Abstract
Receptor editing is believed to play the major role in purging newly formed B cell compartments of autoreactivity by the induction of secondary V(D)J rearrangements. In the process of immunoglobulin heavy (H) chain editing, these secondary rearrangements are mediated by direct VH-to-JH joining or cryptic recombination signals (cRSs) within VH gene segments. Using a statistical model of RS, we have identified potential cRSs within VH gene segments at conserved sites flanking complementarity-determining regions 1 and 2. These cRSs are active in extrachromosomal recombination assays and cleaved during normal B cell development. Cleavage of multiple VH cRSs was observed in the bone marrow of C57BL/6 and RAG2:GFP and μMT congenic animals, and we determined that cRS cleavage efficiencies are 30–50-fold lower than a physiological RS. cRS signal ends are abundant in pro–B cells, including those recovered from μMT mice, but undetectable in pre– or immature B cells. Thus, VH cRS cleavage regularly occurs before the generation of functional preBCR and BCR. Conservation of cRSs distal from the 3′ end of VH gene segments suggests a function for these cryptic signals other than VH gene replacement.
Developmentally immature B cells expressing autoreactive antigen receptors are tolerized by three mechanisms: anergy, clonal deletion, and receptor editing. Whereas anergy and deletion inactivate or remove self-reactive clones, receptor editing alters clonal specificity through secondary rearrangements of the Igκ and -λ loci, or VH gene replacement (1). VH gene replacement represents an atypical V(D)J recombination event mediated by a physiological recombination signal (RS) adjacent to an upstream germline VH gene segment and a cryptic RS (cRS) located near the 3′ end of a rearranged VH gene segment (2–4). In the Igh locus, the D gene segments located between the VH and JH gene clusters are doubly flanked by RSs containing 12-bp spacers (12-RS); these mediate recombination with the 23-RS of JH and VH gene segments (5). VH→DJH rearrangements that complete IgH assembly in pro–B cells deplete the Igh locus of 12-RS (6) and preclude subsequent rearrangements that follow the 12/23 rule (5).
VH replacement alters the specificity of the B cell antigen receptor (BCR) and can rescue developing B cells that would otherwise be eliminated by apoptosis. Such replacements were first noted in mice with autoreactive, site-directed transgene (SDT) receptors (3, 7), but replacement of innocent (8, 9) or nonproductive (10) VDJ SDT has been observed as well. Presumably, VH replacement in the absence of self-reactivity is the consequence of strong selection for a diverse B cell repertoire. Under an antigen-dependent model of receptor editing, binding of an autoantigen to an antigen receptor is required, but pressure to diversify the B cell repertoire via VH gene replacement is presumably antigen independent (3, 11, 12).
It is difficult to predict whether mouse VH replacements are antigen dependent or independent because the stage of normal B cell development at which VH replacements are initiated in vivo is unknown. Recently, signal ends (SEs) at VH cRSs were noted in human immature B cells, but the cloned human VH replacements included N-nucleotide additions, which are characteristic of IgH rearrangement in pro–B cells (11, 13). N-nucleotides are also noted (3) in mouse VH replacements, providing further evidence that VH replacements may be induced at the pro–B cell stage.
In this study, we use a rigorous statistical method to demonstrate conserved cRSs in mouse VH gene segments and find that these cRSs exhibit an orientation and spacer length that facilitates VH→VH rearrangements. We demonstrate RAG1-dependent cleavage of mouse VH cRSs at multiple locations, including conserved sites in FW1 and -2 during normal B cell development. We speculate that these anterior cRSs may create hybrid VH gene segments (14, 15). Although VH cRS SEs have been detected in the BM and spleen of genetically modified mice (16), we show that VH cRS SEs are routinely generated by normal mouse pro–B cells, but are undetectable in pre and immature B cells. This observation is in contrast to that reported for human B cell development (11), and suggests a model of B cell development characterized by stochastic rearrangements of RSs and cRSs, followed by selection for functional heavy chain. This random rearrangement hypothesis implies that VH cRSs are conserved to increase VH genetic diversity (2), rather than for receptor editing in response to self-antigens.
RESULTS
Identification of potential cRSs in VH gene segments
We used a probabilistic model of mouse RSs (17–19) to scan 390 mouse VH gene segments for cRSs by computing the RS information content (RIC) (19) for all possible 28- (RIC12) and 39-bp signals (RIC23). The RIC12 and RIC23 algorithms are capable of identifying and evaluating physiological RSs and cRSs directly from DNA sequence (17–19). A 212-kbp region of chromosome 8 (AC084823) that is not subject to physiological V(D)J recombination was similarly analyzed. RIC scores approaching zero indicate increasing similarity to physiological mouse RSs and higher recombination potentials (17). All 28- and 39-bp DNA sequences beginning with a CA dinucleotide have a finite RIC score and are potential cRSs.
We previously determined that RIC12 ≥ −40 constituted a threshold value for physiological RS activity, and therefore we expect RIC values for 12-cRSs to be lower (18). Indeed, a known mouse VH cRS (3) has a RIC12 of −45.3 (18). Thus, we set a preliminary detection threshold for 12-cRSs as RIC12 > −45. Physiological 23-RSs are identified by RIC23 > −60 (18); we selected a correspondingly lower threshold of RIC23 > −65 for identification of 23-cRSs.
Our scan identified potential cRSs in both DNA strands. cRSs in VH gene segments that share orientation with the physiological VH RSs are defined to be in orientation 1 (O1). VH cRSs in the opposite orientation are O2 cRSs. In our analyses of AC084823, we arbitrarily defined putative cRSs in the listed sequence as O1 cRSs and those in the complementary strand as O2 cRSs. The analyzed VH gene segments contained 8,647 potential 12-cRSs and 8,312 potential 23-cRSs in the O1 orientation (Table S1, available at http://www.jem.org/cgi/content/full/jem.20071244/DC1). In O2, these VH gene segments contained 8,976 and 8,109 potential 12- and 23-cRSs, respectively. Of these, only 223 (O1) and 299 (O2) had RIC12 > −45, and a smaller subset, 135 (O1) and 302 (O2), had RIC23 > −65 (Table S1). In the larger AC084823 sequence, 15,401 (O1) and 17,480 (O2) potential 12-cRSs and 15,401 (O1) and 17,478 (O2) potential 23-cRSs were identified (Table S1). Among these, 259 (O1) and 321 (O2) 12-cRSs had RIC12 > −45, and 701 (O1) and 837 (O2) 23-cRSs had RIC23 > −65 (Table S1). Potential 12-cRSs with RIC12 > −45 were significantly less frequent in the AC084823 sequence than in VH gene segments (0.017 vs. 0.026 [O1]; P = 10−6 and 0.018 vs. 0.033 [O2]; P = 10−14; Table S1). In contrast, the relative frequencies of potential 23-cRSs with RIC23 > −65 in the AC084823 sequence (0.05 [O1 and O2]) were significantly higher than in VH gene segments (0.02 [O1] and 0.04 [O2]; P = 10−31 and P = 10−4, respectively; Table S1).
Whereas VH gene segments and the control AC084823 sequence have similar relative frequencies of potential cRSs with low RIC scores, as RIC scores increase toward threshold values, these frequencies diverge. Potential 12-cRSs with RIC12 > −50 are more common in VH gene segments, and potential 23-cRSs with RIC23 > −70 are present at higher frequencies in the AC084823 control (Fig. 1, A and B). Given that physiological V(D)J recombination does not occur within the AC084823 region of chromosome 8, we interpret these divergent frequencies to indicate evolutionary enrichment for 12-cRSs in VH gene segments, accompanied by the selective removal of potential 23-cRSs.
Figure 1.
The proportion of RS-length sequences with RIC scores above a given threshold. The number of RS-length sequences with RIC greater than the score indicated on the x axis was divided by the number of all RS-length segments begining with CA. The resulting proportion is indicated on the y axis. Results for 28-bp segments (top) and for 39-bp segments are shown (bottom). Filled circles (•) indicate the proportion for segments in the orientation of physiological RS (O1), and open triangles (▵) indicate the proportion in the opposite orientation (O2). Orientation was assigned arbitrarily for the chromosome 8 sequence. Solid lines indicate proportions computed on VH gene segments; dashed lines indicate segments from chromosome 8.
VH 12-cRSs are conserved in O2
To determine if VH 12-cRSs with RIC12 > −45 are conserved in a preferred orientation, we compared the frequencies of O1 and O2 putative 12-cRSs in VH gene segments and in AC084823. Whereas the relative frequencies of O1 and O2 12-cRSs in the AC084823 sequence are virtually identical (0.017 and 0.018, respectively; P = 0.288), the relative frequency of O2 12-cRSs is significantly higher (0.033) than O1 12-cRSs (0.026; P = 0.003) in VH gene segments (Table S1 and Fig. 1 A). VH 12-cRSs with lower RIC12 scores (≤−55) have similar relative frequencies in O1 and O2. As RIC12 scores increase (≥−55), however, O2 12-cRSs become more common than those in O1 orientation (P = 0.052). This bias for VH 12-cRSs in the O2 orientation suggests selection for VH gene segments containing 12-cRSs oriented opposite upstream physiological VH 23-RSs.
Conservation of multiple cRSs in diverse VH gene segments
Of the 299 VH 12-cRSs with RIC12 > −45 and O2 orientation, ∼80% were located at nucleotide 57 (51/299) or at nucleotide 313 (189/299; Fig. 2). We identify these most highly conserved cRSs as sites I (nt 54–63) and V (nt 310–313), respectively (Fig. 2). Locations of the remaining 59 potential 12-cRSs were less well conserved, but the majority (71%; 42/59) cluster into three regions (sites II [nt 100–126], III [nt 148–184], and IV [nt 190–205]) that mark the borders of complementarity-determining region (CDR) 1 and 2 (Fig. 2).
Figure 2.
O2 cRSs are found at multiple locations within mouse VH gene segments. RIC12 was computed for all 28-bp segments embedded in mouse VH gene segments. RIC12 scores of potential cRSs (RIC 12 > −55) are plotted against the segment's nucleotide position (IMGT numbering). Open circles (○) indicate potential 12-cRSs and filled circles (•) indicate those segments with RIC12> −48.2, the lowest RIC12 for which we have detected extrachromosomal recombination (18). Locations of CDR1, -2, and -3 are shown by the shaded areas of the graphs. Roman numerals indicate clusters of cRSs that are conserved across VH gene families. Site I spans amino acid residues 18–22 (nt 54–63); site II spans amino acid residues 34–42 (nt 100–126); site III spans residues 50–62 (nt 148–184); site IV spans residues 64–69 (nt 190–205); and site V is amino acid residue 105 (nt 313).
Previously, we identified a 12-cRS with RIC12 = −48.2 that was active in extrachromosomal rearrangement (18). The numbers of O2 12-cRSs in VH gene segments with RIC12 ≥ −48.2 are double that for RIC12 = −45 (631 vs. 299). Nonetheless, 95% (599/631) remain clustered within sites I−V (Fig. 2). That the distributions of both stringent and relaxed 12-cRSs are highly similar suggests a common mechanism for their conservation.
Our search revealed that potential 12-cRSs were broadly distributed among VH gene families; at least one VH segment from 12 of the 15 VH gene families contained 1 or more 12-cRSs. 12-cRSs were, however, most abundant in the VH1 and VH5 gene families, with 151 and 56 cRSs, respectively. The presence and conserved locations of multiple 12-cRSs in many VH gene families suggests that natural selection maintains VH 12-cRSs, even in locations that cannot support VH→VDJ replacements of the type mediated by site V cRSs (Fig. 2).
Selection for VH cRSs is independent of amino acid conservation
The presence in VH gene segments of near consensus heptamers without obvious nonamers led Wu et al. to speculate that embedded heptamers at sites I and V (Fig. 2) might reflect the conservation of cysteine (Cys) residues critical for BCR structure rather than selection for recombinogenic potential (2).
Cysteine is encoded by TGT or TGC codons. Heptamer motifs within the highly conserved site V 12-cRS result from the combination of codons 104 and 105 (IMGT numbering; Fig. 2). In the great majority (86%) of mouse VH gene segments, cysteine (TGT) at residue 104 is followed by Ala (GCN; 225/283), Val (GTN; 15/283), Glu (GAA/G; 1/283), or Gly (GGN; 1/283; IMGT database). These conserved associations generate the GTG (or CAC) motif (...TGTGNN...) required for cRS heptamers (18).
Substitution of TGC for TGT at position 104 would maintain the cysteine residue, but abrogate any recombination activity at site V cRSs by destroying the heptamer motif (...TGCGNN...). To determine if the TGT codon necessary for site V 12-cRS is conserved independently of the Cys104 residue, we compared codon usage at this and another highly conserved cysteine residue in VH, Cys23 (Table I). In contrast to Cys104, Cys23 does not overlap any potential 12-cRSs (Fig. 2), although both are necessary for IgV domain structure (20). Whereas 98% (287/292) of Cys104 residues are encoded by TGT, only 38% (113/297) of Cys23 codons are encoded by TGT (Table I). The highly significant (P < 10−54) preference for TGT codons at Cys104, but not Cys23, suggests selection for site V recombinogenic sequence that is independent of the amino acids necessary for BCR structure.
Table I.
Biased codon usage associated with VH 12-cRS at sites I and V
| VH cysteine codon usage | |||||
|---|---|---|---|---|---|
| TGT | TGC | Totals | |||
| Cys 23 | 129 (34%) | 253 | 382 | ||
| Cys 104 | 287 (98%) | 5 | 292 | ||
| VH valine codon usage | |||||
| GTG | GTC | GTA | GTT | Totals | |
| Val 2 | 131 (40%) | 148 | 10 | 37 | 326 |
| Val 13 | 292 (89%) | 3 | 26 | 7 | 328 |
| Val 19 | 183 (84%) | 24 | 9 | 2 | 218 |
| Val 42 | 173 (54%) | 42 | 28 | 75 | 318 |
| Val 80 | 0 (0%) | 0 | 139 | 2 | 141 |
VH cRSs are frequently conserved within degenerate codons; site V cRSs are associated with a conserved Cys104 residue (TGY codon); and a common site I cRS requires Val19 (GTN). The frequency of T and C nucleotides in the third position of cysteine codons (TGY) at VH gene segment codons 23 and 104 (top), and the frequency of nucleotides in valine codons (GTN) at VH gene segment codons 2, 13, 19, 42, and 80 (bottom) were compared. Numbers of relevant cysteine (TGY) or valine (GTN) codons in the mouse VH gene segments (n = 390) analyzed are shown, and frequencies of TGT (Cys) and GTG (Val) codons are in parentheses. Totals represent the numbers of VH gene segments containing a conserved residue.
Similarly, the conserved site I 12-cRSs at nt 57 (Fig. 2) are associated with a Val residue (amino acid residue 19) present in 56% (218/390) of VH gene segments screened in our analyses. Of these 218 Val residues, 183 (84%) are encoded by a GTG codon that initiates the cryptic heptamer; of the other Val19 residues, 24 (11%) are encoded by GTC and 11 (5%) by GTA or GTT (Table I). Other Val residues are conserved in mouse VH gene segments, e.g., Val2 (nt 6; 326/390), Val13 (nt 39; 328/390), Val42 (nt 126; 318/390), and Val80 (nt 240; 141/390). Although a majority (54% [596/1,113]; range 0–89%) of these conserved Val residues are encoded by GTG, codon bias at Val19 is significantly (P = 10−16) higher. Importantly, none of the other conserved Val residues, including those encoded by GTG, is associated with O2 12-cRS (Fig. 2). We conclude that conservation of the structural motifs implicit in conserved Val residues is insufficient to specify cRSs.
VH 12-cRSs function in extrachromosomal recombination assays
Replacement rearrangements at cRSs upstream of site V 12-cRSs would result in significantly longer VH domains and might result in suboptimal or nonfunctional IgH polypeptides. If this were the case, we would expect evolutionary selection to suppress the recombinational capacity of the potential cRSs in sites I–IV (Fig. 2). Therefore, we compared the recombinational capacities of 8 12-cRSs at sites I, III, IV, and V in an extrachromosomal recombination assay (see Materials and methods) (18). Additionally, we measured the recombination potential of a single 12-cRS located in FW3 (Table II). As a control, a known (site V) cRS present in the 3H9 H-chain transgene (3) was included in our analysis (Table II). Recombinational activities of these cRSs were normalized to a physiological standard, the Jβ2-2 12-RS (18).
Table II.
Activity of VH 12-cRS in an extrachromosomal recombination assay
| cRS Site
|
Vector
|
Position
|
cRS sequence
|
VH family
|
R [RIC12] |
Efficiency
|
|---|---|---|---|---|---|---|
| % | % Jβ2-2 | |||||
| I | p290-VH/199 | 57 | CACTGAA GCCCCAGGCTTC ACCAGTTCA | 1 | 0.02 ± 0.01 [−42.5] |
2.2% |
| I | p290-VH/241 | 57 | CACTGAA GCCCCAGGCTTC ACAAGCTCA | 1 | <0.005 [−38.8] |
<0.6% |
| III | p290-VH/87 | 181 | CACTATT AGGATCAATCCT TCAAATCCA | 1 | 0.01 ± 0.01 [−44.0] |
1.1% |
| IV | p290-VH/69 | 205 | CACTGTA CTTAATA T CA C T ATAAGGATC | 1 | <0.005 [−42.4] |
<0.6% |
| IV | p290-VH/118 | 198 | CACAGTA TAACCAT T TC C A GGATAAATA | 1 | 0.02 ± 0.02 [−43.8] |
2.2% |
| V | p290-VH/06 | 313 | CACAGTA ATAGACCGCAGA GTCCTCAGA | 1 | 0.01 ± 0.01 [−38.9] |
0.9% |
| V | p290-VH/09 | 313 | CACAATA ATAGACCGCAGA GTCCTCAGA | 1 | <0.005 [−41.2] |
<0.6% |
| FW3 | p290-VH/60 | 259 | CACTGCT TTTTGAATCATC TCTTGAAAT | 13 | 0.02 ± 0.01 [−43.0] |
2.2% |
| VCNTL | p290-3H9 | 313 | CACAGAA GTAGACCGCAGA GTCCTCAGA | 1 | <0.002 [−45.3] |
<0.2% |
The recombination efficiencies of several VH cRSs were calculated by a standard extrachromosomal assay (18). All cRS sequences were embedded in VH1 gene segments, except for p290-VH/60, which comes from the VH13 gene family. The nt position of each cRS is noted. R was calculated as the normalized ratio of amp'cam' to amp' bacterial colonies (see Materials and methods). cRS spacer sequences (italicized) are flanked by cryptic heptamers (left) and nonamers (right). Sequence differences between cRSs from the same sites (I, IV, and V) are in bold. The p290-3H9 substrate was included because this cRS has been observed to be functional in vivo (3).
Of the 8 12-cRSs tested, five supported low, but detectable, levels of recombination (Table II). 12-cRSs from sites I, III, IV, and V performed comparably, with one signal from each cohort mediating recombination at efficiencies of 1–2% of that observed for the Jβ2-2 signal (18). One cRS (p290-VH/60) not located in the conserved sites, but in FW3, also exhibited detectable recombinational activity. In contrast, activity by the 3H9 12-cRS control could not be detected, even though this cRS is active in vivo (3). Failure to detect recombination of the 3H9 cRS indicates that the extrachromosomal recombination assay underestimates cRS activity.
Interestingly, the ability of cRSs to support detectable levels of recombination could be determined by only a few nucleotides, although the site I cRSs (p290-VH/199 and p290-VH/241) differ by only two nonamer nucleotides, only one (p290-VH/199) supported detectable levels of recombination (Table II). Similarly, the two cRSs from site V differed by only a single exchange in the heptamer, but this difference was sufficient to abrogate activity in p290-VH/09 (Table II).
VH cRS SEs are detectable only in pro–B cells
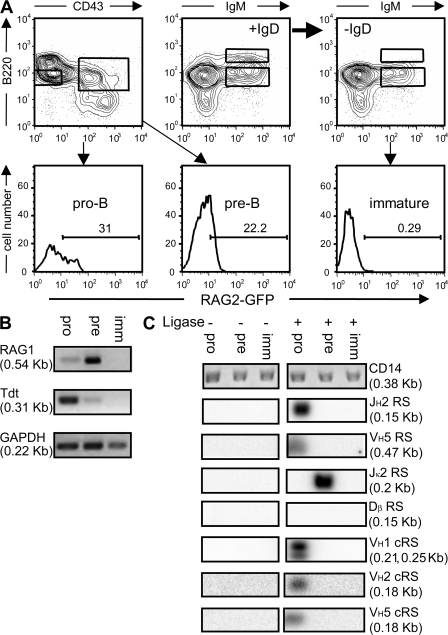
To determine whether the VH 12-cRSs identified by our screen are cleaved during normal B cell development, we used a ligation-mediated PCR (LM-PCR) to amplify RS and cRS SEs (11, 18) in pro–B (B220loIgM−IgD−CD43+GFP+), pre–B (B220loIgM−IgD−CD43−GFP+), and immature B cells (B220+IgM+IgD−CD43−) from RAG2:GFP+/+ mice (Fig. 3 A) (21). RAG2:GFP mice express a RAG2:GFP fusion protein that supports V(D)J recombination and exhibits the kinetics of authentic RAG2 (21). To enrich these developing populations for recombinase activity, we isolated GFP+ pro- and pre–B cells; GFP+ immature B cells were sufficiently rare (<1%) to require the pooling of GFP+ and GFP− immature B cells. In addition, Tdt and RAG1 expression in the sorted cell cohorts were determined by RT-PCR (Fig. 3 B).
Figure 3.
VH cRS cleavage is detected only in pro–B cells from RAG2:GFP knock-in mice (21). (A) Sorted pro-, pre, and immature B cells were analyzed by RT- and LM-PCR. Mature, recirculating B cells (+IgD) were removed by incubation with anti-IgD (-IgD). RAG2:GFP fluorescence was detected in pro- and pre–B cells, but not in immature B cells. (B) RT-PCR for Rag1, Tdt, and GAPDH transcripts revealed RAG1 message in both pro–B and pre–B cells; Tdt expression was detected only in sorted pro–B cells. (C) LM-PCR for primary JH, VH, Jκ, and Dβ rearrangements demonstrate the lineage and developmental restriction of VH cRS SEs, and confirms the purity of the sorted cell populations. VH1, VH2, and VH5 cRS SEs were detected only in pro–B cells. CD14 PCR demonstrated the equivalence of genomic template.
As previously reported, significant GFP fluorescence was detected in both pro- and pre–B cells (21), as was the message for RAG1 and Tdt (Fig. 3) (22, 23). In contrast, immature B cells did not express detectable levels of Tdt or RAG1 (Fig. 3) (21–23). RAG2:GFP fluorescence could be ordered among the sorted B cell populations with GFP+ pro–B cells being brightest and immature B cells being dullest (Fig. 3) (21).
Detection of RS and cRS SE was restricted by lineage- and stage of development. SEs from the physiological RS of VH5 and JH2 were detected only in pro–B cells; Jκ SEs were detected in pre–B cells, but not in pro–B or immature B cells; and Tcr Dβ SEs were not present in any B cell population (Fig. 3 C). LM-PCR products of the size predicted for VH 12-cRS SEs could be amplified from the DNA of pro–B cells and hybridized with 32P-labeled VH-specific probes (Fig. 3). In support of our computational screen for VH cRSs, we detected cRS SEs with primers specific for the VH1, VH2, and VH5 gene families (Fig. 3).
VH cRS SEs require recombinase activity
To demonstrate that the cRS SEs were dependent on RAG1/2 activity, we amplified VH1 cRS SEs from IgH transgenic mice that did or did not express RAG1. Pro–B (B220loCD43+IgM−) and pre–B cells (B220loCD43−IgM−) were isolated from sibling IgH transgenic (H50G) mice (24) that were RAG1 sufficient (H50G+/−Rag1+/−) or deficient (H50G+/−Rag1−/−; Fig. 4 A). Flow cytometric resolution of pro- and pre–B cells in H50G mice was less distinct than in RAG2:GFP animals, presumably because the H50G transgene accelerates preBCR expression and transition to the pre–B cell phenotype (compare Figs. 3 and Figs.4).
Figure 4.
VH cRS cleavage is dependent on Rag1. (A) BM B cells with a pro–B phenotype (B220loCD43+IgM−Lin−) and a pre–B phenotype (B220loCD43−IgM−Lin−) were sorted from sibling H50G+/−Rag1+/− (middle) or H50G+/−Rag1−/− (right) mice. Resolution of pro- and pre–B cells in H50G transgenic mice was not as complete as in RAG2:GFP mice (Fig. 3 A). Mature and immature B cells (+IgM) were excluded when anti-IgM was used as a negative marker (-IgM). (B) LM-PCR demonstrated SEs at both physiological RSs (JH2, Jκ2) and cRSs (J558) were present only in RAG1/2-sufficient cells.
JH2 SEs were readily demonstrated in both pro- and pre–B cells from RAG1-sufficient mice, but could not be amplified from the DNA of RAG-deficient animals (Fig. 4 B). Significantly, even though strong allelic exclusion is observed in H50G mice (24), the H50G IgH transgene does not abrogate RS cleavage in the endogenous loci (Fig. 4 B), a finding similar to that of Chang et al. (25). We presume that the generation of JH2 SEs results from RAG1/2 expression that is incompletely suppressed by the H50G transgene. Similarly, Jκ2 SEs were recovered from H50G+/−Rag1+/− pre–B cells, but not from the analogous phenotypic compartment of H50G+/−Rag1−/− mice (Fig. 4 B).
The presence of JH and Jκ SEs demonstrates that the endogenous Igh and Igκ loci of H50G+/−Rag1+/− mice are accessible to recombinase activity; and, accordingly, we were able to recover VH1 cRS SEs from pro–B and pre–B cells from H50G+/−Rag1+/−, but not H50G+/−Rag1−/−, mice (Fig. 4 B). Thus, LM-PCR amplification of both RS and cRS SE product is equivalently dependent on V(D)J recombinase activity.
VH cRS SEs in μMT pro–B cells
μMT mice cannot generate functional preBCR and are unable to support B cell development beyond the pro–B cell stage (26).To determine whether the preBCR is required for the generation of VH cRS SEs, we subjected genomic DNA from pro–B cells (B220+CD43+IgM−IgD−) from μMT mice and from pro–, pre– (B220+CD43−IgM−IgD−), and immature (B220+CD43−IgM+IgD−) B cells from C57BL/6 controls to LM-PCR for VH cRS SEs (Fig. 5 A).
Figure 5.
VH cRS SEs from C57BL/6 and μMT pro–B cells. (A) BM B cells were recovered from C57BL/6 and μMT mice and pro-, pre, and immature B cells (from C57BL/6), or pro–B cells (from μMT) mice were sorted as in Fig. 3. Genomic DNA was isolated and ligated to the BW linker. (B) LM-PCR on genomic DNA of sorted B cell populations from C57BL/6 or μMT mice was performed to detect VH cRS SEs. cRS SEs were found only in cells with a pro–B phenotype. CD14 PCR was used to normalize template DNA.
VH1 cRS SEs were easily demonstrated in the pro–B cell compartments of both μMT and C57BL/6 mice (Fig. 5 B). In contrast, VH1 cRS SEs were undetectable in equivalent samples of pre–B or immature B cells from C57BL/6 controls (Fig. 5 B). These findings demonstrate that, at least in mice, VH cRS SEs are generated in the absence of normal preBCR signaling and without the possibility of retrograde differentiation by pre– or immature B cells (3, 11).
In vivo, VH cRS SE formation is ∼5% the efficiency of JH2 RS SEs
In our hands, the LM-PCR for VH cRS SEs is capable of detecting SE products in as few as 103 pro–B cells (Fig. 4), even though the efficiencies of selected VH cRSs are low in extrachromosomal recombination assays (Table II). To estimate the recombination efficiencies of VH cRSs in the context of normal B cell development, we compared the relative amounts of VH1 cRS SEs and JH2 RS SEs in pro–B cells (B220loCD43+IgM−IgD−) from C57BL/6 mice (Fig. 6).
Figure 6.
VH1 cRSs are cleaved at 1–7% the efficiency of the JH2 RS. (A) Titration of nested LM-PCR cycle numbers (15–35) to optimize comparison of JH2 RS SE (top) and VH1 cRS SE (bottom) products. LM-PCR of JH2 RS SEs and VH1 cRS SEs were resolved by gel electrophoresis and hybridized with gene-specific probes. (B) Densitometric analysis of the hybridized LM-PCR products showed that 25 cycles constitute the linear phase of amplification. Circles (•) and triangles (▴) indicate the densitometry of JH2 RS SEs and VH1 cRS SEs, respectively. (C) Serial threefold template dilution demonstrates that JH2 RS SEs (top) are less than or equal to threefold more abundant than VH1 cRS SEs (bottom). (D) Relative abundance of JH2 RS SEs and VH cRS SEs determined by real-time quantitative PCR. JH2 RS SEs and VH1 and VH5 cRS SEs were amplified from the BM cells of C57BL/6 mice in a series of quantitative LM-PCR. cRS SE products were normalized to JH2 RS SE product (the mean ± the SD) from the same sample by the comparative threshold cycle method. Subsequently, VH cRS cleavage efficiencies were adjusted to template numbers (JH2 = 1; VH1 = 34; VH5 = 7). The mean efficiency of VH1 cRSs was determined to be 4.9 ± 0.2% of the JH2 RS, whereas VH5 cRSs were 7.6 ± 0.3% as efficient.
Using our standard LM-PCR, JH2 RS SE and VH1 cRS SE products from 103 pro–B cells appeared linear between 20 and 30 amplification cycles (Fig. 6, A and B). Accordingly, samples of VH1 cRS and JH2 RS SE product generated by 25 rounds of amplification were titrated by serial threefold dilutions and blotted for hybridization (Fig. 6 C). This semiquantitative approach indicates that in genomic DNA isolated from pro–B cells, JH2 RS SEs are greater than or equal to threefold more abundant than VH1 cRS SEs. The primer set for JH2 is, however, specific for only one gene segment, whereas the VH1 primer set amplifies ∼34 distinct VH1 gene segments listed in the IMGT database (unpublished data). Therefore, we estimate that the abundance of any single VH1 cRS SE is 1–3% ([1.00 to 0.33 ÷ 34] × 100%) of JH2 RS SE, an estimate that is comparable to VH cRS efficiencies determined in extrachromosomal assays (Table II).
Estimates for the abundance of VH cRS SEs in pro–B cells were also obtained by quantitative LM-PCR amplifications using the JH2-, VH1-, and VH5-specific primer sets (Fig. 6 D). In three independent experiments, the averaged threshold cycle numbers (CT) for JH2 (n = 11), VH1, and VH5 (n = 6 for both) were 26.30 (± 0.59), 25.57 (± 0.98), and 27.20 (± 1.33), respectively. Measured in this way, the abundance of cRS SEs from individual VH1 ([20.73 ÷ 34] × 100% = 4.9%) and VH5 ([2−0.90÷7] × 100% = 7.6%) gene segments was 5–8% of the JH2 RS.
Recombinase cleavage at multiple VH cRSs
To identify specific VH cRS cleavage events and to compare their frequencies, we cloned and sequenced VH cRS SE PCR products recovered from pro–B cells of μMT, RAG2:GFP, and C57BL/6 mice. 33 unique and independent VH cRS SE fragments representing 28 VH1 and a single VH5 gene segment were obtained; 94% (31/33) of these represented cleavage events at cRS sites I, II, III, or V (Table III and Fig. 2). Two SE products, one from RAG2:GFP and another from μMT pro–B cells, represented cleavage at a conserved FW3 site; no cleavage products from the predicted cRSs at site IV were recovered (Table III). Two VH gene segments were shown to contain two functional cRSs; VH1S2*01 produced site I and III SE products and VH1S130*01 supported cleavage at site III and in FW3 (Table III). Thus, in addition to the site V cRSs that are thought to be conserved for receptor editing (3, 27), we have identified other VH cRSs at nucleotide positions 57 (site I), −122 (site II), −155 (site III), −181 (site III), and −267 (FW3), that are cleaved during normal B cell development (Table III). Unexpectedly, site III SE products were most frequently recovered; site III cRS SE products were threefold more common than site V SE products (18 vs. 6, respectively), the next largest group.
Table III.
Conserved cRSs from multiple VH gene segments are cleaved in pro–B cells from RAG2:GFP, C57BL/6, and μMT mice
| Position (nt) |
cRS site |
VH gene | VH cRS sequence | Mouse strain |
|---|---|---|---|---|
| 57 | I | IGHV1S2*01 | TCCAACTGCAGCAGCCTGGGGC TGAGCTTGT GAAGCCTGGGGC TTCAGTG | RAG2:GFP |
| 122 | II | IGHV1-56*02 | ATATCCTGCAAGGCTCCTGGCT ACACCTTCA CCAGCCACTGGA TGCAGTG | μMT |
| 122 | II | IGHV1S132*01 | CTGTCCTGCAAGACTTCTGGCT ACACCTTCA CCAGCTACTGGA TTCAGTG | μMT |
| 155 | III | IGHV1-14*01 | AGCTATGTTATGCACTGGGTGA AGCAGAAGC CTGGGCAGGGCC TTGAGTG | μMT |
| 155 | III | IGHV1-22*01 | GACTACAACATGCACTGGGTGA AGCAGAGCC ATGGAAAGAGCC TTGAGTG | C57BL/6 |
| 155 | III | IGHV1-39*01 | GGCTACACCATGAACTGGGTGA AGCAGAGCC ATGGAAAGAACC TTGAGTG | C57BL/6 |
| 155 | III | IGHV1-54*01 | AATTACTTGATAGAGTGGGTAA AGCAGAGGC CTGGACAGGGCC TTGAGTG | μMT, C57BL/6 |
| 155 | III | IGHV1-55*01 | AGCTACTGGATAAACTGGGTGA AGCTGAGGC CTGGACAAGGCC TTGAGTG | RAG2:GFP |
| 155 | III | IGHV1-63*01 | AACTACTGGATAGGTTGGGTAA AGCAGAGGT CTGGACATATAC ATGGGTG | RAG2:GFP |
| 155 | III | IGHV1-67*02 | GATTATGCTATGCACTGGGTGA AGCAGAGTC ATGCAAAGAGTC TAGAGTG | μMT |
| 155 | III | IGHV1S1*01 | AGCTACTGGATGCACTGGGTGA AGCAGAGGC CTGGACGAGGCC TCGAGTG | RAG2:GFP |
| 155 | III | IGHV1S2*01 | AGCTACTGGATGCACTGGGTGA AGCAGAGGC CTGGACGAGGCC TTGAGTG | RAG2:GFP |
| 155 | III | IGHV1S10*01 | ACCTACTGGATGAACTGGGTGA AGTAGATGC CTGGACAGGGCC TTGAGTG | μMT, C57BL/6 |
| 155 | III | IGHV1S25*01 | GAGTATATTATACACTGGGTAA AGCAGAGGT CTGGACAGGGTC TTGAGTG | μMT |
| 155 | III | IGHV1S30*01 | AGCTACTACATGCACTGGGTGA AGCAGAGCC ATGGAAAGAGCC TTGAGTG | μMT |
| 155 | III | IGHV1S32*01 | AGCTACTATATACACTGGGTGA AGCAGAGGC CTGGACAGGGAC TTGAGTG | μMT |
| 155 | III | IGHV1S55*01 | AGCTCCTGGATGAACTGGGTGA AGCAGAGGC CTGGACAGGGAC TTGAGTG | C57BL/6 |
| 155 | III | IGHV1S70*01 | AGCTACTGGATAAACTGGGTGA AGCAGAGGC CTGGACAAGGCC TTGAGTG | μMT |
| 155 | III | IGHV1S95*01 | AGCTACTGGATGCACTGGGTGA AGCAGAGGC CTGGACAAGGCC TTGAGTG | μMT, RAG2:GFP |
| 155 | III | IGHV1S130*01 | AGCTCCTGGATGCACTGGGCGA AGCAGAGGC CTGGACAAGGCC TTGAGTG | μMT |
| 181 | III | IGHV1-64*01 | GAGGCCTGGACAAGGCCTTGAG TGGATTGGA ATGATTCATCCT AATAGTG | μMT, C57BL/6 |
| 267 | - | IGHV1S14*01 | AGGGCAAGGCCACAATGACTGT AGACACATC CTCCAGCACAGC CTACGTG | RAG2:GFP |
| 267 | - | IGHV1S130*01 | AGGGCAAGGCCACACTGACTGT AGACACATC CTCCAGCACAGC CTACGTG | μMT |
| 313 | V | IGHV1-4*01 | CATGCAACTGAGCAGCCTGACA TCTGAGGAC TCTGCAGTCTAT TACTGTG | C57BL/6 |
| 313 | V | IGHV1-19*01 | CATGGAGCTCAACAGCCTGACA TCTGAGGAC TCTGCGGTCTAT TACTGTG | μMT |
| 313 | V | IGHV1-48*02 | CATGGAGCTCAGCAGCCTGACA TCTGAGGAC TCTGCAGTCTAT TACTGTG | C57BL/6 |
| 313 | V | IGHV1-60*01 | CATGCAGCTCAGCAGCATGACA TCTGAAGCC TCTGATGACTAT TACTGTG | C57BL/6 |
| 313 | V | IGHV1S126*01 | CATGCAGCTCAGCAGCCTGACA TCTGAGGAC TCTGCGGTCTAT TACTGTG | C57BL/6 |
| 313 | V | IGHV5-6-2*01 | CCTGCAAATGAGCAGTCTGAAG TCTGAGGAC ACAGCCTTGTAT TACTGTG | RAG2:GFP |
The positions and sequences of VH 12-cRS SEs recovered from pro–B cells sorted from the BM of μMT, RAG2:GFP, or C57BL/6 mice are listed. Recovered VH1 and VH5 SE LM-PCR products were cloned into the pCR2.1TOPO vector and sequenced. Ligation of the BW-LC linker directly to the cRS heptamer confirmed RAG1/2-mediated cleavage. The sequences were processed in Vector NTI and analyzed by the IMGT database (http://imgt.cines.fr) and Immunoglobulin BLAST (http://www.ncbi.nlm.nih.gov/igblast) for gene identification. A variety of VH gene segments contain functional cRSs, and most SE products represent site III cRSs. Heptamer and nonamer sequences of VH 12-cRS are in bold.
The cRS SE products were readily recovered from μMT mice, demonstrating that preBCR signaling is not required for cRS cleavage (Table III). Indeed, the SE products from preBCR-deficient μMT mice (n = 15) and mice that express preBCR (C57BL/6 and RAG:GFP; n = 18) mice were similarly distributed, with site III SE products predominant in both (11/15 and 11/18, respectively). Comparable numbers of site I/II, FW3, and site V cleavage products were recovered from preBCR-deficient and -sufficient pro–B cells as well, suggesting that VH cRS cleavage may be active during the VH→ DHJH stage of B cell development (Table III). V→VDJ replacements at the site V cRSs (Fig. 2) retain only 1–3 amino acids from the edited VH gene segment (3, 27). In contrast, a V→VDJ replacement using a site III cRS would result in an IgH variable region lengthened by as many as 50 amino acids and containing 4, not 3, hypervariable regions. We doubt that such IgH polypeptides could generate functional preBCR or BCR. Nonetheless, site III cRSs are conserved in mouse VH gene segments (Fig. 2) and are the most frequently cleaved during normal B cell development (Table III). We conclude that the conservation of VH cRSs is not solely driven by their ability to mediate functional V→VDJ replacements.
DISCUSSION
VH replacement is the insertion of a VH gene segment into a formed VHDJH joint (28) by the RAG1/2 recombinase acting on the physiological 23-RS of the invading VH segment and a so-called cryptic heptamer present near the 3′ border (position 313) of many VH gene segments (2). Whereas this specific form of IgH editing has been observed in vivo (3, 10), additional forms of VH replacement and/or secondary rearrangement have been observed in cell lines (14, 29, 30). To determine whether the complex editing events observed in cell lines might also take place in vivo, we screened 390 mouse VH gene segment sequences with the RIC algorithm to identify potentially functional 12- or 23-cRSs (18, 19). RIC is capable of identifying RSs and cRSs in DNA sequences (18); whereas RIC scores for RSs are highly correlated with recombination efficiencies, RIC scores for cRSs are less so, in part because of the narrow range of RIC scores and measured recombination efficiencies that are often below the detection threshold (17, 18). Nevertheless, the measured extrachromosomal recombination efficiencies (R = 0.01–0.02%; Table II) of a small subset of site I, III, and IV VH 12-cRSs fell at the lower range of recombinational activities (R = 0.03–0.6%) of 4 site V VH cRSs that were previously determined (18).
In vivo generation of (site III) VH cRS SEs was more efficient, with quantities of VH cRS SEs ranging from 1 (VH1) to 8% (VH5) of that observed for JH2 SEs (Fig. 6). These higher values are consistent with our ability to detect cRS SEs in as few as 103 pro–B cells, and they imply that rearrangements of VH cRSs may occur as often as rearrangements of the recombining sequence cRS that flank Cκ in mice (31). Frequencies of VH cRS cleavage vary between different VH gene segments/families; quantitative LM-PCR indicated that VH5 cRS SEs were almost twice as abundant as VH1 cRS SEs after correcting for template number (Fig. 6). Although we cannot rule out the possibility that the covalent sealing or degradation of cRS SEs is not uniform, increased abundance of certain cRS SEs suggests that cRSs in some VH may be preferred recipients for upstream VH RSs (3, 32).
Almost half (108/299) of O2 12-cRSs in VH gene segments are not located at the 3′ end of VH gene segments (Fig. 2); e.g., a cluster of 51 VH cRSs is located at nt 57 (Fig. 2). These and other 5′ cRSs are functional at low efficiency, both in vitro (Table II) and in vivo (Table III). Sequence analysis of VH cRS SEs from pro–B cells of RAG2:GFP, μMT, and C57BL/6 mice revealed cleavage at 33 unique 12-cRSs, one in a VH5 gene segment and 32 in VH1 genes (Table III). These VH 12-cRSs comprised 29 unique cRS sites from 27 germline VH genes (Table III). The utility of RIC analyses is supported by the location of these functional 12-cRSs; >90% (31/33) correspond to the predicted VH 12-cRSs (Fig. 2).
To our surprise, we recovered only 6 VH cRS SEs at the well-known site V cRS (313 bp; Table III) commonly observed in IgH replacements (3, 11). Instead, the most common (22/33) VH cRS SEs we recovered represented site III (nt 155) 12-cRSs located near the middle of the VH gene segments (Table III and Fig. 2). VH cRS SEs from sites I–IV comprised ∼80% (27/33) of our sample, indicating that RAG-mediated cleavage at site V cRSs is not favored. At least 2 VH gene segments, VH1S2*01 and VH1S130*01, contain 2 functional 12-cRSs at sites I and III and at an unpredicted position at 267 bp, respectively (Table III). Thus, RIC scores effectively predicted the location of VH cRSs active in vivo and their recombination potential in extrachromosomal assays, but not the frequency of VH cRS SEs recovered from pro–B cells. We conclude that factors absent from extrachromosomal recombination assays significantly affect cRS cleavage in situ.
VH cRS SEs are RAG1 dependent (Fig. 4), and they are independent of preBCR signaling (Fig. 5). The presence of VH cRS SEs in the pro–B cells of μMT mice demonstrates that IgH replacement can occur well before the developing B cell is capable of recognizing antigen in any form (3). Similarly, in BL/6 and RAG2:GFP mice, VH cRS SEs could be detected only in pro–B cells (Fig. 3). In H50G transgenic mice, cRS SEs were present in both pro–B and pre–B cells (Fig. 4), but we note that JH2 SEs were also abundant in the pre–B cells of these IgH transgenic animals (24). We conclude that the phenotypic pre–B compartment of H50G mice includes cells that actively rearrange the endogenous Igh loci. In no case were we able to detect VH cRS SEs in immature B cells, the earliest B lineage cell that expresses mature BCR (Figs. 3–5).
These results are different from those reported by Zhang et al. (11), who did not detect VH cRS SEs in human pro–B cells, but did find them in immature B lymphocytes. Zhang et al. suggested that recovery of VH cRS SEs from pro–B cells might be hindered by rapid cell proliferation and efficient DNA repair. Although these factors may be important in the analysis of human B cell populations, we readily detected VH cRS SEs in mouse pro–B cells (Fig. 4). It is possible that the populations of B cells we analyzed differ somewhat from those sorted by Zhang et al.; however, the cytometric parameters used by both groups were similarly based on IgM expression and an early B cell marker (CD34 for human [11] and CD43 for mouse [Fig. 3]). In addition, we characterized our B cell populations by developmentally regulated gene expression and Igh and Igκ rearrangement (Fig. 3). We are, therefore, confident that the pro–B cells analyzed in our study, B220loCD43+ cells expressing RAG1, RAG2:GFP, Tdt, and Igh rearrangements, contain VH cRS SEs. In our hands, these VH cRS SEs do not persist and/or reform at detectable levels in immature B cells (Figs. 3 and Figs.5). The differences between our results and those reported by Zhang et al. (11) presumably reflect the distinct physiologies of mice and humans.
The presence of VH cRS SEs, which are the molecular intermediates of Igh replacements, in pro–B cells from μMT mice unable to assemble a BCR (26) is inconsistent with any IgH editing process driven by the recognition of self-antigen. It is significant that VH cRS SEs were also abundant in the pro–B cells of H50G+/−Rag1+/− mice (Fig. 4), even though this IgH transgenic line exhibits stringent (≥98%) allelic exclusion (24). Similarly, Rajewsky et al. have observed frequent IgH editing events in the presence of a productive and functional VHDJH SDT (33). Given the presence of VH cRS SEs in normal pro–B cells and efficient IgH editing in genetically modified pro–B cells (10), earlier conclusions that IgH editing is driven by self-antigen (11) merit reconsideration. Instead, we propose that VH→VHDJH replacement occurs spontaneously, albeit at low frequency, in pro–B cells. In mice bearing autoreactive VHDJH SDT, replacement by endogenous VH gene segments would relieve the autoreactive phenotype and permit the “edited” B cells to mature beyond the small pre–B cell stage. In this scenario, self-antigen does not drive receptor editing, but rather selects for mutant cells that are no longer autoreactive.
VH replacements from mice and humans are frequently characterized by N-nucleotide additions (3, 10, 11). Although N-sequence additions imply VH replacement in Tdt+ pro–B cells (13), Chen et al. (3) have proposed that Tdt may be reexpressed in immature, autoreactive B cells after encounter with self-antigen. This seems unlikely, given that little or no Tdt expression has been detected in the pre–B and immature B cell compartments (Fig. 3 and [13]), even though a substantial fraction (≥20%) of late small pre–B- and immature B cells are thought to be autoreactive and edit their L-chains (34, 35). But what of IgH replacements that lack N-nucleotides? Are they evidence for IgH editing in more mature Tdt− developmental compartments?
Recently, Koralov et al. generated genetically modified mice to study VH replacement (10, 36). In these animals, antibody and B cell production depends on the replacement of a nonproductive VHDJH rearrangement that takes place in pro–B cells via two mechanisms: VH→VHDJH replacement; and, less frequently, direct VH-to-JH joining (10). These mice exhibit diverse and substantial B cell populations, and the majority of VH→VHDJH replacement events analyzed did not contain N-nucleotide additions, presumably because secondary cRS rearrangements were facilitated by local sequence homologies. Koralov et al. conclude that Igh replacements in pro–B cells is relatively efficient and that its impact on the antibody repertoire may be greater than is currently thought, as it often leaves no molecular footprint (10).
Given that the conserved site I–IV cRSs in VH gene segments could not mediate VH→VHDJH replacements (3, 11), what other purpose might these signals serve? Taki et al. (8), have reported an inactivating rearrangement involving a 5′ cRS in an Igh SDT. This replacement, a DH→VDJH invasion (8), followed by a physiological rearrangement to an upstream VH (VH→DVDJH) (8) was nonfunctional, as it was isolated only from B cells expressing BCR encoded by an endogenous Igh rearrangements (8). These results suggest that 5′ cRS might function to end V(D)J rearrangements on one allele, as well as an analogy to abrogation of Igκ rearrangements by Cκ-deleting signals (37–41). Indeed, even open-and-shut reactions (42) at FW cRSs would likely produce inactivating frame-shift mutations.
Alternatively, conserved VH cRSs at sites I–IV might interact to create novel, hybrid VH gene segments. We demonstrated frequent cRS cleavage sites between CDR1 and CDR2 (Fig. 2 and Table III), and showed that VH 12-cRSs are strongly conserved in the O2 orientation, i.e., opposite of the physiological 23-RS (Fig. 7 and Table S1). This arrangement facilitates VH→VHDJH fusions at site V 12-cRS, but also allows other recombination events including VH→VH and intra-VH rearrangements (Fig. 7). Both VH→VHDJH and VH→VH fusions lengthen the acceptor VH genes in proportion to cRS location (site I > site V; Fig. 7). In contrast, intra-VH rearrangements between VH 12-cRS and downstream 23-RS would produce VH genes shortened by deletion of the intervening DNA and terminated by a signal joint (Fig. 7). Reopening of this terminal signal joint would allow the 23-RS to form a new signal joint with downstream 12-cRSs, thereby fusing the shortened VH fragment to a truncated VH acceptor (Fig. 7). This double reaction intra-VH rearrangement followed by insertion into a downstream VH/VHDJH would undoubtedly be rare, but could result in hybrid VH genes of nearly normal length. For example, intra-VH rearrangements at site III cRS (Table III), followed by insertion at another site III cRS in a downstream VH gene segment, would produce a novel VH sequence of normal length carrying the CDR1 of the upstream donor and the CDR2 of the downstream acceptor. Hybrid VH gene segments created by this process would contain a signal-to-coding joint at the fusion site (Fig. 7).
Figure 7.
Rearrangement of site I–V VH cRSs in pro–B cells could increase the IgH repertoire by VH replacement or hybrid rearrangement. The primary VDJ germline configuration and the outcomes of secondary cRS rearrangements are depicted. VH replacement (V→V or V→VDJ) is mediated by a cRS (gray triangle in VH gene segment) to an upstream VH RS (white triangle). (Intra-V)→VDJ rearrangement via a 59 cRS to a 59 cRSs in another VH gene segment forms a hybrid rearrangement and creates a hybrid joint (between a CE and a SE) between two VH gene segments. Note that hybrid rearrangements and VH replacements depicted can increase the diversity of VH gene segments by novel CDR combinations.
The hypothesis that cRSs are conserved in VH gene segments to promote genetic diversity implies that cRSs should be conserved in other V gene families as well. We have tested this prediction by scanning all mouse Vκ gene segments with the RIC algorithms. Our search revealed a highly significant conservation of 23-cRSs oriented to interact with upstream Vκ 12-RSs (unpublished data). We do not wish to overemphasize this finding; the presence of conserved cRSs in Vκ gene segments is consistent with, but does not prove, a role for VH cRSs in amplifying V region diversity. Nonetheless, these Vκ cRSs demonstrate that cRSs can arise in Ig loci capable of repeated physiological rearrangements.
Although the reports are controversial (43, 44), several groups have recovered hybrid VH genes of normal length from human B cells (14, 15) and B cell tumors (45–47) that could be generated by recombination at site I–IV cRSs. These reports propose that hybrid VH genes arise as products of secondary rearrangements between 23- and 12-cRSs centrally embedded in VH gene segments or by recombination between like cRSs in violation of the 12/23 rule. The LM-PCR used in our studies does not detect cRS SEs in the O1 orientation, and we have no direct evidence regarding interaction of VH 23- and 12-cRSs. Our RIC scans, however, did not detect conserved, bidirectional 12-/23-cRS in mouse VH gene segments (unpublished data). Intra-VH→VHDJH rearrangements (Fig. 7) allow, at least in theory, for the generation of VH hybrids at sites of conserved O2 12-cRSs. Generation of VH hybrids by this mechanism predicts specific genomic intermediates (intra-V; Fig. 7); their demonstration and frequency would provide a significant test for the significance of site I–IV cRS.
If they do not represent PCR artifacts, hybrid VH gene segments might result from AID- rather than RAG1/2 activity, given the germinal center/post–germinal center origins of many hybrid VH genes (44, 48). AID activity can result in both single- and double-stranded breaks in DNA, and subsequent repair by homologous recombination could produce hybrid VH genes (48). On the other hand, RAG1/2 also introduces single-strand nicks at RSs that could be subject to homologous recombination (49).
In their seminal work, Chen et al. (3) considered, and then dismissed, the possibility that IgH replacements might not represent receptor editing, but rather the continuing activity of RAG1/2 on functional VHDJH templates. This conclusion was supported by the findings of Zhang et al. (11) who observed cRS SEs only in human immature B cells, the earliest developmental compartment capable of antigen recognition. In contrast, we demonstrate abundant VH cRS SEs in the pro–B cell compartment of normal mice; pro–B cells do not express L-chain and are incapable of binding self- or exogenous antigens. Recovery of cRS SEs from the pro–B cells of μMT mice rules out any possibility that these RAG1-dependent SEs reflect antigen-induced editing. Nonetheless, functional VH cRSs are evolutionarily conserved independently of the amino acids necessary for IgH structure. Are these conserved cRSs accidents of evolution, or do they have physiological significance? Combinatorial diversity in IgH rearrangements is determined by the evolutionary concatenation of CDR1 and 2 in distinct VH gene segments and the random, somatic generation of CDR3 during the fusion of VH, D, and JH gene segments. This process generates a set of primary antigen receptors of remarkable breadth, but also one that is focused on the assembly of the CDR3. We suggest that site I–V VH cRSs are conserved, as their rearrangements offer the possibility of (a) greater diversity in CDR3 (VH→VHDJH) and (b) the somatic reassortment of CDR1 and CDR2 associations (intra-VH→VHDJH) otherwise fixed by evolution.
MATERIALS AND METHODS
RS models.
The computational models of RSs assign a RIC value to 28- or 39-bp sequences. We demonstrated that sequences with nucleotide combinations strongly conserved in physiological RSs are efficient at recombination and have high RIC values (18, 19). We used RIC12 or RIC23 to determine the location and the number of 12- or 23-cRSs in VH gene segments. RIC was computed for all 28- and 39-bp sequences in the 390 mouse VH gene segments (excluding leader sequences) in the Immunogenetics Information System (IMGT) reference set (http://imgt.cines.fr). As controls for the identification of VH cRSs, we computed RIC for all 28- and 39-bp sequences in a 212-kb region of mouse chromosome 8 (GenBank accession no. AC084823).
Mice.
All mice were housed in specific pathogen–free conditions at the Duke University Medical Center Vivarium. RAG2:GFP mice (21) were obtained from F.W. Alt (Harvard University, Boston, MA); μMT (26) and C57BL/6 mice were purchased from The Jackson Laboratory. H50G+/− (IgH) transgenic mice (24) were bred with Rag1−/− mice (The Jackson Laboratory) to produce sibling H50G+/−Rag1+/− and H50G+/−Rag1−/− mice. All experiments using animals were reviewed and approved by the Institutional Animal Use and Care Committee of Duke University.
Flow cytometry.
BM was isolated from femurs and tibias of mice. Red blood cells were lysed in ACK buffer, and BM cells were washed and resuspended in HBSS (Invitrogen) supplemented with 2% FCS (Sigma-Aldrich). BM B cells from RAG2:GFP, H50G+/−Rag1+/−, and H50G+/−Rag1−/− mice were stained with lineage (Lin) markers (IgD, Gr-1, Mac-1, TER-119, CD4, and CD8) conjugated with biotin, washed twice with HBSS (2% FCS), incubated with streptavidin conjugated to magnetic MicroBeads (Miltenyi Biotech), washed with HBSS (2% FCS), and depleted by autoMACS (Miltenyi Biotech). After depletion, cell samples were labeled with anti-B220 (APC), anti-CD43 (PE or FITC), anti-IgM (Texas red), 7-amino actinomycin D (Invitrogen), and streptavidin (Cychrome). To obtain B cells from μMT and C57BL/6 mice, single-cell suspensions were stained with anti-IgD (FITC), anti-CD43 (PE), anti-B220 (PE-Cy7), anti-IgM (Texas red), and PE-Cy5-conjugated Lin markers (Gr-1, CD11b, CD4, CD8, and TER-119). After staining and washing, BM samples were sorted on a FACSVantage cell sorter (BD Biosciences). All antibodies and markers are from BD Biosciences or eBioscience, except for 7-amino actinomycin D and anti-IgM (SouthernBiotech).
Cell culture.
103/BCL2 cells (50) were cultured for use in extrachromosomal recombination assays, as previously described (18).
Extrachromosomal recombination assay.
12-cRSs were cloned into pJH290 and electroporated into 103/BCL2 cells (18). Cells were subsequently incubated at 34°C for 2 d and incubated at 39.5°C for 2 d to induce V(D)J rearrangement (18). Recombination plasmids were extracted, digested with DpnI, and used to transform Escherichia coli. Transformed bacteria were incubated on LB-agar plates supplemented with 50 μg/ml ampicillin, 11 μg/ml chloramphenicol, or both. The bacterial colonies on each plate were quantified and normalized to equivalent incubation volumes. The R of various 12-cRSs was estimated as the ratio of amprcamr to ampr bacterial colonies, as previously described (18). R was calculated as the mean of ≥ 3 independent electroporations. The sensitivity limit of extrachromosomal assay was established with a pJH290 plasmid modified by deletion of the 12-RS, leaving the 23-RS as the only physiological RS. Extrachromosomal recombination assays with this plasmid did not produce a single amprcamr bacterial colony out of 98,730 Ar bacterial colonies that harbored a bona fide rearrangement. Thus, the sensitivity limit for the detection of 12- and 23-RS rearrangement was ∼0.001% (1/98,730).
PCR.
Amplification of CD14 was performed to quantify genomic DNA template (51). LM-PCR was used to amplify SEs ligated to a BW-LC linker, as previously described (18). VH cRS SEs from cells isolated from RAG2:GFP, sibling H50G, μMT, and C57BL/6 mice were amplified by a seminested LM-PCR. Primary amplification of VH cRS SEs included a VH family-specific outside primer (VHout) and BW-LCH primer (5′-ACGTGGAATCGCCAGACCAC-3′), using ThermalAce DNA Polymerase (Invitrogen). Primary amplification was mediated by 12 cycles of melting at 98°C for 30 s, annealing at 65°C for 30 s, and extending at 72°C for 30 s, and was finally terminated after a 10-min incubation at 72°C. 10% of the primary reaction was amplified with a VH family–specific inside primer (VHin) and BW-LCH primer. Nested amplification used the same program with 26 cycles. Amplified SEs were detected by hybridization to γ-P32–labeled probes with a Storm PhosphorImager (GE Healthcare). LM-PCR products were cloned and sequenced (18). LM-PCR of Dβ SEs and Jκ SEs were performed as previously described (51, 52). PCR primers for CD14 followed (51).
The relative abundance of JH2 RS SEs and VH cRS SEs in DNA recovered from nucleated BM cells of C57BL/6 mice was estimated by real-time quantitative PCR on an ABI PRISM 7700 Sequence Detector using SYBR green PCR core reagents (Applied Biosystems) according to the manufacturer's instructions. The relative abundance of RS/cRS SEs was calculated by the comparative CT (threshold cycle) method recommended by the manufacturer (Applied Biosystems) normalized to JH2 SE product from the same sample. In brief, ΔCT values were determined by subtracting CT (cRS SE) from CT (RS SE). Expression levels relative to JH2 RS SEs were defined as 2ΔCT .
Sequences for LM-PCR primers and probes are: (a) JH2out, 5′-TACTTCGATGTCTGGGGCACAG-3′, JH2in, 5′-AAAGAGGCAGTCAGAGGCTAGCTG-3′, JH2probe, and 5′-AAATAGGCATTTACATTGTTAGGC-3′ for JH RS SE; (b) VH1(J558)out.1, 5′-AGGTCCAACTGCAGCAGCCTG-3′, VH1(J558)in.1, 5′-CCTGCAAGGCTTCTGGCTACA-3′, VH1(J558)out.2, 5′-CAGGTTCAGCTSCAGCAGTCTG-3′, VH1(J558)in.2, 5′-TRTCCTGCAAGGCTTCTGGCTACAC-3′; VH1(J558)out.3, 5′-AGGTCCAGCTGCAGCAGTCTG-3′, VH1(J558)in.3, 5′-TCAGTGAAGATGTCCTGCAA-3′ (where S = C/G, R = A/G), VH1(J558)cRSprobe, and 5′-TGCCTTTCTCTCCACAGGTGTCCA-3′ for VH1 (J558) cRS SE; (c) VH2(Q52)out, 5′-TGTCCATCACCTGCACAGTCTCTG-3′, VH2(Q52)in, 5′-TCTGGAGTGGCTGGGAGTGATATG-3′, VH2(Q52)cRSprobe, 5′-CCAGAC TGAGCATCAGCAAGGACAA-3′ for VH2 (Q52) cRS SE; VH5(7183)out, 5′- GAGGGTCCCTGAAACTCTCCTG-3′, (d) VH5(7183)in, 5′-GGAGTTGGTC GCAGCCATTAATAG-3′, VH5(7183)cRSprobe, 5′-CTCCAGAGACAATA CCAAGAAGACC-3′ for VH5 (7183) cRS SE. Amplification of the physiological RS SEs of the VH7183 gene segment made use of this additional primer ([7183RS], 5′-ATGTGTGCCAGGAGCCTCTGACCAG-3′). The VH primer sets (VH1[J558].1, VH2, and VH5[7183]) used in this study amplify ∼34 VH1 gene segments, 4 VH2 gene segments, and 7 VH5 gene segments from the C57BL/6 genome.
Online supplemental material.
Table S1 provides the numbers of RS-length segments with a finite RIC score and the numbers of cRSs (RIC > −45 or −65 for 12- and 23-cRSs, respectively) in VH gene segments and a 212-kb region of mouse chromosome 8 (GenBank accession no. AC084823). Relative frequencies of 12- and 23-cRSs are shown in parentheses. VH cRSs are conserved with 12-bp spacers and in the O2 orientation. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20071244/DC1.
Supplemental Material
Acknowledgments
We are grateful for the comments and suggestions of Drs. T.F. Tedder (Duke University) and M.D. Cooper (University of Alabama). This work was supported in part by grants from the National Institutes of Health (AI 24335, AI 56363, and AI 67854), the Bill and Melinda Gates Foundation (to G. Kelsoe.) and the Burroughs-Wellcome Fund (to L.G. Cowell).
The authors have no conflicting financial interests.
Abbreviations used: BCR, B cell antigen receptor; CDR, complementarity determining region; cRS, cryptic recombination signal; Cys, cysteine; FW, Ig frame work region; LM, ligation-mediated; RIC, RS information content; RS, physiological recombination signal; SDT, site-directed transgene; SE, signal end.
M. Davila and F. Liu contributed equally to this paper.
References
- 1.Nemazee, D., and M. Weigert. 2000. Revising B cell receptors. J. Exp. Med. 191:1813–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fanning, L., F.E. Bertrand, C. Steinberg, and G.E. Wu. 1998. Molecular mechanisms involved in receptor editing at the Ig heavy chain locus. Int. Immunol. 10:241–246. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C., Z. Nagy, E.L. Prak, and M. Weigert. 1995. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 3:747–755. [DOI] [PubMed] [Google Scholar]
- 4.Kleinfield, R.W., and M.G. Weigert. 1989. Analysis of VH gene replacement events in a B cell lymphoma. J. Immunol. 142:4475–4482. [PubMed] [Google Scholar]
- 5.Sakano, H., Y. Kurosawa, M. Weigert, and S. Tonegawa. 1981. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature. 290:562–565. [DOI] [PubMed] [Google Scholar]
- 6.Alt, F.W., G.D. Yancopoulos, T.K. Blackwell, C. Wood, E. Thomas, M. Boss, R. Coffman, N. Rosenberg, S. Tonegawa, and D. Baltimore. 1984. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 3:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C., E.L. Prak, and M. Weigert. 1997. Editing disease-associated autoantibodies. Immunity. 6:97–105. [DOI] [PubMed] [Google Scholar]
- 8.Taki, S., F. Schwenk, and K. Rajewsky. 1995. Rearrangement of upstream DH and VH genes to a rearranged immunoglobulin variable region gene inserted into the DQ52-JH region of the immunoglobulin heavy chain locus. Eur. J. Immunol. 25:1888–1896. [DOI] [PubMed] [Google Scholar]
- 9.Cascalho, M., A. Ma, S. Lee, L. Masat, and M. Wabl. 1996. A quasi-monoclonal mouse. Science. 272:1649–1652. [DOI] [PubMed] [Google Scholar]
- 10.Koralov, S.B., T.I. Novobrantseva, J. Konigsmann, A. Ehlich, and K. Rajewsky. 2006. Antibody repertoires generated by VH replacement and direct VH to JH joining. Immunity. 25:43–53. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, Z., M. Zemlin, Y.H. Wang, D. Munfus, L.E. Huye, H.W. Findley, S.L. Bridges, D.B. Roth, P.D. Burrows, and M.D. Cooper. 2003. Contribution of v(h) gene replacement to the primary B cell repertoire. Immunity. 19:21–31. [DOI] [PubMed] [Google Scholar]
- 12.Cascalho, M., J. Wong, and M. Wabl. 1997. VH gene replacement in hyperselected B cells of the quasimonoclonal mouse. J. Immunol. 159:5795–5801. [PubMed] [Google Scholar]
- 13.Li, Y.S., K. Hayakawa, and R.R. Hardy. 1993. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 178:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh, K., E. Meffre, E. Albesiano, A. Farber, D. Dines, P. Stein, S.E. Asnis, R.A. Furie, R.I. Jain, and N. Chiorazzi. 2000. Immunoglobulin heavy chain variable region gene replacement as a mechanism for receptor revision in rheumatoid arthritis synovial tissue B lymphocytes. J. Exp. Med. 192:1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson, P.C., K. Wilson, Y.J. Liu, J. Banchereau, V. Pascual, and J.D. Capra. 2000. Receptor revision of immunoglobulin heavy chain variable region genes in normal human B lymphocytes. J. Exp. Med. 191:1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand, F.E., R. Golub, and G.E. Wu. 1998. V(H) gene replacement occurs in the spleen and bone marrow of non-autoimmune quasi-monoclonal mice. Eur. J. Immunol. 28:3362–3370. [DOI] [PubMed] [Google Scholar]
- 17.Lee, A.I., S.D. Fugmann, L.G. Cowell, L.M. Ptaszek, G. Kelsoe, and D.G. Schatz. 2003. A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol. 1:E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowell, L.G., M. Davila, K. Yang, T.B. Kepler, and G. Kelsoe. 2003. Prospective estimation of recombination signal efficiency and identification of functional cryptic signals in the genome by statistical modeling. J. Exp. Med. 197:207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowell, L.G., M. Davila, T.B. Kepler, and G. Kelsoe. 2002. Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldsby, R.A., T.J. Kindt, B.A. Osborne, and J. Kuby. 2003. Immunology. W. H. Freeman and Company, New York.
- 21.Monroe, R.J., K.J. Seidl, F. Gaertner, S. Han, F. Chen, J. Sekiguchi, J. Wang, R. Ferrini, L. Davidson, G. Kelsoe, and F.W. Alt. 1999. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 11:201–212. [DOI] [PubMed] [Google Scholar]
- 22.Osmond, D.G., A. Rolink, and F. Melchers. 1998. Murine B lymphopoiesis: towards a unified model. Immunol. Today. 19:65–68. [DOI] [PubMed] [Google Scholar]
- 23.Yu, W., H. Nagaoka, M. Jankovic, Z. Misulovin, H. Suh, A. Rolink, F. Melchers, E. Meffre, and M.C. Nussenzweig. 1999. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 400:682–687. [DOI] [PubMed] [Google Scholar]
- 24.Dal Porto, J.M., A.M. Haberman, G. Kelsoe, and M.J. Shlomchik. 2002. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J. Exp. Med. 195:1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang, Y., M.J. Bosma, and G.C. Bosma. 1999. Extended duration of DH–JH rearrangement in immunoglobulin heavy chain transgenic mice: implications for regulation of allelic exclusion. J. Exp. Med. 189:1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitamura, D., J. Roes, R. Kuhn, and K. Rajewsky. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 350:423–426. [DOI] [PubMed] [Google Scholar]
- 27.Kleinfield, R., R.R. Hardy, D. Tarlinton, J. Dangl, L.A. Herzenberg, and M. Weigert. 1986. Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. Nature. 322:843–846. [DOI] [PubMed] [Google Scholar]
- 28.Jung, D., C. Giallourakis, R. Mostoslavsky, and F.W. Alt. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, S.M., E. Agard, S. Suh, and L. Czyzyk. 1997. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol. 17:3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Usuda, S., T. Takemori, M. Matsuoka, T. Shirasawa, K. Yoshida, A. Mori, K. Ishizaka, and H. Sakano. 1992. Immunoglobulin V gene replacement is caused by the intramolecular DNA deletion mechanism. EMBO J. 11:611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siminovitch, K.A., M.W. Moore, J. Durdik, and E. Selsing. 1987. The human kappa deleting element and the mouse recombining segment share DNA sequence homology. Nucleic Acids Res. 15:2699–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connor, A.M., L.J. Fanning, J.W. Celler, L.K. Hicks, D.A. Ramsden, and G.E. Wu. 1995. Mouse VH7183 recombination signal sequences mediate recombination more frequently than those of VHJ558. J. Immunol. 155:5268–5272. [PubMed] [Google Scholar]
- 33.Taki, S., M. Meiering, and K. Rajewsky. 1993. Targeted insertion of a variable region gene into the immunoglobulin heavy chain locus. Science. 262:1268–1271. [DOI] [PubMed] [Google Scholar]
- 34.Casellas, R., T.A. Shih, M. Kleinewietfeld, J. Rakonjac, D. Nemazee, K. Rajewsky, and M.C. Nussenzweig. 2001. Contribution of receptor editing to the antibody repertoire. Science. 291:1541–1544. [DOI] [PubMed] [Google Scholar]
- 35.Wardemann, H., S. Yurasov, A. Schaefer, J.W. Young, E. Meffre, and M.C. Nussenzweig. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. [DOI] [PubMed] [Google Scholar]
- 36.Koralov, S.B., T.I. Novobrantseva, K. Hochedlinger, R. Jaenisch, and K. Rajewsky. 2005. Direct in vivo VH to JH rearrangement violating the 12/23 rule. J. Exp. Med. 201:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, M.W., J. Durdik, D.M. Persiani, and E. Selsing. 1985. Deletions of kappa chain constant region genes in mouse lambda chain-producing B cells involve intrachromosomal DNA recombinations similar to V-J joining. Proc. Natl. Acad. Sci. USA. 82:6211–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu, T., T. Iwasato, and H. Yamagishi. 1991. Deletions of immunoglobulin Cκ region characterized by the circular excision products in mouse splenocytes. J. Exp. Med. 173:1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durdik, J., M.W. Moore, and E. Selsing. 1984. Novel kappa light-chain gene rearrangements in mouse lambda light chain-producing B lymphocytes. Nature. 307:749–752. [DOI] [PubMed] [Google Scholar]
- 40.Siminovitch, K.A., A. Bakhshi, P. Goldman, and S.J. Korsmeyer. 1985. A uniform deleting element mediates the loss of kappa genes in human B cells. Nature. 316:260–262. [DOI] [PubMed] [Google Scholar]
- 41.Graninger, W.B., P.L. Goldman, C.C. Morton, S.J. O'Brien, and S.J. Korsmeyer. 1988. The κ-deleting element. Germline and rearranged, duplicated, and dispersed forms. J. Exp. Med. 167:488–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis, S.M., and J.E. Hesse. 1991. Cutting and closing without recombination in V(D)J joining. EMBO J. 10:3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goossens, T., U. Klein, and R. Kuppers. 1998. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc. Natl. Acad. Sci. USA. 95:2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darlow, J.M., and D.I. Stott. 2005. V(H) replacement in rearranged immunoglobulin genes. Immunology. 114:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deane, M., and J.D. Norton. 1990. Immunoglobulin heavy chain gene rearrangement involving V-V region recombination. Nucleic Acids Res. 18:1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brokaw, J.L., S.M. Wetzel, and B.A. Pollok. 1992. Conserved patterns of somatic mutation and secondary VH gene rearrangement create aberrant Ig-encoding genes in Epstein-Barr virus-transformed and normal human B lymphocytes. Int. Immunol. 4:197–206. [DOI] [PubMed] [Google Scholar]
- 47.Lenze, D., A. Greiner, C. Knorr, I. Anagnostopoulos, H. Stein, and M. Hummel. 2003. Receptor revision of immunoglobulin heavy chain genes in human MALT lymphomas. Mol. Pathol. 56:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darlow, J.M., and D.I. Stott. 2006. Gene conversion in human rearranged immunoglobulin genes. Immunogenetics. 58:511–522. [DOI] [PubMed] [Google Scholar]
- 49.Lee, G.S., M.B. Neiditch, S.S. Salus, and D.B. Roth. 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 117:171–184. [DOI] [PubMed] [Google Scholar]
- 50.Chen, Y.Y., L.C. Wang, M.S. Huang, and N. Rosenberg. 1994. An active v-abl protein tyrosine kinase blocks immunoglobulin light-chain gene rearrangement. Genes Dev. 8:688–697. [DOI] [PubMed] [Google Scholar]
- 51.Han, S., S.R. Dillon, B. Zheng, M. Shimoda, M.S. Schlissel, and G. Kelsoe. 1997. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 278:301–305. [DOI] [PubMed] [Google Scholar]
- 52.McMahan, C.J., and P.J. Fink. 1998. RAG reexpression and DNA recombination at T cell receptor loci in peripheral CD4+ T cells. Immunity. 9:637–647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.