
In situ cross-linking and subsequent immunoprecipitation of plasma membrane-associated protein complexes were performed as described in Hüttelmaier et al. (1998). For the analysis of presumptive nuclear protein complexes, either endogenous proteins were precipitated from C2C12 cells, or the cells were transfected with FLAG-tagged proteins. Extracts were prepared without prior cross-linking in 50 mM Tris, 150 mM KCl, 5 mM MgCl2, 0.25% NP-40, 0.05% DOC, 5% glycerol, 1 mM EDTA, 1 mM EGTA, pH 8.0, containing either RNAsin (100 U/ml; Roche) or RNAse A (1 μg/ml; Roche). Subsequently, precipitates were obtained with either PTB/hnRNPI antibody and protein G–Sepharose beads (Sigma-Aldrich) or a monoclonal FLAG antibody coupled to agarose beads (M2 or M2 beads; Sigma-Aldrich), respectively. Immunoprecipitates were subjected to SDS-PAGE and immunoblotting as described (Hüttelmaier et al., 1998).
For single or double immunofluorescence, cultured cells were fixed and extracted with 2% formaldehyde and 0.2% Triton X-100 before the incubation with the respective monoclonal antibodies followed by either FITC-, Cy2-, or Cy3-conjugated goat anti–mouse antibodies. Colocalization of raver1 with PTB/hnRNPI in C2C12 nuclei was analysed in an Olympus microscope equipped with a Piezo drive for optical sectioning and a cooled CCD camera (Roper Scientific). Skeletal muscle (m. soleus) was prepared from adult mice and immediately shock-frozen in isopentane cooled by liquid nitrogen. Tissue sections (6–8 μm) were obtained by cryosectioning and fixed with 4% PFA or ice-cold acetone, blocked with 2% BSA in PBS for 1 h, and processed for double immunofluorescence. They were incubated with the indicated monoclonal or polyclonal antibodies followed by either FITC-conjugated, Cy2-conjugated, or Cy3-conjugated goat anti–mouse or anti–rabbit antibodies. Counterstaining for vinculin was performed using the polyclonal antivinculin antibody and an FITC-conjugated goat anti–rabbit secondary antibody. Fluorescence analysis of myogenic cells and skeletal muscle sections was essentially as described (Harbeck et al., 2000).
Heterokaryon formation
Heterokaryons of HeLa and mouse C2C12 cells were formed essentially as described in Fan and Seitz (1998) with the following modifications. HeLa cells were seeded onto collagen-coated coverslips at 105 cells per coverslip and transiently transfected as described (Hüttelmaier et al., 1998). After an overnight incubation, C2C12 cells that had been incubated for 45 min in the presence of 75 μg/ml cycloheximide (Sigma-Aldrich) were seeded onto the same coverslip at 2 × 105 cells per coverslip. Subsequently, cells were incubated for an additional 4 h in the presence of 75 μg/ml cycloheximide, fused with 50% polyethylene glycol (PEG 3350; Sigma-Aldrich) for 2 min, washed twice with PBS, and returned to medium supplemented with 100 μg/ml cycloheximide for another 4-h incubation. Cells were fixed by paraformaldehyde and processed for immunostaining as described (Harbeck et al., 2000).
Acknowledgments
We thank B. Kleinhenz and S. Witt for experimental help, and S. Buchmeier, S. Froese and T. Geisler for expert technical assistance. We are grateful to Drs. C.W. Smith for antibodies and clones of murine and human PTB, K. Farina for antibodies, and A. Bershadsky and M. Shtutman (Weizmann Institute, Israel) and H. Jockusch (University of Bielefeld, Germany) for their interest in and support of this study. We acknowledge Dr. J.C. Norman's and D.R. Critchley's (University of Leicester, Leicester, UK) permission to quote their unpublished data. We also thank the Resource Center of the German Human Genome Project at the Max-Planck-Institute of Molecular Genetics (Berlin, Germany) for providing a cDNA clone (ICRF p523J246).
We are grateful to the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie for financial support.
M. Rüdiger's present address is Cardion, Max-Planck-Str. 15a, D-40699 Erkrath, Germany.
Footnotes
Abbreviations used in this paper: hnRNP, heterogeneous nuclear RNP; mvt, metavinculin tail; NES, nuclear export sequence; NLS, nuclear location signal; PIP2, phosphatidylinositol 4,5-bisphosphate; PTB, polypyrimidine tract binding protein; RRM, RNA recognition motif; vh, vinculin head; vt, vinculin tail; ZBP, zip code binding protein.
References
- Bakolitsa, C., J.M. de Pereda, C.R. Bagshaw, D.R. Critchley, and R.C. Liddington. 1999. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 99:603–613. [DOI] [PubMed] [Google Scholar]
- Barth, A.I., I.S. Nathke, and W.J. Nelson. 1997. Cadherins, catenins, and APC protein: interplay between cytoskeletal complexes and signaling pathways. Curr. Opin. Cell Biol. 9:683–690. [DOI] [PubMed] [Google Scholar]
- Belkin, A.M., O.I. Ornatsky, A.E. Kabakov, M.A. Glukhova, and V.E. Koteliansky. 1988. Diversity of vinculin/meta-vinculin in human tissues and cultivated cells. Expression of muscle specific variants of vinculin in human aorta smooth muscle cells. J. Biol. Chem. 263:6631–6635. [PubMed] [Google Scholar]
- Ben Ze'ev, A., and B. Geiger. 1998. Differential molecular interactions of beta-catenin and plakoglobin in adhesion, signaling and cancer. Curr. Opin. Cell Biol. 10:629–639. [DOI] [PubMed] [Google Scholar]
- Bonne, S., J. van Hengel, F. Nollet, P. Kools, and F. van Roy. 1999. Plakophilin-3, a novel armadillo-like protein present in nuclei and desmosomes of epithelial cells. J. Cell Sci. 112:2265–2276. [DOI] [PubMed] [Google Scholar]
- Conte, M.R., T. Grune, J. Ghuman, G. Kelly, A. Ladas, S. Matthews, and S. Curry. 2000. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19:3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote, C.A., D. Gautreau, J.M. Denegre, T.L. Kress, N.A. Terry, and K.L. Mowry. 1999. A Xenopus protein related to hnRNPi has a role in cytoplasmic RNA localization. Mol. Cell. 4:431–437. [DOI] [PubMed] [Google Scholar]
- Critchley, D.R. 2000. Focal adhesions—the cytoskeletal connection. Curr. Opin. Cell Biol. 12:133–139. [DOI] [PubMed] [Google Scholar]
- Danowski, B.A., K. Imanaka-Yoshida, J.M. Sanger, and J.W. Sanger. 1992. Costameres are sites of force transmission to the substratum in adult rat cardiomyocytes. J. Cell Biol. 118:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X.C., and J.A. Seitz. 1998. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA. 95:15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feramisco, J.R., and K. Burridge. 1980. A rapid purification of α-actinin, filamin, and a 130,000-dalton protein from smooth muscle. J. Biol. Chem. 255:1194–1199. [PubMed] [Google Scholar]
- Flint S.J., K. Leong, and G.A. Beltz. 1985. The expression of the SV40 early region in the transformed human cell line SV80. Virus Res. 2:359–373. [DOI] [PubMed] [Google Scholar]
- Ghetti, A., S. Pinol-Roma, W.M. Michael, C. Morandi, and G. Dreyfuss. 1992. hnRNPI, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20:3671–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, A.P., and K. Burridge. 1996. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 381:531–535. [DOI] [PubMed] [Google Scholar]
- Gimona, M., D.O. Fürst, and J.V. Small. 1987. Metavinculin and vinculin from mammalian smooth muscle: bulk isolation and characterization. J. Muscle Res. Cell Motil. 8:329–341. [DOI] [PubMed] [Google Scholar]
- Gonsior, S.M., S. Platz, S. Buchmeier, U. Scheer, B.M. Jockusch, and H. Hinssen. 1999. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J. Cell Sci. 112:797–809. [DOI] [PubMed] [Google Scholar]
- Gooding, C., G.C. Roberts, and C.W. Smith. 1998. Role of inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 4:85–100. [PMC free article] [PubMed] [Google Scholar]
- Hannan, A.J., G. Schevzos, P. Gunning, P.L. Jeffrey, and R.P. Weinberger. 1995. Intracellular localization of tropomyosin mRNA and protein is associated with development of neuronal polarity. Mol. Cell. Neurosci. 6:397–412. [DOI] [PubMed] [Google Scholar]
- Harbeck, B., S. Hüttelmaier, K. Schlüter, B.M. Jockusch, and S. Illenberger. 2000. Phosphorylation of the vasodilator-stimulated phosphoprotein regulates its interaction with actin. J. Biol. Chem. 275:30817–30825. [DOI] [PubMed] [Google Scholar]
- Hill, M.A., and P. Gunning. 1993. Beta and gamma actin mRNAs are differentially located within myoblasts. J. Cell Biol. 122:825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, W., B. Reichart, A. Ewald, E. Müller, I. Schmitt, R.H. Stauber, F. Lottspeich, B.M. Jockusch, U. Scheer, J. Hauber, and M.C. Dabauvalle. 2001. Cofactor requirements for nuclear export of rev response element (RRE)– and constitutive transport element (CTE)–containing retroviral RNAs: an unexpected role for actin. J. Cell Biol. 152:895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. 2000. Perinucleolar structures. J. Struct. Biol. 129:233–240. [DOI] [PubMed] [Google Scholar]
- Hüttelmaier, S., P. Bubeck, M. Rüdiger, and B.M. Jockusch. 1997. Characterization of two F-actin-binding and oligomerization sites in the cell contact protein vinculin. Eur. J. Biochem. 247:1136–1142. [DOI] [PubMed] [Google Scholar]
- Hüttelmaier, S., O. Mayboroda, B. Harbeck, T. Jarchau, B.M. Jockusch, and M. Rüdiger. 1998. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr. Biol. 8:479–488. [DOI] [PubMed] [Google Scholar]
- Jansen, R.P. 1999. RNA-cytoskeletal associations. FASEB J. 13:455–466. [PubMed] [Google Scholar]
- Jansen, R.P. 2001. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2:247–256. [DOI] [PubMed] [Google Scholar]
- Krecic, A.M., and M.S. Swanson. 1999. hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol. 11:363–371. [DOI] [PubMed] [Google Scholar]
- Kroemker, M., A.H. Rüdiger, B.M. Jockusch, and M. Rüdiger. 1994. Intramolecular interactions in vinculin control alpha-actinin binding to the vinculin head. FEBS Lett. 355:259–262. [DOI] [PubMed] [Google Scholar]
- McKenna, N.M., C.S. Johnson, and Y.L. Wang. 1986. Formation and alignment of Z lines in living chick myotubes microinjected with rhodamine-labeled alpha-actinin. J. Cell Biol. 103:2163–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, C., I. Hofman, Z. Wang, M. Teichmann, S. Sepheri Chong, M. Schnolzer, and W.W. Franke. 2001. Nuclear particles containing RNA polymerase III complexes associated with the junctional plaque protein plakophilin 2. Proc. Natl. Acad. Sci. USA. 98:7795–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, W.M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 83:415–422. [DOI] [PubMed] [Google Scholar]
- Moore, M.S. 1996. Protein translocation: nuclear export pathway. Curr. Biol. 6:137–140. [DOI] [PubMed] [Google Scholar]
- Morris, E.J., and A.B. Fulton. 1994. Rearrangement of mRNAs for costamere proteins during costamere development in cultured skeletal muscle from chicken. J. Cell Sci. 107:377–386. [DOI] [PubMed] [Google Scholar]
- Mulligan, G.J., W. Guo, S. Wormsley, and D.M. Helfman. 1992. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J. Biol. Chem. 267:25480–25487. [PubMed] [Google Scholar]
- Munro, T.P., G.J. Magee, G.J. Kidd, J.H. Carson, E. Barbarese, L.M. Smith, and R. Smith. 1999. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 274:34389–34395. [DOI] [PubMed] [Google Scholar]
- Nathke, I.S., L. Hinck, J.R. Swedlow, J. Papkoff, and W.J. Nelson. 1994. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J. Cell Biol. 125:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix, D.A., and M.C. Beckerle. 1997. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 138:1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov, Y., and R.H. Singer. 1998. RNA localization: different zipcodes, same postman? Trends Cell Biol. 8:381–383. [DOI] [PubMed] [Google Scholar]
- Ornatsky, O.I., J.J. Andreucci, and J.C. McDermott. 1997. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272:33271–33278. [DOI] [PubMed] [Google Scholar]
- Ozawa, M., H. Baribault, and R. Kemler. 1989. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 8:1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, J.V., J.D. Siciliano, and S.W. Craig. 1983. Vinculin is a component of an extensive network of myofibril-sarcolemma attachment regions in cardiac muscle fibers. J. Cell Biol. 97:1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.G., E.B. Porro, J. Galceran, P. Tempst, and B. Nadal-Ginard. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7:393–406. [DOI] [PubMed] [Google Scholar]
- Rando, O., K. Zhao, and G.R. Crabtree. 2000. Searching for a function for nuclear actin. Trends Cell Biol. 10:92–97. [DOI] [PubMed] [Google Scholar]
- Ross, A.F., Y. Oleynikov, E.H. Kislauskis, K.L. Taneja, and R.H. Singer. 1997. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell Biol. 17:2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger, M., B.M. Jockusch, and M. Rothkegel. 1997. An epitope tag-antibody combination useful for the detection of protein expression in procaryotic and eukaryotic cells. Biotechniques. 23:96–97. [DOI] [PubMed] [Google Scholar]
- Scheer, U., H. Hinssen, W.W. Franke, and B.M. Jockusch. 1984. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 39:111–122. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., S. Langbein, M. Rode, S. Pratzel, R. Zimbelmann, and W.W. Franke. 1997. Plakophilins 1a and 1b: widespread nuclear proteins recruited in specific epithelial cells as desmosomal plaque components. Cell Tissue Res. 290:481–499. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., L. Langbein, S. Pratzel, M. Rode, H.R. Rackwitz, and W.W. Franke. 1999. Plakophilin 3—a novel cell-type-specific desmosomal plaque protein. Differentiation. 64:291–306. [DOI] [PubMed] [Google Scholar]
- Shamoo, Y., N. Abdul-Mana, and K.R. Williams. 1995. Multiple RNA binding domains (RBDs) just don't add up. Nucleic Acid Res. 23:725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu, A., and M.F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell. 102:135–138. [DOI] [PubMed] [Google Scholar]
- Siomi, H., and G. Dreyfuss. 1997. RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev. 7:345–353. [DOI] [PubMed] [Google Scholar]
- Southby, J., C. Gooding, and C.W. Smith. 1999. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of alpha-actinin mutually exclusive exons. Mol. Cell. Biol. 19:2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D.L. 2001. Nuclear domains. J. Cell Sci. 114:2891–2893. [DOI] [PubMed] [Google Scholar]
- Temm-Grove, C.J., B.M. Jockusch, R.P. Weinberger, G. Schevzov, and D.M. Helfman. 1998. Distinct localizations of tropomyosin isoforms in LLC-PK1 epithelial cells suggests specialized function at cell-cell adhesions. Cell Motil. Cytoskeleton. 40:393–407. [DOI] [PubMed] [Google Scholar]
- Terracio, L., D.G. Simpson, L. Hilenski, W. Carver, R.S. Decker, N. Vinson, and T.K. Borg. 1990. Distribution of vinculin in the Z-disk of striated muscle: analysis by laser scanning confocal microscopy. J. Cell Physiol. 145:78–87. [DOI] [PubMed] [Google Scholar]
- Valcarcel, J., and F. Gebauer. 1997. Post-transcriptional regulation: the dawn of PTB. Curr. Biol. 7:R705–R708. [DOI] [PubMed] [Google Scholar]
- Weekes, J., S.T. Barry, and D.R. Critchley. 1996. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem. J. 314:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg, E., M. Sattler, D.S. Ewaniuk, and R. Salgia. 1997. Role of focal adhesion proteins in signal transduction and oncogenesis. Crit. Rev. Oncog. 8:343–358. [DOI] [PubMed] [Google Scholar]
- Williamson, D.J., S. Banik-Maiti, J. DeGregori, and H.E. Ruley. 2000. hnRNP C is required for postimplantation mouse development but is dispensable fro cell viability. Mol. Cell. Biol. 20:4094–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H.K., K. Tsukahara, Y. Kanaoka, S. Jinno, and H. Okayama. 1999. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 19:3829–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H.L., T. Eom, Y. Oleynikov, S.M. Shenoy, D.A. Liebelt, J.B. Dictenberg, R.H. Singer, and G.J. Bassell. 2001. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 31:261–275. [DOI] [PubMed] [Google Scholar]