Abstract
The broad range of biological responses elicited by transforming growth factor-β (TGF-β) in various types of tissues and cells is mainly determined by the expression level and activity of the effector proteins Smad2 and Smad3. It is not fully understood how the baseline properties of Smad3 are regulated, although this molecule is in complex with many other proteins at the steady state. Here we show that nonactivated Smad3, but not Smad2, undergoes proteasome-dependent degradation due to the concerted action of the scaffolding protein Axin and its associated kinase, glycogen synthase kinase 3-β (GSK3-β). Smad3 physically interacts with Axin and GSK3-β only in the absence of TGF-β. Reduction in the expression or activity of Axin/GSK3-β leads to increased Smad3 stability and transcriptional activity without affecting TGF-β receptors or Smad2, whereas overexpression of these proteins promotes Smad3 basal degradation and desensitizes cells to TGF-β. Mechanistically, Axin facilitates GSK3-β-mediated phosphorylation of Smad3 at Thr66, which triggers Smad3 ubiquitination and degradation. Thr66 mutants of Smad3 show altered protein stability and hence transcriptional activity. These results indicate that the steady-state stability of Smad3 is an important determinant of cellular sensitivity to TGF-β, and suggest a new function of the Axin/GSK3-β complex in modulating critical TGF-β/Smad3-regulated processes during development and tumor progression.
[Keywords: Smad3, TGF-β, Axin, GSK3-β, ubiquitination, protein stability]
The transforming growth factor-β (TGF-β) family of cytokines, including TGF-β, activin/inhibin, and bone morphogenetic proteins (BMPs), plays essential roles in regulating a broad spectrum of biological processes such as cell proliferation, differentiation, apoptosis, migration, and extracellular matrix (ECM) remodeling (Siegel and Massagué 2003). Perturbations of TGF-β signaling are often associated with diseases such as cancer, where TGF-β plays a dual role as both a tumor suppressor and a metastasis promoter at different stages of tumor development (Bierie and Moses 2006).
The canonical TGF-β signaling pathway begins with the TGF-β ligand binding to the type I and type II TGF-β receptors (TβRI and TβRII), which are both serine/threonine kinases. The activated receptor complex phosphorylates downstream transcription factors called receptor-regulated Smad proteins (R-Smads), Smad2 and Smad3, leading to the formation of a complex between the R-Smads and the common partner, Smad4. The Smad complex then becomes concentrated in the nucleus to regulate the expression of a great number of target genes (Shi and Massagué 2003).
As the primary mediators of TGF-β signaling, Smad2 and Smad3 share >90% homology in amino acid sequence. Both Smads consist of two highly conserved domains (MH1 and MH2) connected by a more divergent linker region, plus a C-terminal SSXS motif that serves as the target of receptor-mediated phosphorylation (Massagué 2000). In the presence of TGF-β, Smad2 and Smad3 undergo a dynamic recycling process involving Smad activation, nuclear translocation, transcriptional control of target genes, and signal termination by Smad dephosphorylation or degradation (Inman et al. 2002; Schmierer and Hill 2005; Lin et al. 2006; for review, see Heldin et al. 1997; Heldin and ten Dijke 1999; Massagué 2000; Schilling et al. 2006). All these steps are tightly controlled to ensure that Smad2/3 transmit signals from the plasma membrane to the nucleus in a manner that faithfully reflects the strength and duration of ligand stimulation. However, a complete understanding is still lacking with respect to how Smad2 and Smad3 are regulated at their steady state prior to receptor-mediated activation, and how such regulations predetermine cellular sensitivity to TGF-β in a cell type- and tissue-specific manner.
Despite the extensive sequence similarity and functional overlaps, Smad2 and Smad3 play distinct roles in the regulation of certain genes and may be differentially regulated. Smad3 controls a broader array of genes than Smad2, and genetic depletions of Smad2 and Smad3 in mice create remarkably different phenotypes (Piek et al. 2001; Kretschmer et al. 2003; Dunn et al. 2005; for review, see Weinstein et al. 2000; Roberts et al. 2006). One explanation for these distinctions lies in the unique structure of the MH1 domain of Smad2, which contains a 30-amino-acid insert (encoded by Exon 3) that is absent in Smad3 (Dennler et al. 1999; Yagi et al. 1999). This sequence prevents Smad2 from directly binding to DNA and many other proteins (Shi et al. 1998; Jayaraman and Massagué 2000).
As a scaffolding protein, Axin has been functionally defined as a negative regulator of the Wnt/β-catenin pathway. In the absence of Wnt ligand, Axin interacts with adenomatous polyposis coli (APC), glycogen synthase kinase 3 (GSK3-α/β), and casein kinase I α (CKIα) to form a β-catenin destruction complex (van Es et al. 2003). Within this complex, GSK3 phosphorylates multiple Ser/Thr residues on β-catenin following CKIα-mediated priming phosphorylation, consequently triggering the ubiquitination and proteasomal degradation of β-catenin. Intriguingly, both Axin and GSK3-β have been found to complex with Smad3 (Furuhashi et al. 2001; Jian et al. 2006; Liu et al. 2006), although the physiologic significance of such interactions remains to be fully elucidated.
Here, we show that nonactivated Smad3, but not Smad2, undergoes ubiquitin-dependent proteasomal degradation. In a manner reminiscent of the β-catenin destruction complex, Axin and GSK3-β cooperatively modulate Smad3 protein turnover and consequently determine cellular sensitivity to the TGF-β signal.
Results
Polyubiquitination and proteasomal degradation of nonactivated Smad3
Previous studies indicate that the basal level of Smad3 protein varies in different cell types, and Smad3 protein stability is subject to regulation at the steady state (Inoue et al. 2004; Waddell et al. 2004; Kim et al. 2005; our unpublished data). To further investigate this issue, we first examined Smad3 protein turnover by blocking protein synthesis and/or proteasomal degradation. Two commonly used TGF-β-responsive cell lines, the human keratinocyte HaCaT and hepatocellular carcinoma (HCC) HepG2 cells, were pretreated with a specific TβRI kinase inhibitor, SB-431542, to block any autocrine TGF-β activity. Under this condition, addition of the protein synthesis inhibitor cycloheximide (CHX) caused a remarkable decrease of Smad3 in both cell types, whereas treatment with the 26S proteasome inhibitor MG-132 led to Smad3 accumulation (Fig. 1A; data not shown). In contrast, Smad2 level was barely affected by these drugs (Fig. 1A; data not shown). Similar results were seen with 35S-labeled Smad2 and Smad3 in a pulse-chase assay (Supplemental Fig. S1A). Consistently, we found that endogenous Smad3 is polyubiquitinated in HaCaT cells treated with MG-132 (Fig. 1B). The results demonstrate that Smad3 is actively degraded by the proteasomes in the absence of TGF-β, and that Smad2 and Smad3 are differentially regulated at the steady state in spite of their high homology.
Figure 1.
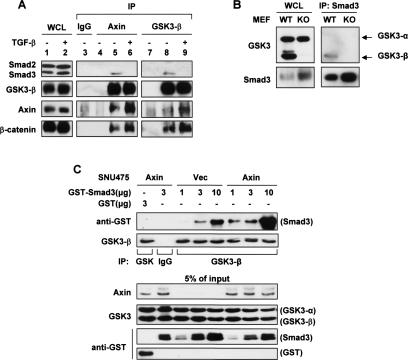
Constitutive proteasomal degradation of steady-state Smad3 is regulated by Axin. (A) HaCaT cells were pretreated with 10 μM SB-431542 for 6 h, and 50 μg/mL CHX or 25 μM MG-132 was then added as indicated. Total cell lysates were probed for endogenous Smad3 and Smad2. γ-Tubulin was used as a loading control. (B) A ubiquitination assay of endogenous Smad3. HaCaT cells were pretreated with either vehicle (ethanol) or 10 μM SB-431542 for 1 h before the addition of 30 μM MG-132. After another 3 h, cells were lysed in SDS lysis buffer for immunoprecipitation (IP) with no antibody (−) or anti-Smad3 (Zymed). (C) Retrovirally infected stable populations of SNU475 cells expressing vector control or wild-type hAxin were pretreated with 10 μM SB-431542 and then treated with 50 μg/mL CHX for the indicated lengths of time. Endogenous proteins were probed. (D) Quantification of endogenous Smad3 level in C and in Alexander and DLD-1 cells tested in similar experiments with the presence of SB-431542. The level of Smad3 in each cell line at time “0” was set as 1.0. (E) Effective knockdown of endogenous Axin in 293T cells by two shRNAs designated as R1 and R2. pSuper-GFP was used as a nontargeting shRNA control. (F,G) Smad3 ubiquitination assays in SNU475 stable lines (F) and in 293T cells (G). pSuper-GFP was used as a control for Axin RNAi (R1 + R2). Cells were pretreated with 10 μM SB-431542 for 1 h followed by MG-132 treatment (25 μM) for 4 h and then lysed in RIPA buffer. Note in G that endogenous hAxin migrates faster than myc-mAxin due to smaller size.
Axin negatively regulates Smad3 basal stability by promoting its ubiquitination
To determine the mechanism through which Smad3 protein stability is regulated at the steady state, we investigated candidate regulators of Smad3 degradation. The scaffolding protein Axin is known to play an important role in β-catenin degradation and was demonstrated to bind Smad3 only in the absence of TGF-β (Furuhashi et al. 2001; Liu et al. 2006). Similarly, we reported that certain Axin-associated kinases, such as CKIs and GSK3-β, also preferentially bind Smad3 in its nonactivated form (Waddell et al. 2004; Jian et al. 2006). These observations suggest that Smad3 basal stability may be regulated by Axin.
To study the role of Axin in Smad3 basal turnover, we first examined three human cancer cell lines that are known to harbor loss-of-function mutations in the AXIN1 gene. SNU475 is a human HCC cell line with homozygous deletions of exons 1 and 2 of AXIN1 and therefore has no Axin expression. Alexander (also termed PLC/PRF/5) is another HCC line in which the exon 4 of AXIN1 (encoding the GSK3-β-binding sequence) is lost (Satoh et al. 2000). The colon cancer cell line DLD-1 has lost one allele of AXIN1, and the remaining allele contains a point mutation, L396M. The resulting mutant Axin is unable to bind GSK3-β and has a dominant-negative effect on β-catenin degradation (Webster et al. 2000). In all three cell lines, the half-life of endogenous Smad3 protein was significantly longer than that observed in cell lines without AXIN1 mutations, such as HaCaT and HepG2 (Fig. 1C,D, cf. A). When exogenous wild-type (WT) Axin was re-expressed in these cells at a physiological level, the efficient turnover of Smad3 was effectively restored (Fig. 1C,D). This change in protein stability was also observed with ectopically expressed Smad3 (data not shown). However, the basal stability of Smad2 was not affected by Axin (Fig. 1C; data not shown), consistent with its high stability observed in HaCaT and HepG2 cells. In addition, strong ubiquitination of Smad3 was detected in Axin-reconstituted SNU475 cells but not in the SNU475-Vec cells (Fig. 1F). To determine the effect of acute loss of Axin on Smad3, we designed two independent shRNAs targeting human Axin (Fig. 1E). Knockdown of the ectopically expressed Axin in SNU475-Axin cells almost completely suppressed Smad3 ubiquitination (Fig. 1F). Conversely, in 293T cells expressing these same shRNAs, the reduced Smad3 ubiquitination was fully restored to its original level by simultaneous expression of mouse Axin, which is functionally interchangeable with human Axin but resistant to the shRNAs (Fig. 1G). These data strongly suggest that the basal ubiquitination and degradation of Smad3 are tightly and specifically controlled by Axin in several different cell lines.
Axin modulates cellular sensitivity to TGF-β
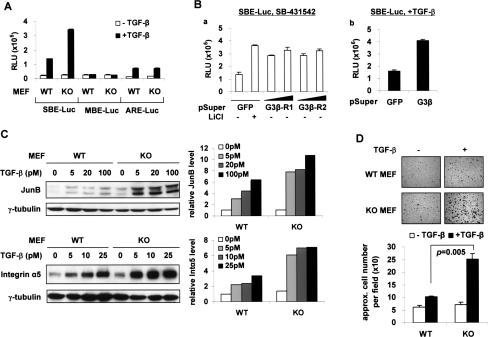
We next evaluated the impact of altering Axin expression on Smad3 transcriptional activity. Down-regulation of Axin by shRNAs in HepG2 and 293T cells increased both the basal and TGF-β-induced luciferase reporter activity of SBE-Luc (Smad3-specific) (Lin et al. 2003) and plasminogen activator inhibitor-1 (PAI-1)-Luc (Smad3-responsive) (Wrana et al. 1992), but not MBE-Luc (the mutant control of SBE-Luc) or ARE-Luc (Smad2-specific when used with FoxH1/Fast-1) (Piek et al. 2001; Fig. 2A; Supplemental Fig. S2D; data not shown). These data are consistent with the ability of Axin to accelerate Smad3 basal turnover and suggest a negative role for Axin in TGF-β signaling, specifically through the regulation of Smad3. Indeed, Axin depletion facilitated Smad3 activation by TGF-β without affecting the level and activity of TβRI or Smad2 (Supplemental Fig. S2A,B,E). Furthermore, TGF-β-induced expression of a series of endogenous Smad3 target genes (fibronectin, NET1, PAI-1, JunB, ATF3, and p15) was further enhanced by Axin RNAi in HaCaT cells (Fig. 2B). Similar results were also observed in the human breast cancer MDA-MB-231 cells (data not shown). Among these genes, fibronectin (FN) and NET1 have been shown to participate in cell migration (Shen et al. 2001; Guo and Giancotti 2004). Correspondingly, in a wound-healing/scratch assay, HaCaT cells with Axin knockdown showed a greater increase in cell motility in response to TGF-β than control cells, especially when treated with low doses of the ligand (Fig. 2C). Other Smad3 target genes such as p15 and ATF3 have been shown to mediate TGF-β/Smad3-induced growth inhibition (Hannon and Beach 1994; Kang et al. 2003a). In agreement with this, Axin RNAi rendered HaCaT cells more sensitive to the cytostatic effect of TGF-β (Fig. 2D). One potential caveat is that lowering Axin expression could simultaneously stabilize Smad3 and β-catenin, which can coregulate certain genes (Labbe et al. 2000, 2007; Jian et al. 2006). To separate the effects of Smad3 from β-catenin on these TGF-β-induced biological responses, we performed parallel experiments in HaCaT cells that were either stably expressing shRNAs against APC or pretreated with Wnt-3A (data not shown). Although the β-catenin level was elevated in both cases, none of the Smad3-mediated responsiveness shown in Figure 2, B and C, was affected. Therefore, the overall sensitization of HaCaT cells to TGF-β by Axin RNAi primarily results from enhancement of Smad3 activity.
Figure 2.
Axin negatively affects Smad3-mediated TGF-β activity. (A) Luciferase reporter assays in HepG2 cells. The indicated luciferase constructs and pCMV-β-galactosidase (0.5 μg each) were cotransfected with a total of 3 μg of the pSuper plasmids into each well of a six-well plate, except that, in panel a, 3 and 6 μg of pSuper-Ax-R1 or pSuper-Ax-R2 were used. In panels b and c, mixtures of the two corresponding pSuper-R1 and pSuper-R2 constructs were used, 1.5 μg each. Twenty-four hours post-transfection, cells in panels b and c were incubated with or without 100 pM TGF-β for another 16–24 h. (B) HaCaT cells were infected twice with pSuperRetro viruses. pSR-GL2 targets luciferase and was used as a negative control. Cells were treated with 50 pM TGF-β for 4 h. The protein level of each Smad3 target gene was quantified on the right. (C) HaCaT cells prepared as in B were tested in a wound-healing assay. Cell migration was quantified on the right. (*) P < 0.01. (D) A cell proliferation assay of the indicated HaCaT cells treated with 0, 2.5, 10, 25, or 50 pM TGF-β for 12 h. The P-values between the two types of cells under each concentration of TGF-β are shown. (E) Axin knockdown in HCT116 + Chr.3 and TGF-β treatment were performed as in B. (F) A wound-healing assay of HCT116 + Chr.3 cells. Cells were scratched at 80% confluence, treated with 200 pM TGF-β, and allowed to migrate for 24 h.
In order to further dissect the role of Axin in restricting TGF-β/Smad3 activity, we extended the functional assays to other cell lines. Both the human colon cancer cell line, HCT116 + Chr.3, and HepG2 cells express constitutively active forms of mutant β-catenin that are beyond the regulation of Axin (Morin et al. 1997; Satoh et al. 2000). In the case of HCT116 cells, reconstitution of Chromosome 3 restores TβRII expression and TGF-β responsiveness (Ilyas et al. 1999; data not shown). In both cell types, Axin down-regulation again facilitated the induction of Smad3 target genes (including PAI-1, JunB, CTGF [connective tissue growth factor], and ATF3), the C-terminal phosphorylation of Smad3 (P-Ser423/425), and/or cell motility by TGF-β (Fig. 2E,F; Supplemental Fig. S2B,C). These results demonstrate that Axin can modulate TGF-β signaling independently of β-catenin. As a complementary approach, we assessed TGF-β/Smad3 activity in Alexander and DLD-1 cells with or without re-expression of Axin. In Alexander cells, restoration of wild-type Axin (human or Xenopus) dampened Smad3 activation and hence TGF-β-induced growth arrest (Supplemental Fig. S3A–C; data not shown). Interestingly, this inhibitory effect was also seen with two XAxin mutants, L473A/D474A and H476A (Xing et al. 2003), which are defective in β-catenin binding but can interact with Smad3 and promote Smad3 ubiquitination (Supplemental Fig. S3B,C; data not shown). Reintroducing wild-type Axin into DLD-1 cells completely blocked TGF-β-stimulated, but not spontaneous, cell migration (Supplemental Fig. S3D). Taken together, results from these functional assays provide strong evidence that Axin imposes a broad, negative control on cellular sensitivity to TGF-β through facilitating Smad3 basal turnover, an event that is separate from β-catenin regulation.
GSK3-β associates with nonactivated Smad3
Since Axin is a scaffolding protein without known catalytic activity, we postulated that Axin regulates Smad3 stability by forming a degradation complex with other proteins such as the kinase GSK3. To test this hypothesis, we examined a human Axin mutant, L392P. This single amino acid mutation, which is similar to that found in the DLD-1 cells (L396M), specifically disrupts the interaction between Axin and GSK3 (Dajani et al. 2003). The results show that hAxin(L392P) failed to promote Smad3 ubiquitination, although it still bound Smad3 (Supplemental Fig. S4A,B). This suggests that Axin depends on GSK3 to modulate Smad3 turnover.
In order to determine the relationship between GSK3 and Smad3, we tested whether these proteins could form a complex. Endogenous Smad3 of HaCaT cells was found to coimmunoprecipitate with GSK3-β as well as Axin, which was detected only if the cells were pretreated with MG-132 due to the relatively low levels of Smad3 and Axin (Fig. 3A, lanes 5,8). Consistent with previous reports (Furuhashi et al. 2001; Jian et al. 2006), TGF-β caused the release of Smad3 from Axin and GSK3-β (Fig. 3A, lanes 6,9) without affecting the integrity of the β-catenin/Axin/GSK3-β complex. In contrast, endogenous Smad2 was not found in complex with either Axin or GSK3-β under the same conditions, regardless of TGF-β treatment (Fig. 3A), which may account for the high stability of Smad2 protein (see Discussion). Furthermore, endogenous Smad3 of the examined cell lines preferentially interacted with GSK3-β as opposed to GSK3-α (Fig. 3B; data not shown), suggesting that GSK3-β is more relevant in Smad3 regulation.
Figure 3.
GSK3-β interacts with nonactivated Smad3. (A) HaCaT cells were pretreated with 50 μM MG-132 for 4 h and with or without 100 pM TGF-β for the last 2 h. Cells were then lysed in ULB+ for endogenous coimmunoprecipitation assays using either preimmune goat IgG or the indicated polyclonal antibodies. Smad2 and Smad3 were probed together with anti-Smad123(H-2). In lanes 4 and 7, ULB+ alone (no lysate) was used as a negative control. (B) Endogenous Smad3 was precipitated from wild-type (WT) and GSK3-β KO MEFs using an anti-Smad3 antibody (Zymed). Coprecipitated GSK3 isoforms were probed. (C) SNU475-Vec or SNU475-Axin cell lysates (1 mg per reaction) were first mixed with 3 μg of GST alone or 1, 3, or 10 μg of GST-Smad3 for 2 h at 4°C and then subjected to anti-GSK3-β immunoprecipitation. GSK3-β did not bind the GST moiety.
To probe the relationships between Smad3, GSK3-β, and Axin, we mixed purified GST-Smad3 with whole-cell lysates of SNU475-Vec or SNU475-Axin cells. Importantly, when Axin was present, the interaction between recombinant Smad3 and endogenous GSK3-β was much stronger (Fig. 3C). This scaffolding function of Axin was further substantiated by gel chromatography analysis of whole-cell extracts from Alexander-Vec and Alexander-Axin cells. In the absence of Axin, GSK3-β exists primarily as monomers. With Axin reconstitution, a considerable amount of GSK3-β shifted to fractions of higher molecular weight, in which Smad3 and/or β-catenin were also detected (data not shown). These results indicate that, in the absence of TGF-β, Axin strengthens the interaction between GSK3-β and Smad3 via direct binding with both proteins, which may be important for facilitating Smad3 ubiquitination.
GSK3-β kinase activity is required for Smad3 basal turnover
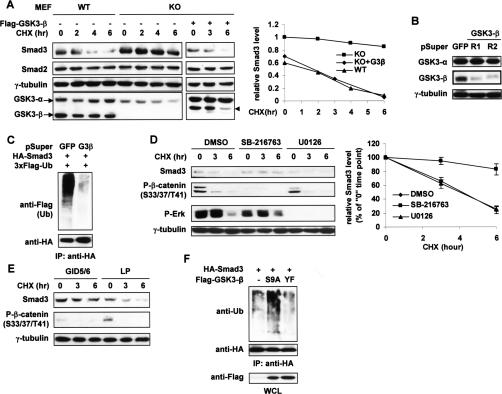
The above results prompted us to define the role of GSK3-β in Smad3 basal turnover. To this end, we compared Smad3 stability in paired mouse embryonic fibroblasts (MEFs) derived from wild-type and GSK3-β knockout (KO) littermates (Hoeflich et al. 2000). Being 100% identical with human Smad3 in amino acid sequence, mouse Smad3 also underwent basal turnover at a similar rate in wild-type MEFs (Fig. 4A). Intriguingly, in GSK3-β-null MEFs, where GSK3-α is abundant and fully active (data not shown), the steady-state level of endogenous Smad3 protein was not only higher than that in wild-type MEFs, but also remained essentially unchanged during the course of CHX treatment (Figs. 4A, 3B). When wild-type GSK3-β was introduced into the null cells, the abundance of endogenous Smad3 decreased, and the rate of its turnover increased, both to the levels seen in the wild-type MEFs (Fig. 4A). The same phenomena were observed with ectopically expressed Smad3 in the paired MEFs (data not shown). These data, together with Figure 3B, clearly suggest that GSK3-β, but not GSK3-α, plays a critical role in maintaining nonactivated Smad3 protein at a specific level by modulating its turnover. Notably, mouse Smad2 (>99% identical with human Smad2) was equally abundant and stable in both MEF lines (Fig. 4A), once again supporting our postulation that the two Smad proteins are differentially regulated at their nonactivated state.
Figure 4.
GSK3-β kinase activity is required for Smad3 basal degradation. (A) Wild-type (WT) and GSK3-β-null MEFs were treated with 50 μg/mL CHX for the indicated time course. Transfected Flag-GSK3-β(WT) is indicated (arrowhead). Protein levels of endogenous Smad3 were analyzed by Western blot and quantified on the right. Smad3 level in the KO cells at time “0” was defined as 1.0. (B) Specific knockdown of GSK3-β in 293T cells by two shRNAs designated as R1 and R2. (C) 293T cells were pretransfected twice with a mixture of the two shRNAs against GSK3-β. After treatment with 10 μM SB-431542 and 25 μM MG-132 for 3 h, cells were lysed in RIPA buffer for the Smad3 ubiquitination assay. (D) HaCaT cells were treated with DMSO, SB-216763 (10 μM), or U0126 (10 μM) for a total of 12 h and with 20 μg/mL CHX for the time course shown. The turnover of endogenous Smad3 was determined by Western blot (left, representative of three independent experiments) and quantified for each treatment group (right). Phospho-β-catenin and phospho-Erk were also examined to show the efficacy of the kinase inhibitors. (E) 293T cells were transfected twice with myc-GID5/6 or the mutant control (myc-GID5/6 LP) before CHX treatment. Endogenous Smad3 and phospho-β-catenin were probed. (F) HA-Smad3 was overexpressed in 293T cells with vector or GSK3-β mutants (S9A or Y216F). After MG-132 treatment (30 μM, 3 h), cells were harvested in RIPA buffer for the Smad3 ubiquitination assay.
We next undertook three different ways to perturb the function of endogenous GSK3-β in human cells and assessed the subsequent impact on Smad3 turnover. First, shRNA-mediated GSK3-β knockdown by two independent targeting sequences significantly reduced Smad3 ubiquitination and almost completely blocked its basal turnover in 293T cells (Fig. 4B,C; data not shown), recapitulating the results from the MEFs. Second, pretreatment of HaCaT cells with a potent and selective GSK3-β inhibitor, SB-216763, prevented Smad3 degradation (Fig. 4D). In contrast, even though Smad3 can be phosphorylated by Erk1/2 and CDKs (Matsuura et al. 2004, 2005), neither the MEK inhibitor U0126 nor the CDK inhibitor Riscovitine had any effect on Smad3 stability (Fig. 4D; data not shown), confirming the specificity of the GSK3-β inhibitor in blocking Smad3 turnover. As a third approach, GID-5/6, a small polypeptide derived from the GSK3-β-binding domain of Axin, was overexpressed in 293T cells to displace Axin from GSK3-β (Hedgepeth et al. 1999). Smad3 was stabilized by this peptide inhibitor but not the control peptide GID-5/6 LP, which contains the aforementioned L392P mutation and is unable to compete for binding with GSK3-β (Fig. 4E). As iterated before, the stability of Smad2 was unaffected by any of these treatments (data not shown). Given that Axin is wild type and functional in all the above cell lines, we conclude that this scaffolding protein is not sufficient but requires the cooperation of GSK3-β to regulate Smad3 turnover. As a complementary approach to the loss-of-function strategy, we also examined the effects of GSK3-β mutants on Smad3 by overexpression. Shown in Figure 4F, Smad3 ubiquitination was enhanced by the constitutively active form, GSK3-β(S9A) (Cross et al. 1995), but not the kinase-deficient mutant, GSK3-β(Y216F) (Hughes et al. 1993). Taken together, these data demonstrate that catalytically active GSK3-β, in conjunction with Axin, is responsible for the constant turnover of nonactivated Smad3.
Selective potentiation of Smad3 activity in GSK3-β−/− MEFs
To gauge the functional consequence of the link between Smad3 and GSK3-β, we next determined whether the GSK3-β−/− MEFs, which have a higher level of Smad3 protein, would show increased responsiveness to TGF-β as compared with the wild-type MEFs. Indeed, upon TGF-β treatment, the C-terminal phosphorylation and nuclear translocation of Smad3 were more prominent in the GSK3-β-null cells than in the wild-type cells, while this difference was not observed with Smad2 (Supplemental Fig. S5). Accordingly, TGF-β-induced SBE-Luc luciferase activity was significantly higher in the absence of GSK3-β, but the Smad2-dependent ARE-Luc activity was induced in a similar pattern in both MEF lines (Fig. 5A). This result was reproduced in HepG2 cells treated with GSK3-β shRNAs or LiCl, a generic inhibitor of GSK3-β (Figs. 5B, 2A, panel c). In light of the undisturbed state of Smad2 signaling, GSK3-β does not appear to interfere with the functions of TGF-β receptors or the Smad2/Smad4 complex, but selectively controls Smad3 activity.
Figure 5.
Selective potentiation of Smad3 activity in GSK3-β−/− MEFs. (A) MEFs were transfected with the indicated reporters (0.3 μg of SBE/MBE or 0.5 μg of ARE + FoxH1 per well of six-well plates) for 24 h followed by 100 pM TGF-β treatment for 12 h. (B) HepG2 cells were transfected as in Figure 2A. Three micrograms and 6 μg of pSuper-GSK3-β-R1 or pSuper-GSK3-β-R2 were used in panel a, and a mixture of both shRNAs (1.5 μg each) was used in panel b. LiCl (20 mM) and TGF-β (100 pM) treatments both lasted for 24 h. (C) Wild-type (WT) and KO MEFs were treated with different concentrations of TGF-β for 2 h and harvested in ULB+. Protein expressions were analyzed by Western blot (left) and quantified (right). (D) A cell adhesion assay of the MEFs. The numbers of attached cells were quantified and presented as mean ± SD.
Subsequently, we assessed the expression of two endogenous Smad3 target genes, JunB and Integrin α5 (Kang et al. 2003a), in the wild-type and GSK3-β−/− MEFs. We found that both genes were considerably more sensitive to TGF-β stimulation in the GSK3-β−/− cells (Fig. 5C). As a functional consequence, and consistent with the important role of Integrin α5 in mediating the cell–ECM interaction (Guo and Giancotti 2004), the GSK3-β−/− MEFs displayed far more efficient reattachment to the substratum than wild-type cells following TGF-β treatment when tested in a cell adhesion assay (Fig. 5D).
All these data strongly argue for the notion that stabilization of Smad3 protein can increase the probability of its activation by the TGF-β receptors, thereby providing a molecular basis for the hypersensitivity to TGF-β signaling exhibited by the GSK3-β-deficient cells.
GSK3-β phosphorylates Smad3 at Thr66
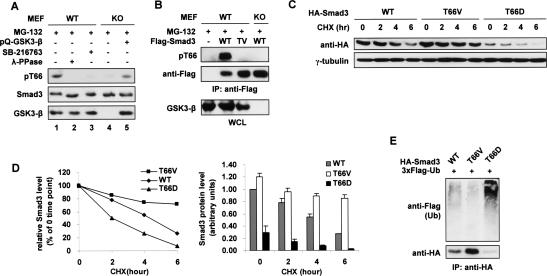
The requirement for GSK3-β kinase activity in the turnover of nonactivated Smad3 and the physical interaction between these two proteins led us to examine whether Smad3 is a direct substrate of this kinase, since GSK3-β has been shown to trigger the ubiquitination of several proteins by direct phosphorylation (Jope and Johnson 2004). In the initial test, in vitro kinase assays indicated that Smad3 is a reasonable substrate of GSK3-β (Supplemental Fig. S6). Subsequent mass spectroscopy (MS) and immunoblotting analyses identified Thr66 as a GSK3-β phosphorylation site both in vitro and in vivo (Fig. 6A,B; data not shown). Thr66 is located in the lysine-rich MH1 domain of Smad3, and its surrounding sequence (TKCITIPRS) conforms to a GSK3-β consensus motif, S/TxxxS/T (Cohen and Frame 2001). In the presence of MG-132, endogenous Smad3 was found to be phosphorylated at Thr66 in GSK3-β+/+ MEFs, HaCaT cells, and HepG2 cells, while this phosphorylation was strongly inhibited by the GSK3-β inhibitor, SB-216763 (Fig. 6A, lanes 1–3; data not shown). In contrast, Thr66 phosphorylation was barely detectable in GSK3-β-null MEFs, but could be restored by introduction of exogenous wild-type GSK3-β (Fig. 6B [lanes 4,5], B). Moreover, phosphorylation of Thr66 was completely abolished when this site was mutated to valine (T66V) (Fig. 6B).
Figure 6.
GSK3-β-mediated Thr66 phosphorylation leads to Smad3 basal degradation. (A) Endogenous Smad3 is phosphorylated at Thr66. GSK3-β wild-type (WT) and KO MEFs were treated with 20 μM MG-132 for 3 h, and cell lysates were probed with the anti-pT66 antiserum. (Lane 3) SB-216763 (10 μM) was added 10 h prior to MG-132 treatment. (Lane 5) pQCXIP-GSK3-β(WT) was transfected 24 h prior to cell lysis. (B) GSK3-β wild-type (WT) and KO MEFs were transfected with 3xFlag-Smad3(WT or T66V) and treated with MG-132 as in A. Cells were harvested in RIPA buffer followed by anti-Flag IP. (C) Equal amounts of HA-tagged Smad3 constructs (WT, T66V, or T66D) were individually expressed in 293T cells for 20 h before the indicated CHX treatment (50 μg/mL). The Western blot is representative of three independent experiments, and the results from these experiments are quantitatively presented in D to show the turnover rate (left) and the protein level (right) of the Smad3 variants. (E) 293T cells transfected with the indicated constructs were treated with 30 μM MG-132 for 3 h and lysed in SDS lysis buffer for the ubiquitination assay.
The phosphorylation status of Thr66 directly correlates with Smad3 stability
In order to define the physiologic significance of Thr66 phosphorylation, we first compared the protein stability of wild-type Smad3, the phospho-deficient mutant Smad3(T66V), and the phospho-mimicking mutant Smad3(T66D). When the same amount of each DNA construct was expressed in 293T cells, the Smad3(T66V) protein was far more stable, hence resulting in a higher basal level than wild-type Smad3. On the other hand, the Smad3(T66D) mutant expressed poorly (∼70% less than wild type) with a significantly shortened half-life (Fig. 6C–E). Consistently, the T66D mutant was strongly ubiquitinated at the steady state, which was in contrast to the very weak ubiquitination of the T66V mutant (Fig. 6E). These observations strongly support our hypothesis that, by phosphorylating Thr66, GSK3-β directly controls Smad3 protein stability, and that Thr66 phosphorylation is necessary and probably sufficient for effective basal degradation of Smad3. Further investigations indicated that Smad3(T66V), as compared with wild-type Smad3, exhibited proper subcellular localization and identical affinity to Axin, GSK3-β, Smad4, and a DNA probe containing the SBE sequence (data not shown). Therefore, Smad3(T66V) appears otherwise indistinguishable from wild-type Smad3 except for its increased protein stability.
Smad3(T66V) is hyperactive in mediating TGF-β effects
We next determined whether these Smad3 variants (WT, T66V, and T66D) with different protein stability would accordingly exhibit different levels of activity. In HepG2 cells, ectopic expression of Smad3(WT) caused growth arrest in a dose-dependent manner. This effect was more evident with Smad3(T66V) expression, even though cells were transfected with lesser amounts of DNA (Fig. 7A). Similar results were obtained from SBE-Luc reporter assays in 293T cells (data not shown). Therefore, an increase of Smad3 protein stability indeed augments its overall transcriptional activity. We then performed the wound-healing assays in HaCaT cells and cell adhesion assays in wild-type MEFs. In both cases, Smad3(T66V)-transduced cells displayed greater responses to TGF-β than those transduced with equivalent amounts of Smad3(WT). As expected, the expression of Smad3(T66D) was always low in these experiments and failed to cause a change in TGF-β responsiveness (data not shown).
Figure 7.
Smad3(T66V) is hyperactive. (A) HepG2 cells seeded in 12-well plates were transfected with the indicated amounts of 3xFlag-Smad3(WT or T66V). Cells were treated with or without TGF-β (200 pM) for 12 h and assessed for proliferation. The data are presented as the percentage of growth inhibition compared with vector-transfected, untreated cells. (B) An equal amount of vector, HA-Smad3(WT), or HA-Smad3(T66V) was transfected into GSK3-β wild-type (WT) and KO MEFs. Twenty-four hours later, cells were treated with 25 pM TGF-β for 4 h. (Right) The protein levels of Integrin α5 and JunB were quantified. (C) SMAD3−/− MEFs were transfected with 1 μg of vector control or 3xFlag-Smad3(WT, T66V, or T66D). Twelve hours post-transfection, 10 μM SB-216763 or DMSO was added. Cells were harvested in ULB+ after another 12 h. (D) An SBE-Luc reporter assay in SMAD3−/− MEFs. After transfection of the luciferase construct, cells were incubated with or without 20 mM LiCl for 24 h. (E) A schematic representation of the Smad3/Axin/GSK3-β complex (right) in comparison with key components of the β-catenin destruction complex (left). Unlike Wnt, TGF-β does not disassemble the Axin/GSK3-β complex. The rapid nuclear translocation of Smad3 is caused by the C-terminal phosphorylation rather than protein accumulation.
We subsequently asked whether such difference in activity between wild-type and mutant Smad3 was primarily determined by the status of GSK3-β, and whether GSK3-β regulates Smad3 stability/activity solely through T66 phosphorylation. To answer the first question, we transfected GSK3-β+/+ and GSK3-β−/− MEFs with identical amounts of either Smad3(WT) or Smad3(T66V) and compared their activity in inducing the expression of JunB and Integrin α5. In the GSK3-β+/+ MEFs, Smad3(T66V) was more abundant (and therfore more active) than Smad3(WT), as seen in other cell types. However, in GSK3-β-null MEFs, these two forms of Smad3 expressed at similar levels with almost identical activity that was comparable with that mediated by Smad3(T66V) in GSK3-β+/+ MEFs (Fig. 7B). This result indicates that the distinction between Smad3(WT) and Smad3(T66V) disappeared in the absence of GSK3-β. To address the second question, we introduced different forms of Smad3 into SMAD3−/− MEFs so that their activity could be assessed without interference from endogenous Smad3. As expected, the activity of Smad3(WT) was increased upon GSK3-β inhibition in these cells, whereas neither Smad3(T66V)- nor Smad3(T66D)-induced responses were affected (Fig. 7C,D). Therefore, GSK3-β-mediated regulation of Smad3 stability and its target genes such as JunB and Integrin α5 is dependent on Thr66 phosphorylation.
Taken together, these data clearly demonstrate that phosphorylation of Smad3-Thr66 by GSK3-β controls Smad3 protein stability at the steady state, which, in turn, predetermines Smad3 activity.
Discussion
Distinct protein stability of Smad2 and Smad3 at the steady state
In this study, we unexpectedly found that Smad3, but not Smad2, is actively degraded by the proteasome at the steady state. The rapid turnover of nonactivated Smad3 may reflect the necessity for the cells to more stringently control the level and activity of this protein, since Smad3 has the intrinsic ability to bind DNA that Smad2 lacks, and Smad3 is generally more versatile than Smad2 in transducing TGF-β signals. In fact, it was recently reported that the protein level of steady-state Smad3 is four to five times lower than that of Smad2 in most mammalian cells (Clarke et al. 2006). At the mechanistic level, such differential control of Smad2 and Smad3 is made possible by the selective interaction between Smad3 and Axin/GSK3-β. Using chimerical Smad mutants, we confirmed that the Smad2-MH1 domain interfered with Smad-GSK3-β binding, while the Smad3-MH1 domain was responsible for rapid protein degradation at the steady state (Supplemental Fig. S8C; data not shown). In particular, a splice isoform of Smad2, Smad2(ΔExon3), which lacks the unique 30-amino-acid insert in its MH1 domain and therefore closely resembles Smad3 (Jayaraman and Massagué 2000; Dunn et al. 2005), showed a similar rate of basal degradation as Smad3 (Supplemental Fig. S8B). These results strongly suggest that the unique structure of the Smad2-MH1 domain is the primary reason for the high protein stability of Smad2 at the steady state.
A novel role for Axin and GSK3-β in Smad3 basal turnover
By functional manipulations of Axin and GSK3-β, we could alter the rate of Smad3 basal turnover and demonstrate its importance in predetermining cellular sensitivity to TGF-β. Axin and GSK3-β do not interfere with Smad2-mediated responses. Thus they should not affect the functions of TGF-β receptors, Smad2/4, and other factors shared by Smad2 and Smad3. In other words, the concerted modulation of Smad3 stability is a major, if not the only, mechanism by which Axin and GSK3-β regulate TGF-β signaling activity.
It has been reported that Axin, when overexpressed in AXIN1 wild-type cells, can play positive roles in the TGF-β pathway by either presenting Smad3 to the activated receptors (Furuhashi et al. 2001) or increasing the degradation of Smad7 (an inhibitory Smad) through recruiting the E3 ubiquitin ligase, Arkadia (Liu et al. 2006). However, the physiological level of Axin protein is usually extremely low, and Axin overproduction or AXIN1 gene amplification has not been reported in normal or cancer cells. A substantial increase of Axin might produce nonphysiological, erratic effects (Lee et al. 2003). In the present study, we rely primarily on loss-of-function assays and convincingly show that Axin is not required for Smad3 activation by the receptors. Rather, Axin negatively influences broad aspects of TGF-β/Smad3 responses. On the other hand, the biological relevance of Axin-mediated Smad7 degradation in TGF-β signaling (Liu et al. 2006) remains debatable. For instance, TβRI and Smad2 are both inhibited by Smad7 (Massagué 2000) but they are not affected by Axin. In addition, the oncoprotein SnoN was recently shown to be the primary target of Arkadia in the TGF-β pathway (Levy et al. 2007). Interestingly, however, Liu et al. (2006) showed that treatment with Wnt-1 reduced Smad7 ubiquitination, while our data indicate that Smad3 stability and activity were not affected by Wnt-3A (Supplemental Fig. S9A,B,E). It is possible that the different conclusions from the study of Liu et al. (2006) and our study may represent different modes of Axin function involving distinct molecular mechanisms. In any event, the previously reported positive effect of Axin in TGF-β signaling is likely to be secondary to the inhibitory function of Axin, which is more physiologically relevant as demonstrated by our extensive biochemical and biological analyses.
Thr66 phosphorylation by GSK3-β
GSK3-β regulates the degradation or proteolysis of a large number of proteins (for review, see Cohen and Frame 2001; Jope and Johnson 2004). Our demonstration that GSK3-β also regulates Smad3 degradation further supports the importance of this kinase as an integrator of signaling networks.
GSK3-β phosphorylates Smad3 at Thr66 in vitro and in vivo, which is necessary and probably sufficient to trigger Smad3 ubiquitination and degradation at the steady state. Thr66 is proximal to several lysine residues in the MH1 domain, which have been postulated to be involved in the basal level ubiquitination of Smad3 (Inoue et al. 2004). It is noteworthy that Thr66 is evolutionarily conserved in Smad3 from different species and in other R-Smads as well, such as Smad1 and Smad2. However, like Smad2, Smad1 was not found to physically interact with Axin or GSK3-β in unstimulated cells, and its stability at the steady state was not affected by SB-216763 (Furuhashi et al. 2001; data not shown). Therefore, the Axin/GSK3-β-mediated proteasomal degradation appears to be Smad3-specific. On the other hand, sequence lineup reveals that Thr76 of Smad2 corresponds to Thr66 of Smad3. Interestingly, phospho-mimetic mutation of Thr76 (T76D) significantly destabilized Smad2, as was seen with Smad3(T66D) (Supplemental Fig. S8B,D). However, due to the presence of the 30-amino-acid insert next to this residue, phosphorylation of Thr76 may not occur in cells, as the T76V mutation had no effect on Smad2 turnover. Taken together, these results further highlight the critical importance of GSK3-β-binding and phosphorylation in controlling Smad3 basal stability.
It should be noted that, besides Thr66, we found that Ser204 in the linker region of Smad3 was also phosphorylated by GSK3-β in vitro and in vivo (Supplemental Fig. S7A,B; data not shown). There is no corresponding GSK3 site of Ser204 in other Smads. Mutation of this site and/or its putative priming site, Ser208, did not block Smad3 basal degradation (Supplemental Fig. S8A; data not shown), consistent with an earlier report using the Smad3-EPSM mutant (Sapkota et al. 2007). Ser204 has been shown to be phosphorylated by multiple kinases but has not been reported to regulate Smad3 stability (Matsuura et al. 2004, 2005; Kamaraju and Roberts 2005). Importantly, Smad3(WT) and Smad3(T66V) are both phosphorylated at Ser204 in both GSK3-β wild-type and KO MEFs (Supplemental Fig. S7C). Therefore, the elevated Smad3 stability and activity seen in GSK3-β−/− cells were not due to loss of Ser204 phosphorylation.
With few exceptions (e.g., Zeng et al. 2005), GSK3-β almost exclusively acts on prime-phosphorylated substrates (Cohen and Frame 2001). Since GSK3-β phosphorylates Smad3 at Thr66 (P), the putative priming site at the P + 4 position would be Ser70. Our preliminary result indicates that phosphorylation of Ser70 is probably required for Thr66 phosphorylation in vivo (our unpublished data). Intriguingly, CKIα, which primes GSK3-β for β-catenin and Gli phosphorylation, also regulates Smad3 basal turnover and TGF-β signaling (our unpublished data). If we determine that CKIα prime-phosphorylates Ser70 of Smad3, the result would further support the notion that the functional coupling of CKIα and GSK3-β, aided by a scaffold protein (e.g., Axin), plays pivotal roles in both embryonic development and adult tissue homeostasis by regulating multiple essential signaling pathways such as Wnt, Shh, and TGF-β (Liu et al. 2002; Price and Kalderon 2002; Zeng et al. 2005).
The E3 ubiquitin ligase and Thr66 phosphatase
Multiple E3 ubiquitin ligases have been found to regulate Smad proteins in various ways (Izzi and Attisano 2006). We examined several of those E3 ligases and found that nonactivated Smad3 was not further ubiquitinated by overexpression of SCFβ-TrCP, Smurf1/2, or Arkadia (data not shown). Levy et al. (2007) recently reported their successful identification of Arkadia as a positive regulator of TGF-β signaling by screening an siRNA library of 289 human E3 ubiquitin ligases. Similar approaches are expected to shed light on the identity of the E3 ligase that ubiquitinates Smad3 at the steady state. In addition, mounting evidence suggests that protein phosphatases are equally critical as kinases in regulating the function of R-Smads. Several phosphatases were recently identified to dephosphorylate R-Smads at the tail and/or linker region (Chen et al. 2006; Duan et al. 2006; Knockaert et al. 2006; Lin et al. 2006; Sapkota et al. 2006). Since Thr66 phosphorylation gradually diminished upon GSK3-β inhibition or TGF-β treatment (Fig. 6A; Supplemental Fig. S9D), a phosphatase or phosphatases might exist to counteract the kinase and protect Smad3 from degradation. Identification of such phosphatase(s) will certainly help to expand our knowledge on the regulation of the TGF-β/Smad3 pathway.
Wnt and APC do not affect Smad3 stability at the steady state
The ability of Axin and GSK3-β to down-regulate nonactivated Smad3 is reminiscent of their negative role in the Wnt/β-catenin pathway. However, Smad3 turnover at the steady state was not affected by Wnt-3A. In addition, unlike Axin shRNAs, Wnt-3A did not sensitize cells to TGF-β (Supplemental Fig. 9A,B,E; data not shown). Thus, the functional cross-talk between the Wnt and TGF-β cascades does not appear to happen at the step of Smad3 turnover in resting cells, and the signaling specificity of the two pathways is well maintained. Consistent with this notion, TGF-β treatment dissociated Smad3 from Axin and GSK3-β without affecting the β-catenin/Axin/GSK3-β interaction or β-catenin stability and transcriptional activity (Fig. 3A; data not shown). Taken together, these observations and results of our gel filtration assays (data not shown) suggest that Smad3 and β-catenin may be controlled by different pools of Axin/GSK3-β-containing protein complexes that are probably differentiated by their molecular composition (Fig. 7E). In fact, we noticed that APC, which is intimately involved in β-catenin degradation, did not regulate Smad3 stability or activity in HaCaT and 293T cells (our unpublished data). The results indicate that β-catenin is not directly involved in the TGF-β/Smad3-induced biological events examined in these cell lines; and the differential requirements for APC suggest mechanistic differences between the ubiquitination of Smad3 and β-catenin.
A new anti-tumor function of Axin/GSK3-β?
The roles of TGF-β/Smad3, Axin, and GSK3-β in human diseases such as cancer have been extensively studied. TGF-β/Smad3 have long been considered as tumor suppressors in cancer development, but can also promote tumor invasion and metastasis in the advanced stage of the disease (Siegel and Massagué 2003; Bierie and Moses 2006). Smad3 target genes, such as the CTGF and PAI-1, have been shown to associate with a high risk of breast cancer metastasis (Kang et al. 2003b; Weigelt et al. 2005). In this study, both of these genes were more profoundly induced by TGF-β in cells with lowered Axin expression (Fig. 2B,E). Generally accepted as a tumor suppressor, AXIN1 has been found to be mutated in several types of cancers, resulting in stabilization of β-catenin (and Smad3) (Satoh et al. 2000; Webster et al. 2000; Salahshor and Woodgett 2005; this study). GSK3-β activity is also frequently suppressed in cancer cells due to abnormal activation of the PI3K/Akt, mTOR/S6K, and Ras/MAPK pathways (Cohen and Frame 2001). Recent investigations have shown that elevation in the expression level of Glut1, a glucose transporter protein, is commonly found in metastatic tumors (Macheda et al. 2005) and also leads to GSK3-β inhibition (Zhao et al. 2007). Indeed, we found that overexpression of either activated Akt or Glut1 caused reduction of Smad3 Thr66 phosphorylation and stabilization of Smad3 protein in a GSK3-β-dependent manner (our unpublished data). These results not only demonstrate that Thr66 phosphorylation of Smad3 is a regulated event, but also suggest a novel mechanism by which loss or reduced activity of the Axin/GSK3-β complex may contribute to tumor progression via enhancing the prometastatic activity of TGF-β/Smad3 in the late stage of cancer development.
Materials and methods
Cell culture, transfection, and retroviral infection
The culture of 293T, HaCaT, and HepG2 cells has been described previously (Waddell et al. 2004). Immortalized GSK3-β+/+ and GSK3-β−/− MEFs (generously provided by James Woodgett, Ontario Cancer Institute, Canada), SMAD3−/− MEFs (kindly provided by Rik Derynck, University of California at San Francisco), and Alexander cells (from American Type Culture Collection [ATCC]) were cultured in DMEM containing 10% FBS (Invitrogen). SNU475 cells (provided by Patrick Casey, Duke University Medical Center) and DLD-1 cells (from ATCC) were grown in RPMI1640 (Invitrogen) containing 10% heat-inactivated FBS. HCT116 + Chr.3 cells (provided by Thomas Kunkel, National Institute of Environmental Health Sciences) were grown in McCoy’s 5A medium containing 10% FBS. Transient transfection in HepG2 and 293T cells was done using the Ca3(PO4)2 precipitation method, while all MEFs and Alexander cells were transfected with Superfect (Qiagen) and SNU475 cells with Fugene 6 (Roche), according to the manufacturers’ protocols. Retroviruses (pBabe-puro, pQCXIP, and pSuperRetro) were packaged in 293T cells by transfection with the desired retroviral constructs and the pCL10A-1 helper genes. The medium was changed 24 h after transfection, and the supernatant was then collected 24 and 48 h later, filtered, and mixed with 4 μg/mL polybrene (Sigma) before addition to target cells. Stable cell populations were maintained under puromycin selection (2 μg/mL).
Plasmids, mutagenesis, and shRNAs
Full-length and fragments of GST-Smad3 were described elsewhere (Waddell et al. 2004). N-terminally tagged HA-Smad3 was generated by subcloning the Smad3 coding sequence into a pcDNA3-HA vector (made in our laboratory). Point mutations of all constructs were introduced using the QuikChange site-directed mutagenesis protocol (Stratagene). The mutagenesis primers are described in the Supplemental Material. 3xFlag-Ub(WT) was a kind gift of Michael Ehlers (Duke University Medical Center) and was originally from Adriano Marcheze and Jeffery Benovic (Thomas Jefferson University). pCAN-hAxin (wild type, myc-tagged) was a gift of Patrick Casey, and hAxin cDNA was subsequently transferred into pBabe-puro and pQCXIP (Clontech) vectors for generating retroviruses. Full-length mAxin as well as the cDNA of human GSK3-β were generously offered by Xi He (Harvard University). GSK3-β was then cloned into the pFlag-CMV-2 vector (Sigma). Axin GID5-6 and the LP control were gifts of Peter Klein (University of Pennsylvania). Design of shRNA sequences was assisted by the Web tools from Dharmacon and GenScript, and the annealed primers were ligated into pSuper or pSuperRetro vectors (Oligoengine). The RNAi targeting sequences (sense) are described in the Supplemental Material.
Antibodies and reagents
The rabbit antiserum against phospho-Thr66 was generated using the synthetic peptide VNTKCI(pT)IPRSLDGR according to the standard procedure. Integrin α5 polyclonal antibody was provided by Jun-Lin Guan (University of Michigan). The following commercially available antibodies were used for immunoblotting: Smad3, Smad2, and Axin (Zymed); Flag(M2) and γ-tubulin (Sigma); P-Smad2, P-Smad3(S423/425), and P-β-catenin (S33/37/T41) (Cell Signaling); and HA(F-7), GST(B-14), c-myc(9E10), β-catenin(E-5), JunB(C-11), PAI-1(C-9), Smad1/2/ 3(H-2), CTGF(L-20), NET1(N-17), ATF3(C-19), Ubiquitin(P4D1), GSK3-α/β(0011-A), Fibronectin(EP5), p15(C-20), and P-Erk(E-4) (Santa Cruz Biotechnology). The antibodies used for immunoprecipitation were Smad3(I-20), Axin(S-20), HA(Y-11), and GSK3-β(L-17) (Santa Cruz Biotechnology), and Smad3 (Zymed). TGF-β1 was obtained from R&D Systems. The following inhibitors were purchased from the indicated companies: SB-431542 and SB-216763 (Tocris); U0126 (Cell Signaling); Riscovitine (Calbiochem); CHX and LiCl (Sigma); and MG-132 (BioMol).
Immunoblotting and immunoprecipitation
The protocols using the universal lysis buffer (ULB+) have been described (Waddell et al. 2004), and the following modifications were made when necessary. For detection of Smad3 ubiquitination, cells were harvested in either RIPA buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, with protease and phosphatase inhibitors) or SDS lysis buffer (Lo and Massagué) containing 10 mM NEM (Sigma). For detection of Smad3 Thr66 phosphorylation, 25 nM microcystin (Sigma) was also included in the cell lysis buffer. λ-Phosphatase (New England Biolabs) treatment was done on cell lysates containing only protease inhibitors but not phosphatase inhibitors following the manufacturer’s protocol. Quantification of results was aided by the ImageJ software (http://rsb.info.nih.gov/ij).
Transcriptional reporter assay
SBE-Luc and MBE-Luc were gifts from Xin-Hua Feng (Baylor College of Medicine). ARE-Luc and FoxH1 were kindly provided by Anita Roberts (National Cancer Institute). PAI-1-Luc (p800LUC) has been reported (Frederick et al. 2004). All experiments were performed in triplicate essentially as described previously (Waddell et al. 2004), and the data were presented as average ± SD.
Cell proliferation assay
Cell proliferation was determined by [3H]-thymidine incorporation as described (Frederick et al. 2004). Specifically, cells were seeded in 12-well plates in triplicate and grown under normal conditions to no more than 40% confluence. The cells were then treated with TGF-β or vehicle for 12–24 h and labeled with 5 μCi/mL [3H]-thymidine for the last 4–5 h of treatment.
Cell migration/wound-healing assay
Cells were plated at high density into 12-well plates and grown to confluence. The scratch was made by a sterile P-200 micropipette in the middle of each well. Cells were then washed three times with PBS and incubated in regular media with or without TGF-β. Photographs of the same fields were taken at the beginning and the end of the experiments following the PBS wash. At least four fields were photographed for each condition each time, and the wound areas were calculated using ImageJ. Cell migration was defined as the reduction of the wound area in each photographed field during the course of treatment. The results were presented as mean ± SD.
Cell adhesion assays
GSK3-β wild-type and KO MEFs were treated with 100 pM TGF-β for 12–24 h, then washed and incubated with EDTA (Versine, 1:5000; Invitrogen) for 20 min at 37°C. The dissociated cells were collected in DMEM(−), centrifuged, and resuspended in DMEM(−). One milliliter of the cell suspension (2 × 105 cells) was added to each well of a 12-well plate and incubated for 15 min at 37°C. After removal of the media, the attached cells were gently washed twice with PBS and fix-stained with Toluidine Blue O (Sigma) in 4% paraformaldehyde for 24 h at room temperature. Cells were then washed three times with PBS, and at least four fields were photographed for each condition. Counting of cell number was done by ImageJ.
Acknowledgments
We thank Drs. Patrick Casey, Zhijian Chen, Michael Ehlers, Xin-Hua Feng, Jun-Lin Guan, Xi He, David Kimelman, Peter Klein, Thomas Kunkel, Joan Massagué, Kohei Miyazono, Anita Roberts, Bert Vogelstein, and James Woodgett for providing crucial materials and reagents. We thank Dr. Yigong Shi (Princeton University) for helpful discussions and suggestions, and Drs. Timothy Haystead and Manabu Kurokawa for excellent technical assistance on phospho-peptide mapping and the gel filtration assays, respectively. We are also grateful to Irwin Liu and Stephen Schilling (Wang laboratory), and Drs. Ann Marie Pendergast and Anita Hjelmeland for critical reading of the manuscript. This work was supported by NIH grants (DK064113 and GM083000) to X.-F.W.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1590908
References
- Bierie B., Moses H.L. Tumour microenvironment: TGFβ: The molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- Chen H.B., Shen J., Ip Y.T., Xu L. Identification of phosphatases for Smad in the BMP/DPP pathway. Genes & Dev. 2006;20:648–653. doi: 10.1101/gad.1384706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D.C., Betterton M.D., Liu X. Systems theory of Smad signalling. IEE Proc. Syst. Biol. 2006;153:412–424. doi: 10.1049/ip-syb:20050055. [DOI] [PubMed] [Google Scholar]
- Cohen P., Frame S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Cross D.A., Alessi D.R., Cohen P., Andjelkovich M., Hemmings B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Dajani R., Fraser E., Roe S.M., Yeo M., Good V.M., Thompson V., Dale T.C., Pearl L.H. Structural basis for recruitment of glycogen synthase kinase 3β to the axin–APC scaffold complex. EMBO J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S., Huet S., Gauthier J.M. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene. 1999;18:1643–1648. doi: 10.1038/sj.onc.1202729. [DOI] [PubMed] [Google Scholar]
- Duan X., Liang Y.-Y., Feng X.-H., Lin X. Protein serine/threonine phosphatase PPM1A dephosphorylates Smad1 in the bone morphogenetic protein signaling pathway. J. Biol. Chem. 2006;281:36526–36532. doi: 10.1074/jbc.M605169200. [DOI] [PubMed] [Google Scholar]
- Dunn N.R., Koonce C.H., Anderson D.C., Islam A., Bikoff E.K., Robertson E.J. Mice exclusively expressing the short isoform of Smad2 develop normally and are viable and fertile. Genes & Dev. 2005;19:152–163. doi: 10.1101/gad.1243205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick J.P., Liberati N.T., Waddell D.S., Shi Y., Wang X.-F. Transforming growth factor β-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 2004;24:2546–2559. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M., Yagi K., Yamamoto H., Furukawa Y., Shimada S., Nakamura Y., Kikuchi A., Miyazono K., Kato M. Axin facilitates Smad3 activation in the transforming growth factor-β signaling pathway. Mol. Cell. Biol. 2001;21:5132–5141. doi: 10.1128/MCB.21.15.5132-5141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Giancotti F.G. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- Hannon G.J., Beach D. pl5INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Hedgepeth C.M., Deardorff M.A., Rankin K., Klein P.S. Regulation of glycogen synthase kinase 3β and downstream Wnt signaling by axin. Mol. Cell. Biol. 1999;19:7147–7157. doi: 10.1128/mcb.19.10.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C.H., ten Dijke P. SMAD destruction turns off signalling. Nat. Cell Biol. 1999;1:E195–E197. doi: 10.1038/70223. [DOI] [PubMed] [Google Scholar]
- Heldin C.-H., Miyazono K., Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hoeflich K.P., Luo J., Rubie E.A., Tsao M.S., Jin O., Woodgett J.R. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- Hughes K., Nikolakaki E., Plyte S.E., Totty N.F., Woodgett J.R. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 1993;12:803–808. doi: 10.1002/j.1460-2075.1993.tb05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas M., Efstathiou J.A., Straub J., Kim H.C., Bodmer W.F. Transforming growth factor β stimulation of colorectal cancer cell lines: Type II receptor bypass and changes in adhesion molecule expression. Proc. Natl. Acad. Sci. 1999;96:3087–3091. doi: 10.1073/pnas.96.6.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman G.J., Nicolas F.J., Hill C.S. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Kitagawa M., Onozaki K., Hayashi H. Contribution of the constitutive and inducible degradation of Smad3 by the ubiquitin–proteasome pathway to transforming growth factor-β signaling. J. Interferon Cytokine Res. 2004;24:43–54. doi: 10.1089/107999004772719909. [DOI] [PubMed] [Google Scholar]
- Izzi L., Attisano L. Ubiquitin-dependent regulation of TGF signaling in cancer. Neoplasia. 2006;8:677–688. doi: 10.1593/neo.06472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman L., Massagué J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-β pathway. J. Biol. Chem. 2000;275:40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- Jian H., Shen X., Liu I., Semenov M., He X., Wang X.F. Smad3-dependent nuclear translocation of β-catenin is required for TGF-β1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes & Dev. 2006;20:666–674. doi: 10.1101/gad.1388806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jope R.S., Johnson G.V. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kamaraju A.K., Roberts A.B. Role of Rho/ROCK and p38 MAP kinase pathways in transforming growth factor-β-mediated Smad-dependent growth inhibition of human breast carcinoma cells in vivo. J. Biol. Chem. 2005;280:1024–1036. doi: 10.1074/jbc.M403960200. [DOI] [PubMed] [Google Scholar]
- Kang Y., Chen C.R., Massagué J. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 2003a;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- Kang Y., Siegel P.M., Shu W., Drobnjak M., Kakonen S.M., Cordon-Cardo C., Guise T.A., Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003b;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kim S.G., Kim H.A., Jong H.S., Park J.H., Kim N.K., Hong S.H., Kim T.Y., Bang Y.J. The endogenous ratio of Smad2 and Smad3 influences the cytostatic function of Smad3. Mol. Biol. Cell. 2005;16:4672–4683. doi: 10.1091/mbc.E05-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockaert M., Sapkota G., Alarcon C., Massague J., Brivanlou A.H. Unique players in the BMP pathway: Small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc. Natl. Acad. Sci. 2006;103:11940–11945. doi: 10.1073/pnas.0605133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer A., Moepert K., Dames S., Sternberger M., Kaufmann J., Klippel A. Differential regulation of TGF-β signaling through Smad2, Smad3 and Smad4. Oncogene. 2003;22:6748–6763. doi: 10.1038/sj.onc.1206791. [DOI] [PubMed] [Google Scholar]
- Labbe E., Letamendia A., Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-β and wnt pathways. Proc. Natl. Acad. Sci. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe E., Lock L., Letamendia A., Gorska A.E., Gryfe R., Gallinger S., Moses H.L., Attisano L. Transcriptional cooperation between the transforming growth factor-β and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- Lee E., Salic A., Krüger R., Heinrich R., Kirschner M.W. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1:e10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L., Howell M., Das D., Harkin S., Episkopou V., Hill C.S. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 2007;27:6068–6083. doi: 10.1128/MCB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Liang M., Liang Y.-Y., Brunicardi F.C., Melchior F., Feng X.-H. Activation of transforming growth factor-β signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J. Biol. Chem. 2003;278:18714–18719. doi: 10.1074/jbc.M302243200. [DOI] [PubMed] [Google Scholar]
- Lin X., Duan X., Liang Y.Y., Su Y., Wrighton K.H., Long J., Hu M., Davis C.M., Wang J., Brunicardi F.C., et al. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li Y., Semenov M., Han C., Baeg G.-H., Tan Y., Zhang Z., Lin X., He X. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Liu W., Rui H., Wang J., Lin S., He Y., Chen M., Li Q., Ye Z., Zhang S., Chan S.C., et al. Axin is a scaffold protein in TGF-β signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 2006;25:1646–1658. doi: 10.1038/sj.emboj.7601057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheda M.L., Rogers S., Best J.D. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J. Cell. Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- Massagué J. How cells read TGF-β signals. Nat. Rev. Mol. Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Matsuura I., Denissova N.G., Wang G., He D., Long J., Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- Matsuura I., Wang G., He D., Liu F. Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry. 2005;44:12546–12553. doi: 10.1021/bi050560g. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Piek E., Ju W.J., Heyer J., Escalante-Alcalde D., Stewart C.L., Weinstein M., Deng C., Kucherlapati R., Bottinger E.P., Roberts A.B. Functional characterization of transforming growth factor β signaling in Smad2- and Smad3-deficient fibroblasts. J. Biol. Chem. 2001;276:19945–19953. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- Price M.A., Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell. 2002;108:823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Roberts A.B., Tian F., Byfield S., DaCosta S.C., Ooshima A., Saika S., Flanders K.C. Smad3 is key to TGF-β-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Salahshor S., Woodgett J.R. The links between axin and carcinogenesis. J. Clin. Pathol. 2005;58:225–236. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G., Knockaert M., Alarcon C., Montalvo E., Brivanlou A.H., Massague J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-β pathways. J. Biol. Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- Sapkota G., Alarcon C., Spagnoli F.M., Brivanlou A.H., Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Satoh S., Daigo Y., Furukawa Y., Kato T., Miwa N., Nishiwaki T., Kawasoe T., Ishiguro H., Fujita M., Tokino T., et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- Schilling S.H., Datto M.B., Wang X.F. A phosphatase controls the fate of receptor-regulated Smads. Cell. 2006;125:838–840. doi: 10.1016/j.cell.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Schmierer B., Hill C.S. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Li J., Hu P.P., Waddell D., Zhang J., Wang X.-F. The activity of guanine exchange factor NET1 is essential for transforming growth factor-β-mediated stress fiber formation. J. Biol. Chem. 2001;276:15362–15368. doi: 10.1074/jbc.M009534200. [DOI] [PubMed] [Google Scholar]
- Shi Y., Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang Y.F., Jayaraman L., Yang H., Massagué J., Pavletich N.P. Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Siegel P.M., Massagué J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–820. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- van Es J.H., Barker N., Clevers H. You Wnt some, you lose some: Oncogenes in the Wnt signaling pathway. Curr. Opin. Genet. Dev. 2003;13:28–33. doi: 10.1016/s0959-437x(02)00012-6. [DOI] [PubMed] [Google Scholar]
- Waddell D.S., Liberati N.T., Guo X., Frederick J.P., Wang X.-F. Casein kinase Iε plays a functional role in the transforming growth factor-β signaling pathway. J. Biol. Chem. 2004;279:29236–29246. doi: 10.1074/jbc.M400880200. [DOI] [PubMed] [Google Scholar]
- Webster M.T., Rozycka M., Sara E., Davis E., Smalley M., Young N., Dale T.C., Wooster R. Sequence variants of the axin gene in breast, colon, and other cancers: An analysis of mutations that interfere with GSK3 binding. Genes Chromosomes Cancer. 2000;28:443–453. [PubMed] [Google Scholar]
- Weigelt B., Peterse J.L., van’t Veer L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- Weinstein M., Yang X., Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- Wrana J.L., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X.-F., Massague J. TGFβ signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Xing Y., Clements W.K., Kimelman D., Xu W. Crystal structure of a β-catenin/Axin complex suggests a mechanism for the β-catenin destruction complex. Genes & Dev. 2003;17:2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K., Goto D., Hamamoto T., Takenoshita S., Kato M., Miyazono K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- Zeng X., Tamai K., Doble B., Li S., Huang H., Habas R., Okamura H., Woodgett J., He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Altman B.J., Coloff J.L., Herman C.E., Jacobs S.R., Wieman H.L., Wofford J.A., Dimascio L.N., Ilkayeva O., Kelekar A., et al. Glycogen synthase kinase 3α and 3β mediate a glucose-sensitive antiapoptotic signaling pathway to stabilize Mcl-1. Mol. Cell. Biol. 2007;27:4328–4339. doi: 10.1128/MCB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]