Abstract
Activation-induced deaminase (AID) catalyses deamination of deoxycytidine to deoxyuridine within immunoglobulin loci, triggering pathways of antibody diversification that are largely dependent on uracil-DNA glycosylase (uracil-N-glycolase [UNG]). Surprisingly efficient class switch recombination is restored to ung−/− B cells through retroviral delivery of active-site mutants of UNG, stimulating discussion about the need for UNG's uracil-excision activity. In this study, however, we find that even with the overexpression achieved through retroviral delivery, switching is only mediated by UNG mutants that retain detectable excision activity, with this switching being especially dependent on MSH2. In contrast to their potentiation of switching, low-activity UNGs are relatively ineffective in restoring transversion mutations at C:G pairs during hypermutation, or in restoring gene conversion in stably transfected DT40 cells. The results indicate that UNG does, indeed, act through uracil excision, but suggest that, in the presence of MSH2, efficient switch recombination requires base excision at only a small proportion of the AID-generated uracils in the S region. Interestingly, enforced expression of thymine-DNA glycosylase (which can excise U from U:G mispairs) does not (unlike enforced UNG or SMUG1 expression) potentiate efficient switching, which is consistent with a need either for specific recruitment of the uracil-excision enzyme or for it to be active on single-stranded DNA.
Functional immunoglobulin genes are assembled by V-(D)-J joining, a process catalysed by the RAG1/RAG2 recombinase. The functionally rearranged Ig variable (IgV) regions can then be diversified by somatic hypermutation or gene conversion; the immunoglobulin constant (IgC) region can be exchanged by switch recombination (allowing the switch from IgM to IgG, IgA, or IgE). Somatic hypermutation, gene conversion, and switch recombination are all dependent on activation-induced deaminase (AID) (1–4).
A variety of lines of evidence (for review see references [5, 6]) reveal that AID acts by deaminating deoxycytidine residues to deoxyuridine (dU) at sites within the immunoglobulin loci, generating dU:deoxyguanosine (dG) lesions. Thus, genetic and biochemical assays show that AID is able to deaminate cytidine in single-stranded DNA with a local sequence preference that accords with the localization of in vivo somatic mutation hotspots; antibody gene diversification pathways are also altered in B cells that carry disruptions in the pathways that process uracil in DNA (7–23).
Uracil excision by the uracil-DNA glycosylase (uracil-N-glycolase [UNG]) is envisioned to play a central role in AID-triggered immunoglobulin gene diversification (7–9, 11, 12, 17, 21–23). The transversion mutations at C:G pairs observed during IgV somatic hypermutation are proposed to be a consequence of DNA synthesis over the UNG-generated abasic site in the IgV; switch recombination is proposed to be, in large part, achieved through UNG-mediated excision of the AID-generated uracils in the vicinity of the immunoglobulin switch region. Although such a DNA deamination scheme for antibody gene diversification enjoys widespread support, the ability of active-site mutants of UNG to restore switch recombination in in vitro assays was a major surprise, and it has triggered controversy about the importance of uracil excision in antibody gene diversification (24–28).
We revisit the ability of UNG mutants to partake in immunoglobulin gene diversification, and find that those UNG mutants that, on retroviral delivery, are capable of restoring switch recombination to UNG-deficient B cells still retain uracil-excision activity both in vivo and in vitro. The results suggest that the ability of UNG mutants that retain <1% of the specific uracil-excision of the wild-type enzyme to potentiate efficient switch recombination likely reflects, in part, the heavy overexpression associated with retroviral delivery, but is also an indication that uracil excision at only a small proportion of AID-generated uracils may suffice for switch recombination, a conclusion that is consistent with the proposals of other studies concerning switch-associated DNA breaks (29–31).
RESULTS
Restoration of class switching by retroviral delivery of mutant UNGs
We first sought to recapitulate the findings of Begum et al. (25) that switching could be effectively restored to ung−/− B cells by retroviral delivery of UNG mutants carrying amino acid substitutions at their active sites. Constructs were generated encoding mutant versions of the nuclear isoform of mouse UNG with the choice of mutants guided both by the identity of the mutants studied by Begum et al. (25), as well as by various structural and kinetic analyses of human UNG, to which mouse UNG is 95% identical.
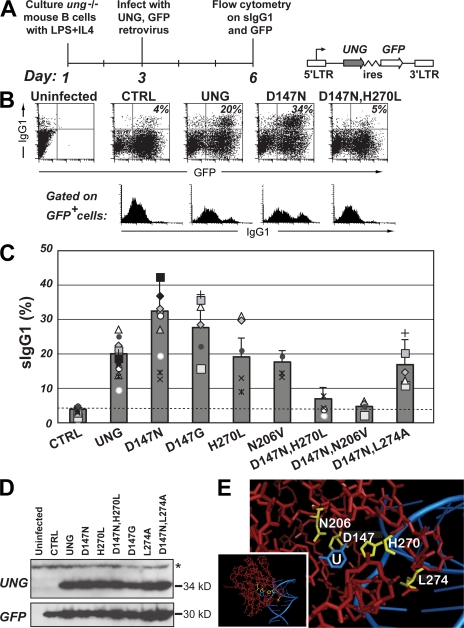
We find that single amino-acid substitution mutants of residues that participate in catalyzing the cleavage of the glycosidic bond (D147N or D147G, H270L, and N206V [32–36]) are all able to complement the switching deficiency of B cells from ung−/− mice in the retroviral reconstitution assay, whereas no evident restoration is obtained with the double UNG mutants (D147N,H270L) and (D147N,N206V) (Fig. 1). This is entirely in keeping with (and extends upon) a previous work (25). Interestingly, combining D147N with mutation of residue L274 still allows efficient restoration of switching. Residue L274 does not partake in the actual hydrolytic catalysis, but instead is inserted into the DNA helix upon uracil flipping, with mutation of this residue significantly diminishing UNG activity by reducing the stability of the DNA–UNG complex (32, 36).
Figure 1.
Class switching in ung−/− B cells after retroviral delivery of mutant UNGs. (A) Experimental strategy with schematic representation of the retroviral vector used for UNG delivery. (B) Representative flow cytometric plots of switching to IgG1 by ung−/− B cells after retroviral delivery of mutant mouse UNGs. The proportion of retrovirally infected (GFP+-positive) cells that have switched to IgG1 is indicated in the top right quadrant of each two-dimensional plot, with the sIgG1 profile of the gated GFP+ cells shown below. (C) Compilation of the results of multiple comparisons of switching to IgG1 achieved with different mutant mouse UNGs. The histogram presents the results of 13 independent experiments represented by the different symbols (each using B cells prepared from a single mouse), with each mutant being analyzed in at least 4 separate experiments. To facilitate comparison of the results obtained in different experiments, a common symbol is used to depict the results obtained in a single experimental set. To facilitate comparison of overall switching proficiency of the different UNG mutants, as deduced from multiple experiments, the means in the histograms are presented as values plus the SD that were normalized to 20% switching by wild-type UNG. The switching achieved using the D147N, D147G, (D147N,H270L), and (D147N,N206V) mutants differs significantly from that achieved with wild-type UNG (P < 0.05 for the single mutants; P < 0.001 for the double mutants). (D) Expression of the different UNG mutants analyzed by Western blot of extracts from the retroviral packaging cells. A Western blot for GFP provides a control. The N206V mutation prevents recognition by the anti-UNG antibody, although the UNG-N206V mutant yields uracil-excision activity and supports switch recombination (C and Fig. 3 C). A nonspecific band recognized by the anti-UNG antibody is indicated by an asterisk. (E) The amino acid positions mutagenized in mouse UNG (yellow) are highlighted on a structure of human UNG (red) cocrystallized with a U-containing DNA fragment (blue; pdb 1SSP [36]) in which U is flipped out of the DNA helix and into the enzyme active site. The area surrounding the active site has been zoomed in from the whole structure (inset). Residue numbering is as in mouse UNG nuclear isoform.
Restoration of switching by mutant UNGs is affected by MSH2 deficiency
Several explanations can be envisioned to account for the efficacy with which the UNGs carrying single active-site mutations are able to restore switching to B cells from ung−/− mice. One is that they retain sufficient uracil-excision activity to allow switch recombination to proceed in vivo. A second (the interpretation favored by Begum et al. (24, 25]) is that UNG fulfils some unidentified role in switch recombination that is dependent neither on its uracil-excision nor on its uracil-binding activity. A third possibility is that mere binding of the catalytically compromised UNG mutants to the dU:dG lesions generated by AID-targeted deamination creates a physical disturbance that is sufficient to potentiate switch recombination by a pathway that does not require uracil excision.
Previous work has shown that, in the absence of UNG, switching can nevertheless proceed (albeit with greatly reduced efficacy) through a pathway that depends on the MSH2 mismatch recognition protein, presumably depending on MSH2/MSH6 recognition of the initiating dU:dG lesion (7, 11). One could imagine that binding of a catalytically compromised UNG mutant to the dU:dG lesions could enhance switch recombination through this MSH2-dependent pathway, which is a possible exemplification of the third type of scenario envisioned in the previous paragraph.
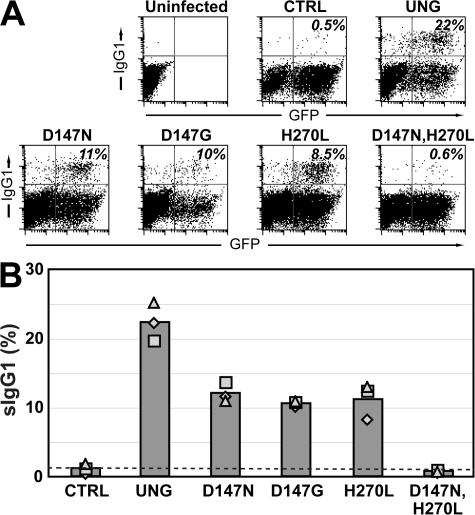
We therefore asked whether the restoration of switching by the mutant UNGs was dependent on MSH2, and we tested this by retroviral delivery to B cells from ung−/− msh2−/− double-knockout mice. It is evident that although the D147N, D147G, and H270L single amino-acid substitution mutants of UNG are all able to reconstitute switching in B cells from these double-knockout mice, they are consistently less active than the wild-type enzyme in this regard (Fig. 2). This is in contrast to what was observed with B cells from the ung−/− single-knockout mice, where the UNG single-point mutants were equally or more potent as compared with the wild-type enzyme in restoring switching in the retroviral infection assay (Fig. 1 C).
Figure 2.
Class switching in ung−/− msh2−/− B cells after retroviral delivery of mutant UNGs. (A) Representative flow cytometric plots presented as described in the legend to Fig. 1 B. (B) Compilation of the results of three separate experiments represented by the different symbols, with the bars representing the averages. Switching by all the UNG mutants differs significantly from that achieved with the wild-type enzyme (P < 0.05).
These results revealed that the restoration of switching by the mutant UNGs was, although more dependent on MSH2 than the switching by wild-type UNG, nevertheless certainly not wholly dependent on an MSH2-dependent backup pathway. This suggested that the switch-potentiating activity of these proteins might be at least partially dependent on their uracil-excision activity. We therefore wished to investigate the extent to which the switch-potentiating activity of the different mutant UNGs correlated with their ability to excise uracil from DNA.
Uracil-excision activity of mutant UNGs
Although Begum et al. (25) failed to detect uracil-excision activity in extracts of mammalian cells expressing their mouse UNG mutants carrying single amino-acid substitutions at their active site, analogous mutants of human UNG have been found to display residual uracil-excision activity (between 0.05 and 0.5% that of the wild type) after expression in Escherichia coli (28, 34, 35, 37). We suspect that this discordance reflects an insensitivity of the uracil-excision assay used by Begum et al. (25).
We find that the D147N mouse UNG mutant does (just like its human counterpart) display residual catalytic activity, as monitored by comparative kinetic analysis of wild-type and recombinant mouse UNGs purified from E. coli. Thus, whereas wild-type mouse UNG gave a KM of 0.39 ± 0.09 μM and kcat of 7.5 ± 0.6 s−1 on a single-stranded U-containing oligodeoxyribonucleotide substrate as judged by a fluorescence-based assay (see Materials and methods), the mouse D147N mutant gave a similar KM (0.37 ± 0.12 μM), but an ∼1,000-fold reduced kcat (6.5 × 10−3 ± 6 × 10−4 s−1). This difference in kcat is similar to that observed between the wild-type and D147N-equivalent mutant of human UNG (34, 37).
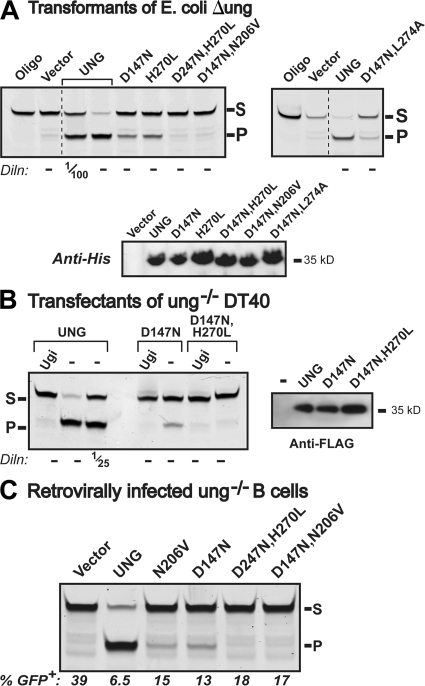
We therefore went on to test whether this uracil-excision activity associated with the mutant UNGs was also detectable in whole-cell extracts using the gel-based oligonucleotide assay (rather than a high-sensitivity fluorescence-based assay of purified protein). Despite its low turnover activity, the mouse D147N mutant was expressed at sufficient levels in whole-cell extracts of ung-deficient transformants of E. coli, in stable transfectants of ung−/− chicken DT40 B cells, and in the retrovirally transduced B cells from ung−/− mice for its catalytic activity to be clearly detectable in such assays (Fig. 3, A–C). Furthermore, the same result was obtained for the mouse UNG mutants carrying single amino-acid substitutions (H270L and N206V; Fig. 3, A and C). However, despite comparable expression levels, no activity could be detected for any of the mutants carrying two mutations in active-site residues, although activity was readily detectable for the [D147N, L274A] in which the L274 residue is not implicated in the excision of the uracil itself. Thus, all the UNG mutants that had been able to potentiate class switching in the retroviral infection assay exhibited uracil-excision activity in the oligonucleotide cleavage assay (Fig. 3), whereas those defective in the uracil-excision assay failed to give detectable switching. Therefore, the most straightforward interpretation of the data is that a low level of uracil-excision activity suffices for efficient (though significantly MSH2-dependent) switching in the retroviral infection assay; however, UNG mutants with undetectable uracil-excision activity in the oligonucleotide cleavage assays are ineffective at potentiating switching.
Figure 3.
Mutant UNGs that support switching retain uracil-excision activity. (A) Assay for uracil-excision activity by recombinant UNGs produced in E. coli. 10 μg extracts of E. coli (Δung) transformants expressing His-tagged versions of the indicated mouse UNG variants were incubated with 0.5 pmol of FITC-labeled, U-containing single-stranded oligonucleotide substrate. After incubation with apyrimidinic endonuclease and subsequent PAGE, uracil excision is evident from conversion of the fluorescently labeled 42-mer substrate (S) into the 26-mer product (P). Undiluted extracts were used, except where indicated. A Western blot using an anti-histidine tag antiserum was used to control for expression levels. (B) Assay for uracil-excision activity in extracts of ung−/− DT40 cells transfectants expressing wild-type or mutant FLAG-tagged mouse UNGs. Samples (5 μg protein, or a dilution where specified) of whole-cell extracts were assayed for uracil-excision activity as described in A, but with the extracts preincubated with the UNG-specific inhibitor Ugi where indicated. (right) An anti-FLAG Western blot to control for expression levels. (C) Assay for uracil-excision activity in extracts of ung−/− mouse B cells that have been transduced with the indicated UNG-expressing retroviral vectors. Extracts (which were assayed undiluted) were prepared 4 d after retroviral transduction, and the proportion of retrovirally infected cells (as judged by GFP fluorescence) in each population is indicated below each lane. Endogenous SMUG1 activity was neutralized by preincubating the extracts with anti-SMUG1 PSM1 antibody (11).
Mutant UNGs are defective in an IgV gene conversion assay
Apart from its role in class switching, UNG is also needed for IgV gene conversion, as well as for the introduction of transversion mutations at C:G pairs during hypermutation (8, 11, 12, 17, 21, 23). We wondered whether the low specific activity D147N mutant could substitute for wild-type UNG in these pathways of antibody diversification as it did in class switching. Chicken DT40 B cells deficient in UNG activity are ineffective at IgV gene conversion, and instead diversify their IgV genes by the introduction of transition mutations at C:G pairs (8, 12). However, although stable transfection of a wild-type UNG construct allowed a restoration of gene conversion and transversion mutations at C:G pairs (as well as some mutations at A:T), no such restoration was obtained with D147N UNG (Table I).
Table I.
IgVλ diversification in UNG-reconstituted ung−/− DT40 cellsa
| Gene conversions |
Mutations at C:G
|
Mutations at A:T |
||
|---|---|---|---|---|
| Total | Tv | |||
| UNG-WT | 14 | 50 | 16 | 13 |
| UNG-D147N | 1b | 56 | 0 | 0 |
The IgVλ genes were sequenced from the sorted sIgM-loss population from 7 independent transfectants expressing wild-type mouse UNG (52 mutated sequences) and 5 independent transfectants expressing the D147N mutant (29 mutated sequences). All events identified in more than one sequence within a given transfectant population were counted only once to avoid overestimation caused by repeat counting of dynastically related events.
This putative gene conversion event comprises two consecutive C→T transitions; it cannot be definitively concluded that these do not reflect separate single-nucleotide substitutions.
Retroviral transduction yields substantial UNG overexpression
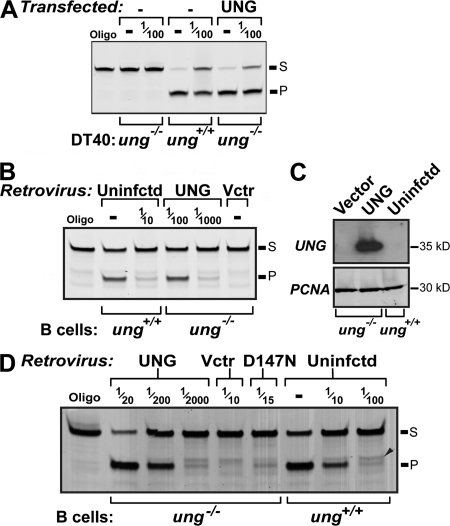
This ability of the D147N mutant to restore class switching in the retroviral infection assay, but not restore IgV gene conversion/transversions at C:G pairs in the stably transfected DT40 cells, might be explained in part by the considerable difference in the assay systems. In whole-cell extracts of ung−/− DT40 cells that have been stably transfected with the pIRESHyg expression plasmid, the level of UNG activity is of a similar order to that in the parental ung+/+ DT40 line (Fig. 4 A). However, in contrast, retroviral infection of mouse ung−/− B cells with the pMX-UNG retrovirus results in a level of UNG activity that is approximately two orders of magnitude higher than that in normal ung+/+ mouse B cells (Fig. 4 B). In fact, whereas we and others (Kavli, B., personal communication) have found that the level of UNG protein in unfractionated whole-cell extracts of mouse B cells is below the limits of detection in Western blots using anti-UNG peptide antiserum, a band is readily detectable in extracts of B cells that have been transduced with the pMX-UNG retrovirus (Fig. 4 C).
Figure 4.
Comparison of endogenous UNG expression with that achieved after gene delivery. (A) uracil-excision activity in whole-cell extracts of pIRES-hygro[FLAG-UNG] transfectants of ung−/− DT40 as compared with that in parental ung+/+ DT40. Extracts were assayed undiluted or with a 10−2 dilution, as indicated. (B) Uracil-excision activity in extracts of B cells from either wild-type (ung+/+) mice or from ung−/− mice that have been transduced with pMX-UNG (UNG) or empty pMX retroviral vector (Vctr). The nominal dilutions of the extracts used are indicated, and have been normalized according to the percentage of GFP+ cells to control for differences in the efficiency of retroviral transduction. (C) UNG expression monitored by Western blot in whole-cell extracts of B cells from ung+/+ or ung−/− mice that had been transduced with either pMX-UNG or empty vector and cultured for 3 d. The filters were subsequently stripped and reprobed with an anti-PCNA antibody to provide a loading control. (D) Uracil-excision activity in extracts of B cells from ung−/− mice that have been transduced with retroviral vectors expressing wild-type or D147N UNG as compared with that in extracts of (LPS + IL4)-activated B cells from wild-type (ung+/+) mice. The nominal dilutions of the extracts used are indicated, normalized as in B. A minor band whose migration position (arrowhead) is slightly slower than the canonical oligonucleotide deamination product (P) reflects a weak activity in the extracts, which cleaves the substrate oligonucleotide just 5′ of the target uracil; after uracil excision, this minor band disappears, comigrating with the product P.
Retrovirally transduced UNGs do not restore C:G transversions in hypermutation
In addition to the switch recombination event itself, the induction of switch recombination in mouse B cells is accompanied by the introduction of point mutations into the Sμ region (38, 39). As with somatic hypermutation in the vicinity of the IgV segment, the transversion mutations at C:G pairs are dependent on UNG-mediated uracil excision, presumably through replication over the abasic site. In wild-type mice, 30–40% of the mutations at C:G pairs are transversions; this falls to <10% in UNG-deficient animals (unpublished data) (19, 25).
It seemed possible that the restoration of switch recombination in B cells from ung−/− mice that is achieved through retroviral transduction of low-activity UNG mutants might simply reflect the fact that the overexpression achieved through retroviral delivery compensates for the low specific activity of the mutant UNG enzyme, thereby allowing a normal level of uracil excision to take place. If that were the case, one would expect that retroviral delivery of low-activity UNGs would, in the same cells, also restore transversion mutations at C:G pairs in the Sμ region. This is not the case. Analysis of mutations in the region flanking the 5′-side of the Sμ repeats (the preSμ region) from UNG-deficient B cells that had been infected with the H270L UNG retrovirus revealed that although class switching had been effectively restored, the preSμ mutations were still restricted to transitions at C:G pairs (all 73 independent mutations at C:G pairs were transitions). Similarly, with the D147N UNG retrovirus (which also fully restored switching in the retrovirally transduced cells), 46 out of 50 independent C:G mutations were transitions. Thus, the level of mutant UNG overexpression that is sufficient to efficiently restore switching in retrovirally infected ung−/− B cells appears insufficient to restore C:G transversions during hypermutation of the preSμ region. Indeed, despite the heavy overexpression associated with retroviral transduction, the level of uracil-excision activity detected in extracts of ung−/− B cells that have been transduced with a retrovirus expressing the D147N mutant is still substantially lower (approximately an order of magnitude on a per cell basis) than in activated B cells from wild-type mice (Fig. 4 D).
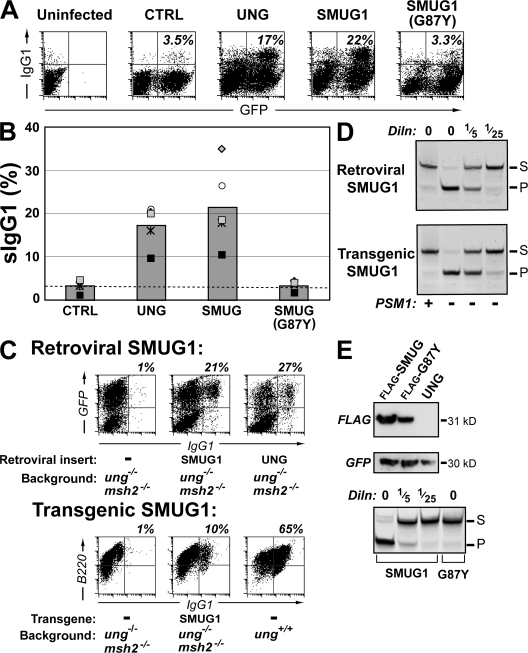
SMUG1 potentiation of switching is enhanced by retroviral delivery
We wondered whether the efficacy with which switching is restored by low-activity uracil-excision enzymes, in part, reflects the sensitivity of the retroviral switching assay. We have previously observed that enforced expression, through transgenesis, of another uracil-excision enzyme (single-stranded monofunctional uracil DNA glycosylase [SMUG] 1) only partially rescues switch recombination in B cells of ung−/− msh2−/− mice (9). We therefore wished to test whether SMUG1 might act more potently in promoting switching if introduced by retroviral infection. This is indeed the case. As with the low-activity UNG mutants, SMUG1 is equally (or slightly more) potent than wild-type UNG in potentiating switching after retroviral delivery to ung−/− B cells (Fig. 5, A and B), although it is marginally less potent than wild-type UNG when assayed using B cells from ung−/− msh2−/− mice (Fig. 5 C). In contrast, transgenically expressed SMUG1, although clearly able to trigger switching, is significantly less potent than endogenous UNG (Fig. 5 C). The greater extent of switching achieved with retrovirally delivered (as opposed to transgenically expressed) SMUG1 could in part be caused by a slightly higher level of SMUG1 activity achieved (Fig. 5 D). However, the issue may not merely be one of the extents of overexpression. It is possible that when ung−/− B cells are cultured for 2 d before delivery of the uracil-excision enzyme by retroviral transduction, the cells have accumulated large numbers of dU:dG lesions that may “prime” the reaction. Indeed, human UNG-deficient B cells have been found to accumulate uracil in their DNA (29). In contrast, in ung+/+ B cells, the dU:dG lesions generated in each cell cycle are recognized for either repair or switch recombination by endogenous UNG, such that large numbers of dU:dG lesions do not have occasion to accumulate.
Figure 5.
Comparison of class switching in ung−/− B cells expressing SMUG1 (or G87Y) through transgenesis versus retroviral delivery. (A) Representative flow cytometric plots showing switching to IgG1 in purified ung−/− mouse B cells that have been retrovirally infected with the pMX retroviral vector encoding the indicated uracil-excision enzymes. Switching was assayed as in Fig. 1, with the proportion of retrovirally infected (GFP+) cells that have switched to IgG1 indicated in the top right quadrants. (B) Comparative efficiency of switching restoration by the indicated uracil-excision enzymes as monitored in multiple experiments using B cells from ung−/− mice. Symbols are used as described in the legend to Fig. 1 so as to facilitate comparisons of the results obtained in individual sets of experiments. There is no significant difference between the extent of switching achieved with UNG and SMUG1 or SMUG[G87Y] and CTRL. (C) Comparison of the efficacy of retrovirally delivered and transgenically expressed SMUG1 in facilitating switch recombination. (top) A flow cytometric comparison of switching to IgG1 in samples of (LPS + IL4)-stimulated splenic B cells from ung−/− msh2−/− mice that have been infected with the pMX vector alone or with derivatives directing expression of human SMUG1 or mouse UNG. The percentages of infected cells (GFP+) that switched to IgG1 are indicated above each plot. (bottom) A flow cytometric comparison of switching to IgG1 as analyzed at day 8 of culture in samples of (LPS + IL4)-stimulated splenic B cells from ung−/− msh2−/− mice that did or did not carry a transgene directing enforced B cell–specific expression of human SMUG1. A parallel analysis performed on cultured B cells from UNG-proficient mice is shown on the right. The proportion of B cells (judged by CD45R[B220] staining) that switched to IgG1 is indicated. (D) Comparison of SMUG1-mediated uracil-excision activity in extracts (10 μg protein) of LPS-activated B cells from ung−/− msh2−/− mice that either carry a SMUG1 transgene (day 4 after stimulation) or that were infected with the SMUG1 retroviral construct (day 3 after infection). Preincubation with the PSM1-neutralizing anti-SMUG1 antibody confirms the origin of the uracil-excision activity. (E) Comparison of SMUG1 and SMUG1-G87Y expression. (top) SMUG1 expression in cells transfected with the indicated pMX-based retroviral vector was assessed by Western blot analysis using an anti-FLAG mAb. (middle) A Western blot for the vector-encoded GFP provides a control for infection efficiency. (bottom) The uracil-excision activity of the SMUG1 and SMUG1-G87Y mutant were assessed in stably transfected DT40 cells (because DT40 cells do not have detectable endogenous SMUG1 activity [9, 23]), as described in Fig. 3, except that all reactions were preincubated in the presence of 0.5 U of Ugi to inhibit endogenous chicken UNG (9).
Analysis of the G87Y active-site mutant of SMUG1 provides further evidence that uracil-excision activity is required for class switching. This mutant has completely lost catalytic activity (Fig. 5 E) (40), and it has simultaneously lost the ability to trigger class switch recombination (Fig. 5 A, B).
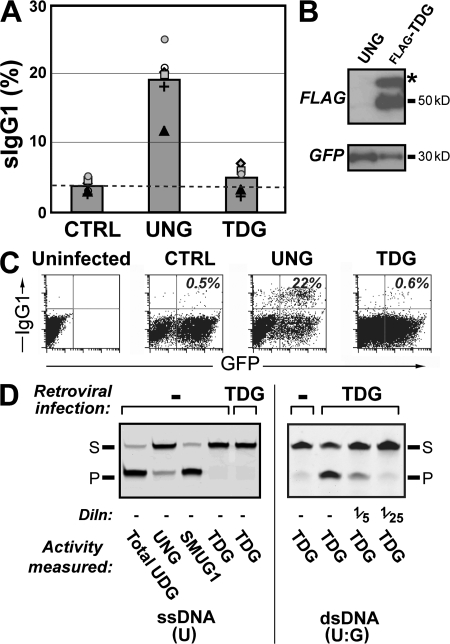
Retroviral delivery of thymine-DNA glycosylase (TDG) does not restore switching
Given the potent restoration of switching by both SMUG1 and catalytically compromised UNGs in the retroviral assay, we were curious to know whether overexpression of any uracil-excision enzyme could potentiate switching in this assay. Strikingly, retroviral delivery of mouse TDG (which excises U efficiently from U:G mispairs, but is ineffective on U in the context of single-stranded DNA) gives little evident switch recombination, although TDG polypeptide and its uracil-excision activity are readily detectable in cell extracts (Fig. 6).
Figure 6.
Retroviral expression of TDG does not restore class switching in ung−/− B cells. (A) Comparison of switching to IgG1 by B cells from ung−/− mice that have been infected with the pMX retroviral vector encoding GFP alone or GFP together with either UNG or FLAG-tagged TDG, as monitored in multiple experiments indicated by the different symbols. The difference between the switching obtained with TDG and that obtained in the CTRL sample is not significant. (B) Analysis of FLAG-TDG expression in retrovirally infected B cells as monitored by Western blot using an anti-FLAG mAb; the band marked by an asterisk is probably sumoylated TDG (37). A Western blot for the vector-encoded GFP provides a control for infection efficiency. (C) Flow cytometric plots showing switching to IgG1 in purified B cells from an ung−/− msh2−/− mouse that has been infected with the pMX retroviral vector encoding GFP alone or GFP together with either UNG or FLAG-tagged TDG. Switching was assayed as in Fig. 1, with the proportion of retrovirally infected (GFP+) cells that have switched to IgG1 indicated in the top right quadrants. (D) Demonstration of TDG-encoded uracil-excision activity in extracts of retrovirus producing cells that have been transfected with the pMX retroviral vector encoding FLAG-tagged TDG. (left) Uracil-excision activity was monitored on a U-containing single-stranded oligonucleotide (which is not a substrate for TDG), whereas a double-stranded oligonucleotide containing a U:G mismatch is used on the right. Because TDG, UNG, and SMUG1 will all act as uracil-DNA glycosylases (UDG) on such a dsDNA oligonucleotide substrate, extracts were preincubated with Ugi (to inhibit UNG) and the PSM1 mAb (to inhibit SMUG1) when wishing to restrict the assay to TDG.
DISCUSSION
The results presented here confirm previous findings that active site mutants of UNG are potent in stimulating class switch recombination in the retroviral infection assay (25). However, our data reveal that those mutants that facilitate switching retain uracil-excision activity that is readily detectable in extracts of transduced B cell, whereas those mutants that show no uracil-excision activity have lost the ability to potentiate switching. Thus, in distinction to Begum et al. (25), our results do not call into doubt the proposal that UNG acts in class switching through uracil excision, but actually provide further support for it, showing a correlation between UNG excision capability and immunoglobulin class switching. This analysis is buttressed by the finding that SMUG1, which is a quite distinct uracil-excision enzyme, but not an enzymatically inactive SMUG[G87Y] mutant, can substitute for UNG in the retroviral switching assay.
It is nevertheless striking that mutants of UNG that retain <1% of the specific activity of the parental enzymes are still as good (and, in some cases, better) than the parental enzymes in potentiating switching in the retroviral infection assay. This probably reflects, in large part, the overexpression of UNG that is achieved by retroviral infection, generating a substantial excess of UNG activity in the infected cells over that which is necessary to achieve switching. Indeed, humans bearing UNG[F255S] are deficient in secondary antibody diversification (17), likely because the mutant protein is mistargeted to the mitochondrion (29). However, the same mutant UNG functions efficiently in the retroviral switching assay (25), with others having previously suggested that one explanation for this discrepancy could be the overexpression associated with the retroviral assay (5, 29, 41).
It is, however, notable that the level of uracil-excision activity in the same cultured B cells that have been infected with mutant UNG retroviruses is not sufficient to achieve a restoration of transversion mutations at C:G pairs in the preS region. Thus, we do not believe that overexpression entirely explains the efficacy of low-activity UNG mutants in these retroviral switching assays. Rather, we suspect that uracil excision at only a small proportion of AID-generated S region-associated dU:dG pairs may suffice for switch recombination. Thus, analysis of S region mutation spectra in B cells from ung−/− msh2−/− mice reveals that several hundred dU:dG lesions are generated in the vicinity of the S regions during 7 d of culture with LPS (42). It may be that uracil excision at only a small proportion of these dU:dG lesions suffices for the initiation of switch recombination. This would certainly be consistent with the data of Zarrin et al. (31), who showed that the introduction of a single, staggered double-strand break in donor plus accepter S regions sufficed for the induction of switch recombination. In contrast, the introduction of transversion mutations at C:G pairs during IgV/Sμ hypermutation will presumably correlate directly with the proportion of dU residues from which the uracil is excised.
It is interesting that the switching triggered by the low-activity UNG mutants is more dependent on MSH2 than that by wild-type UNG. There are several possible interpretations of this finding. A variety of studies have suggested that, quite apart from providing a backup mechanism for recognizing the dU:dG lesion when UNG is absent, MSH2 likely plays an important role in resolving the dU:dG lesions into switch recombination in normal mice (43–46). Work from the Stavnezer and Selsing laboratories has revealed that when Sμ is deleted so as to greatly reduce the number of target sites for AID-mediated deoxycytidine deamination, the switching becomes much more dependent on MSH2 (44, 45, 47). This has led to the proposition that MSH2 acts to convert AID-instigated single-strand DNA nicks into the double-strand breaks necessary for switch recombination (29, 30). In terms of this model, one could well envision that the low-activity UNG mutants would only act to generate relatively few single-strand nicks in the S regions; resolution to switch recombination might therefore be highly dependent on MSH2. In contrast, with wild-type UNG, uracil excision is likely to occur at a much higher proportion of sites, possibly leading directly to the creation of staggered double-strand breaks, and thus resulting in the diminished dependence on MSH2.
Although the results reveal that the retroviral delivery of even very low-activity UNG mutants (as well as of wild-type SMUG1) is sufficient to yield efficient switching, the same effect is not achieved through overexpression of TDG, despite the sensitivity of the retroviral assay. Although TDG acts on deoxythymidine:dG mispairs, biochemical characterization of TDG from various species reveals that it is a very potent uracil-excision enzyme when monitored on dU:dG, in fact, showing a preference for dU:dG over deoxythymidine:dG (48, 49). The issue therefore arises as to why TDG cannot substitute effectively for UNG or SMUG1 in switch recombination; infection with the retroviral TDG construct certainly results in plenty of uracil-excision activity, as monitored biochemically on a dU:dG-containing oligonucleotide substrate (Fig. 6 D). One possible explanation is that UNG might harbor a motif that enables it to be recruited to the dU:dG lesion in the immunoglobulin locus (24), and that such a motif is shared by SMUG1, but not TDG. A distinct possibility is that the critical substrate for uracil excision to achieve switch recombination is not a dU:dG mispair, but rather dU in the context of single-stranded DNA. Thus, like the AID-mediated deamination event itself, the uracil excision may need to occur on a single-stranded DNA substrate to trigger switching. Certainly, both UNG and SMUG1 (but not TDG) are active on dU in the context of single-stranded DNA. Further analysis of mutant uracil-excision enzymes should shed light on this issue.
In conclusion, the results presented in this study provide direct evidence that class switch recombination requires uracil excision, with the role of UNG in antibody gene diversification hinging on the protein's catalytic activity rather than just the mere presence of the enzyme. The demonstration that some, but not all, uracil-excision enzymes (UNG and SMUG1, but not TDG) can function in antibody diversification may provide insight into the nature of the uracilated DNA substrate (single- versus doubled-stranded), as well as the circumstances necessary for its access (e.g., during replication, as previously suggested [9]).
MATERIALS AND METHODS
Expression vectors for UNGs.
The full-length cDNA for the nuclear isoform of mouse UNG (UNG2) was obtained from the Integrated Molecular Analysis of Genomes and their Expression (I.M.A.G.E.) consortium (I.M.A.G.E. clone 4009947) through Geneservice, Ltd. The coding region was PCR-amplified using oligonucleotides CATGATCGGCCAGAAGACCCT and GTCACAGCTCCTTCCAGTTGA and subcloned into pCR blunt II TOPO (Invitrogen). Point mutations were introduced into this construct by the QuickChange strategy (Stratagene) using the following oligonucleotides for each indicated mutation: D147N (GTTGTCATTCTGGGACAGAATCCCTATCACGGACCTAATC and GATTAGGTCCGTGATAGGGATTCTGTCCCAGAATGACAAC); N206V (GGTGTCCTCCTCCTCGTCGCCGTCCTCACTG and CAGTGAGGACGGCGACGAGGAGGAGGACACC); D147G (GTTGTCATTCTGGGACAGGGTCCCTATCACGGACCTAATC and GATTAGGTCCGTGATAGGGACCCTGTCCCAGAATGACAAC); H270L (CCATGTTCTGCAGACAGCTCTCCCCTCCCCGCTGTCGGTG and CACCGACAGCGGGGAGGGGAGAGCTGTCTGCAGAACATGG); and L274A (GACAGCTCACCCCTCCCCGGCGTCGGTGCACAGAGGGTTCC and GGAACCCTCTGTGCACCGACGCCGGGGAGGGGTGAGCTGTC). The UNG variants were excised from this vector using flanking EcoRI sites and subcloned into the retroviral vector pMXs-ires-GFP (gift from T. Kitamura, University of Tokyo, Tokyo, Japan; abbreviated here as pMX) (50). The correct orientation of the insert was determined by digestion with ApaLI. For expression in DT40 cells, mouse UNG, D147N, and (D147N,H270L) variants were subcloned into the eukaryotic expression vector pIRESHyg3 (Clontech Laboratories, Inc.) by introducing an NheI site at the N terminus and using the EcoRV site from the pCR-blunt II TOPO vector. A FLAG tag was then brought in at the N terminus by introducing the annealed oligonucleotides GTACAATGGACTACAAGGACGATGATGACAAGG and CTAGCCTTGTCATCATCGTCCTTGTAGTCCATT into the BsrGI and NheI sites of pIRESHyg3. For bacterial expression, the indicated mouse UNG mutants were subcloned as NheI–EcoRI fragments into pTrcHis (Invitrogen), resulting in the 6xHis-tagged versions of the enzymes.
Cloning of full-length human SMUG1 into pEGFP-N3 (Clontech Laboratories, Inc.) has been previously described (9). A derivative containing the G87Y SMUG1 mutation that had been created by oligonucleotide mutagenesis was generously provided by Z. Yang (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK). The EGFP coding region in these vectors was then replaced with a FLAG tag by introducing the annealed oligonucleotides GATCCGACTACAAGGACGATGACGACAAGTGAGC and GGCCGCTCACTTGTCGTCATCGTCCTTGTAGTCG into the BamHI–NotI-digested SMUG and G87Y vectors. The C-terminally tagged SMUG and G87Y were then subcloned EcoRI–NotI into pMXs-ires-GFP.
I.M.A.G.E. clone 6416434 containing the full-length mouse TDG cDNA was obtained from Geneservice, Ltd. The coding sequence was PCR-amplified using oligonucleotides AAGAATTCATGGACGCAGAGGCCGCGCGCA and ACACGCGGCCGCTCCTAAGCGTGGCTCTCTTCTT containing EcoRI and NotI sites, respectively, and subcloned into the corresponding sites of pCDNA3.1(+). An N-terminal FLAG tag was introduced using the annealed oligonucleotides GATCCATGGACTACAAGGACGATGATGACAAGG and AATTCCTTGTCATCATCGTCCTTGTAGTCCATG into BamHI and EcoRI sites. The BamHI–NotI insert was then subcloned into pMXs-ires-GFP.
Mouse B cell cultures and retroviral infection.
Lymphocytes from C57B/6, ung−/−, and ung−/− msh2−/− mice were purified from homogenized spleens by centrifugation over a density cushion of Lympholyte-M (Cedarlane Laboratories, Ltd.). Naive B cells were then enriched by depletion of CD43+ cells using anti-CD43–coupled magnetic microbeads and LD columns in a MACS separator (Miltenyi Biotech) according to the manufacturer's instructions. B cells (106) were seeded in 24-well plates in 1 ml RPMI 10% FBS, 0.05 mM 2-mercaptethanol in the presence of 50 μg/ml LPS (Sigma-Aldrich) and 50 ng/ml recombinant mouse IL4 (R&D Systems). Ecotropic retroviruses were produced by transfecting 2 μg of the pMXs-ires-GFP constructs into Plat-E packaging cells (51). Transfections were performed in 6-well plates using Fugene (Roche), and the medium was replaced by fresh medium after 24 h. Transfection efficiency was similar for all the constructs (80–90%), as judged by the proportion of GFP+ cells. B cells were infected 48 h after plating by the addition of 1 ml of culture supernatant from the packaging cells that had been supplemented to 20 mM in Hepes, pH 7.5, and 16 μg/ml Polybrene (Sigma-Aldrich), followed by centrifugation for 1.5 h at 650 g at 30°C in a tabletop centrifuge. The plates were incubated at 37°C for 4 h before changing the medium. Infection efficiency was typically ∼25%, as judged by GFP fluorescence in flow cytometry, but ranged from 5 to 80% depending on the experiment and the construct used. Isotype switching was analyzed 3 d after infection (day 6 after stimulation) by flow cytometry after staining with anti-IgG1–biotin, followed by PE-conjugated streptavidin (BD Biosciences) and propidium iodide (to exclude dead cells). Statistical analysis of the comparative efficiency of switching achieved in multiple experiments with different uracil-excision enzymes was performed using the two-tailed Student's t test.
Analysis of IgV gene conversion in ung−/− DT40 transfectants.
The cDNAs encoding mouse UNG, D147N, and D147N,H270L were cloned into pIRESHyg3 vectors as described above and transfected into ung−/− DT40 cells (8) with selection for hygromycin-resistance. For scoring gene conversion, the surface IgM-loss populations from independent subclones of UNG and D147N transfectants were sorted by flow cytometry after 4 wk of clonal expansion, and the Vλ region was PCR amplified as previously described (23), cloned in pBluescript, and sequenced. Gene conversion and mutation events were scored as described using the Staden sequence analysis package and Muthr gene conversion software (23, 52).
Uracil-excision assays.
Total cell extracts of vertebrate cells were prepared by sonication and samples (2–20 μg of protein) assayed for their ability to excise uracil from a FITC-labeled oligonucleotide substrate essentially as previously described (23), except that 0.2 U of apyrimidinic endonuclease (New England Biolabs) was included in each reaction. When measuring SMUG1 activity, the endogenous UNG enzyme (if present) was inhibited using 0.5 U of Ugi (New England Biolabs, Inc.) where indicated. For measuring overexpressed TDG activity, endogenous SMUG1 activity was additionally inhibited using the neutralizing antibody PSM1 (gift from G. Slupphaug, Norwegian University of Science and Technology, Trondheim, Norway; reference 53). To monitor uracil excision from single-stranded DNA, the PAGE-purified oligonucleotide ATTATTATTATTCCGUGGATTTATTTATTTATTTATTTATTTFITC (Operon) was used as substrate. To monitor excision from a double-stranded DNA substrate, the same FITC-labeled oligonucleotide was used, but after annealing at a 1:2 ratio with AAATAAATAAATAAATAAATAAATCCGCGGAATAATAATAAT. In the case of the single-stranded DNA substrate, 10 mM EDTA was included in the assay mixture to prevent nonspecific degradation of the oligonucleotide by nucleases in the extract.
Similar assays were performed using extracts of E. coli BW319 [ung] transformants expressing various UNG variants. E. coli extracts were prepared from BW130 transformants that had been induced for 5 h with 1 mM-IPTG, and uracil-excision activity was monitored on a single-stranded oligonucleotide substrate, as above.
Immunoblots.
After SDS-PAGE of whole-cell lysates, proteins were transferred to PVDF and Western blotted using HRP-conjugated anti-FLAG mAb (Sigma-Aldrich), HRP-conjugated anti-GFP antiserum (Abcam), rabbit anti-His tag antiserum (Santa Cruz Biotechnology, Inc.), anti-PCNA (Ab29 from Abcam), or rabbit anti-UNG peptide antiserum (Abcam).
Kinetic analysis of mouse UNGs.
The mouse UNG and D147N were overexpressed from pTrcHIS in the Rosetta strain of E. coli (Novagen) by inducing with IPTG for 5 h and purified by affinity chromatography as previously described for human UNG (34). Steady-state kinetic parameters k cat and K M for a single-stranded substrate were determined by measuring fluorescence spectra using oligonucleotide substrates containing an internal 2-aminopurine analogue adjacent to the target uracil, as previously described (34). All assays were performed in reaction buffer (50 mM Tris, pH 7.5, 50 mM NaCl, and 0.1 mg/ml BSA) at 25°C after determining optimal conditions for mouse UNG.
Acknowledgments
We are indebted to Dr. Toshio Kitamura for providing the pMX-ires-GFP retroviral vector and Plat-E cells, Dr. Geir Slupphaug for the PSM1 antibody, and Dr. Zizhen Yang for constructing the SMUG1 G87Y mutant.
J. Din was supported in part by a fellowship from the Karn Fund.
The authors have no conflicting financial interests.
Abbreviations used: AID, activation-induced deaminase; dG, deoxyguanosine; dU, deoxyuridine; SMUG, single-stranded monofunctional uracil DNA glycosylase; TDG, thymine-DNA glycosylase; UNG, uracil-N-glycolase.
J.M. Di Noia's present address is Institut de Recherches Cliniques de Montréal, H2W 1R7 Montreal, QC, Canada.
References
- 1.Arakawa, H., J. Hauschild, and J.M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 295:1301–1306. [DOI] [PubMed] [Google Scholar]
- 2.Harris, R.S., J.E. Sale, S.K. Petersen-Mahrt, and M.S. Neuberger. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12:435–438. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 4.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri, J., U. Basu, A. Zarrin, C. Yan, S. Franco, T. Perlot, B. Vuong, J. Wang, R.T. Phan, A. Datta, et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157–214. [DOI] [PubMed] [Google Scholar]
- 6.Di Noia, J.M., and M.S. Neuberger. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22. [DOI] [PubMed] [Google Scholar]
- 7.Shen, H.M., A. Tanaka, G. Bozek, D. Nicolae, and U. Storb. 2006. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J. Immunol. 177:5386–5392. [DOI] [PubMed] [Google Scholar]
- 8.Saribasak, H., N.N. Saribasak, F.M. Ipek, J.W. Ellwart, H. Arakawa, and J.M. Buerstedde. 2006. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J. Immunol. 176:365–371. [DOI] [PubMed] [Google Scholar]
- 9.Di Noia, J.M., C. Rada, and M.S. Neuberger. 2006. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 25:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen, H.M., and U. Storb. 2004. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc. Natl. Acad. Sci. USA. 101:12997–13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rada, C., J.M. Di Noia, and M.S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. [DOI] [PubMed] [Google Scholar]
- 12.Di Noia, J.M., and M.S. Neuberger. 2004. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur. J. Immunol. 34:504–508. [DOI] [PubMed] [Google Scholar]
- 13.Beale, R.C., S.K. Petersen-Mahrt, I.N. Watt, R.S. Harris, C. Rada, and M.S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585–596. [DOI] [PubMed] [Google Scholar]
- 14.Sohail, A., J. Klapacz, M. Samaranayake, A. Ullah, and A.S. Bhagwat. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 16.Pham, P., R. Bransteitter, J. Petruska, and M.F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107. [DOI] [PubMed] [Google Scholar]
- 17.Imai, K., G. Slupphaug, W.I. Lee, P. Revy, S. Nonoyama, N. Catalan, L. Yel, M. Forveille, B. Kavli, H.E. Krokan, et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4:1023–1028. [DOI] [PubMed] [Google Scholar]
- 18.Dickerson, S.K., E. Market, E. Besmer, and F.N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. [DOI] [PubMed] [Google Scholar]
- 20.Bransteitter, R., P. Pham, M.D. Scharff, and M.F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 100:4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 22.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103. [DOI] [PubMed] [Google Scholar]
- 23.Di Noia, J., and M.S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. [DOI] [PubMed] [Google Scholar]
- 24.Begum, N.A., N. Izumi, M. Nishikori, H. Nagaoka, R. Shinkura, and T. Honjo. 2007. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J. Biol. Chem. 282:731–742. [DOI] [PubMed] [Google Scholar]
- 25.Begum, N.A., K. Kinoshita, N. Kakazu, M. Muramatsu, H. Nagaoka, R. Shinkura, D. Biniszkiewicz, L.A. Boyer, R. Jaenisch, and T. Honjo. 2004. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 305:1160–1163. [DOI] [PubMed] [Google Scholar]
- 26.Begum, N.A., K. Kinoshita, N. Kakazu, M. Muramatsu, H. Nagaoka, R. Shinkura, D. Biniszkiewicz, L.A. Boyer, R. Jaenisch, and T. Honjo. 2004. Comment on “Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch” author's reply. Science. 306:2042. [DOI] [PubMed] [Google Scholar]
- 27.Longerich, S., and U. Storb. 2005. The contested role of uracil DNA glycosylase in immunoglobulin gene diversification. Trends Genet. 21:253–256. [DOI] [PubMed] [Google Scholar]
- 28.Stivers, J.T. 2004. Comment on “Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch.” Science. 306:2042. [DOI] [PubMed] [Google Scholar]
- 29.Kavli, B., S. Andersen, M. Otterlei, N.B. Liabakk, K. Imai, A. Fischer, A. Durandy, H.E. Krokan, and G. Slupphaug. 2005. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J. Exp. Med. 201:2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stavnezer, J., and C.E. Schrader. 2006. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 22:23–28. [DOI] [PubMed] [Google Scholar]
- 31.Zarrin, A.A., C. Del Vecchio, E. Tseng, M. Gleason, P. Zarin, M. Tian, and F.W. Alt. 2007. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 315:377–381. [DOI] [PubMed] [Google Scholar]
- 32.Elder, R.T., X. Zhu, S. Priet, M. Chen, M. Yu, J.M. Navarro, J. Sire, and Y. Zhao. 2003. A fission yeast homologue of the human uracil-DNA-glycosylase and their roles in causing DNA damage after overexpression. Biochem. Biophys. Res. Commun. 306:693–700. [DOI] [PubMed] [Google Scholar]
- 33.Handa, P., S. Roy, and U. Varshney. 2001. The role of leucine 191 of Escherichia coli uracil DNA glycosylase in the formation of a highly stable complex with the substrate mimic, ugi, and in uracil excision from the synthetic substrates. J. Biol. Chem. 276:17324–17331. [DOI] [PubMed] [Google Scholar]
- 34.Krusong, K., E.P. Carpenter, S.R. Bellamy, R. Savva, and G.S. Baldwin. 2006. A comparative study of uracil-DNA glycosylases from human and herpes simplex virus type 1. J. Biol. Chem. 281:4983–4992. [DOI] [PubMed] [Google Scholar]
- 35.Mol, C.D., A.S. Arvai, G. Slupphaug, B. Kavli, I. Alseth, H.E. Krokan, and J.A. Tainer. 1995. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 80:869–878. [DOI] [PubMed] [Google Scholar]
- 36.Parikh, S.S., C.D. Mol, G. Slupphaug, S. Bharati, H.E. Krokan, and J.A. Tainer. 1998. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 17:5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavli, B., G. Slupphaug, C.D. Mol, A.S. Arvai, S.B. Peterson, J.A. Tainer, and H.E. Krokan. 1996. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 38.Dunnick, W., M. Wilson, and J. Stavnezer. 1989. Mutations, duplication, and deletion of recombined switch regions suggest a role for DNA replication in the immunoglobulin heavy-chain switch. Mol. Cell. Biol. 9:1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen, S., R. Casellas, B. Reina-San-Martin, H.T. Chen, M.J. Difilippantonio, P.C. Wilson, L. Hanitsch, A. Celeste, M. Muramatsu, D.R. Pilch, et al. 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen, H.S., O. Sundheim, K.M. Gilljam, G. Slupphaug, H.E. Krokan, and B. Kavli. 2007. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 35:3879–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durandy, A., N. Taubenheim, S. Peron, and A. Fischer. 2007. Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv. Immunol. 94:275–306. [DOI] [PubMed] [Google Scholar]
- 42.Xue, K., C. Rada, and M.S. Neuberger. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 203:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehrenstein, M.R., and M.S. Neuberger. 1999. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 18:3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min, I.M., L.R. Rothlein, C.E. Schrader, J. Stavnezer, and E. Selsing. 2005. Shifts in targeting of class switch recombination sites in mice that lack μ switch region tandem repeats or Msh2. J. Exp. Med. 201:1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Min, I.M., C.E. Schrader, J. Vardo, T.M. Luby, N. D'Avirro, J. Stavnezer, and E. Selsing. 2003. The Smu tandem repeat region is critical for Ig isotype switching in the absence of Msh2. Immunity. 19:515–524. [DOI] [PubMed] [Google Scholar]
- 46.Schrader, C.E., J. Vardo, and J. Stavnezer. 2002. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med. 195:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luby, T.M., C.E. Schrader, J. Stavnezer, and E. Selsing. 2001. The μ switch region tandem repeats are important, but not required, for antibody class switch recombination. J. Exp. Med. 193:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardeland, U., M. Bentele, J. Jiricny, and P. Schar. 2003. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 31:2261–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardeland, U., R. Steinacher, J. Jiricny, and P. Schar. 2002. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21:1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitamura, T., Y. Koshino, F. Shibata, T. Oki, H. Nakajima, T. Nosaka, and H. Kumagai. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014. [PubMed] [Google Scholar]
- 51.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. [DOI] [PubMed] [Google Scholar]
- 52.Sale, J.E., D.M. Calandrini, M. Takata, S. Takeda, and M.S. Neuberger. 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 412:921–926. [DOI] [PubMed] [Google Scholar]
- 53.Kavli, B., O. Sundheim, M. Akbari, M. Otterlei, H. Nilsen, F. Skorpen, P.A. Aas, L. Hagen, H.E. Krokan, and G. Slupphaug. 2002. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 277:39926–39936. [DOI] [PubMed] [Google Scholar]