Abstract
Many microRNA (miRNA) loci exhibit compelling hairpin structures on both sense and antisense strands; however, the possibility that a miRNA gene might produce functional species from its antisense strand has not been examined. We report here that antisense transcription of the Hox miRNA locus mir-iab-4 generates the novel pre-miRNA hairpin mir-iab-8, which is then processed into endogenous mature miRNAs. Sense and antisense iab-4/iab-8 miRNAs are functionally distinguished by their distinct domains of expression and targeting capabilities. We find that miR-iab-8-5p, like miR-iab-4-5p, is also relevant to Hox gene regulation. Ectopic mir-iab-8 can strongly repress the Hox genes Ultrabithorax and abdominal-A via extensive arrays of conserved target sites, and can induce a dramatic homeotic transformation of halteres into wings. We generalize the antisense miRNA principle by showing that several other loci in both invertebrates and vertebrates are endogenously processed on their antisense strands into mature miRNAs with distinct seeds. These findings demonstrate that antisense transcription and processing contributes to the functional diversification of miRNA genes.
[Keywords: BX-C, antisense, homeotic gene, microRNA]
Nearly 100 years of genetic, molecular, and biochemical studies of the Drosophila bithorax complex (BX-C) have revealed myriad insights into fundamental mechanisms of gene regulation, molecular evolution, and developmental patterning (Maeda and Karch 2006). Its most well-studied residents are the homeobox genes Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B) (Fig. 1A), which are responsible for the specification of thoracic and abdominal segments. These genes encode some of the founding members of the Hox family, which are highly conserved regulators of anterior–posterior axis development in all animals.
Figure 1.
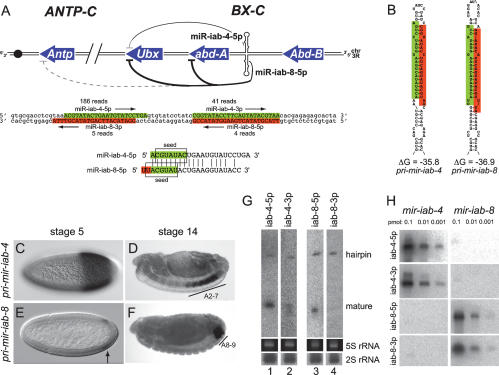
Sense and antisense transcription of the iab-4 locus in the BX-C produces mature miRNAs. (A) Map of the BX-C depicting the homeobox genes Ubx, abd-A, and Abd-B, and sense and antisense iab-4 hairpins; the Antp gene of the antennapedia complex (ANTP-C) is also shown. Targets with highly conserved “2–8” seed + t1A pairing to miR-iab-4-5p or miR-iab-8-5p are indicated; see Figure 3 and Supplemental Figures S1–S3 for details of target site pairing and conservation. Darker lines represent stronger regulatory relationships, as evidenced by multiple sites and experimental validation. Below this is the DNA sequence of the iab-4 hairpin region, with the sense-strand miRNAs shaded in green and antisense-strand miRNAs shaded in red. Alignment of mature miR-iab-4-5p and miR-iab-8-5p shows that the latter has a 2-nt “UU” extension at its 5′ end. (B) Proposed secondary structures of the mir-iab-4 and mir-iab-8 hairpins. (C–F) In situ hybridization demonstrates temporally and spatially distinct expression patterns of pri-mir-iab-4 and pri-mir-iab-8. (G) Northern analysis of RNA from 6- to 10-h embryos reveals processed pre-miRNA hairpins for mir-iab-4 and mir-iab-8, and mature miRNAs for iab-4-5p, iab4-3p, and iab-8-5p; iab-4-5p and iab-8-5p are predominant. Ethidium stains of 5S rRNA and Northern analysis of 2S rRNA species are shown as loading and transferring controls. (H) Tests of probe specificity. Titration series of synthetic mir-iab-4 and mir-iab-8 hairpin RNAs were analyzed using probes for all four of the iab-4 locus miRNAs. Both iab-4-5p and iab-4-3p probes recognize only the mir-iab-4 hairpin, while the iab-8-5p and iab-8-3p probes recognize only the mir-iab-8 hairpin.
Despite its long history of study, fundamental aspects of the BX-C remain poorly understood. Prime among these regards the role of noncoding RNAs (ncRNAs), some of which were discovered >20 years ago (Lipshitz et al. 1987). As with the homeobox genes, BX-C ncRNAs are expressed in spatially discrete domains along the anterior–posterior axis that suggest their participation in developmental regulation. One of these is encoded by iab-4, which is located between abd-A and Abd-B (Fig. 1A). Originally recognized by Cumberledge (Cumberledge et al. 1990), who cloned its spliced and polyadenylated messages, iab-4 was later recognized by the Tuschl group as a likely primary-miRNA (pri-miRNA) transcript that generates the mir-iab-4 hairpin and mature ∼22-nucleotide (nt) RNAs, termed miR-iab-4-5p and miR-iab-4-3p (Aravin et al. 2003). miRNAs comprise an extensive class of regulatory RNAs that are inferred to have profound influence on the activity of thousands of transcripts (Lai 2003). However, the in vivo activities and phenotypically relevant targets of few miRNAs are currently known.
We recently demonstrated that ectopic mir-iab-4 directly represses Ubx and induces a partial Ubx loss-of-function phenotype, namely the transformation of haltere toward wing identity (Ronshaugen et al. 2005). We also extended the previous finding that both sense and antisense strands of the iab-4 region are transcriptionally active in temporally and spatially distinct domains (Bae et al. 2002; Drewell et al. 2002; Ronshaugen et al. 2005). Specifically, transcription of the antisense strand initiates later and more posteriorly relative to iab-4 (Fig. 1C–F). Since the sequence antisense to mir-iab-4 is also predicted to adopt an extended hairpin (Fig. 1B), we hypothesized that the antisense iab-4 transcript might independently produce miRNAs whose activity might be relevant to Hox gene regulation.
In this study, we demonstrate that the antisense strand of iab-4 produces the novel miRNA precursor mir-iab-8, which in turn, generates functional miRNAs whose regulatory activity is distinct from that of their sense counterparts. In particular, while ectopic miR-iab-4-5p weakly represses Ultrabithorax and has no effect on abdominal-A, ectopic miR-iab-8-5p could potently silence Ultrabithorax and abdominal-A. This is true of both sensor transgenes and the endogenous gene products. Thus, sense and antisense miRNAs generated from the iab-4 locus have overlapping and distinct activities with respect to BX-C gene regulation. Accompanying studies corroborate our findings regarding the regulatory activity of antisense iab-4 miRNAs, and demonstrate their endogenous requirement for Ubx regulation and normal development (Bender 2008; Stark et al. 2008). We then generalize this principle by showing that Drosophila mir-307 and mammalian mir-338 are endogenously processed on their antisense strands into mature miRNAs, whose seeds differ from their sense counterparts. Therefore, antisense transcription and processing contributes to the functional diversification of animal miRNA genes.
Results
Both strands of the mir-iab-4 hairpin are processed in vivo into mature miRNAs
Although both sense and antisense strands of the mir-iab-4 locus are transcriptionally active in Drosophila embryos, this fact alone does not imply the existence of antisense iab-4 miRNAs. We anticipated that the detection of such putative antisense miRNAs might be challenging, since far fewer cells express primary transcripts of the antisense mir-iab-8 strand than express mir-iab-4 (Fig. 1C–F). In fact, previously published Northern analyses failed to detect the mir-iab-4 hairpin, or even its mature products miR-iab-4-5p and miR-iab-4-3p (Leaman et al. 2005).
Locked nucleic acid probes proved efficacious for Northern analysis. On the sense strand, both miR-iab-4-5p and miR-iab-4-3p probes detected endogenous hairpin precursor and mature miRNAs in embryos (Fig. 1G). The level of mature miR-iab-4-5p was much higher than miR-iab-4-3p, as judged by the relative ratio between hairpin and mature RNA signals with these probes. On the antisense strand, a miR-iab-8-5p probe similarly detected both pre-miRNA and mature species, whereas a miR-iab-8-3p probe detected only the pre-miRNA (Fig. 1G). Thus, the major products of the sense and antisense iab-4 hairpins are their respective “5p” species. We wished to rule out that these positive Northern signals might be confounded by probe cross-hybridization. To do so, we transcribed mir-iab-4 and mir-iab-8 hairpin RNAs in vitro, and analyzed titration series of these RNAs against all four iab-4/iab-8 miRNA probes. As shown in Figure 1H, synthetic mir-iab-4 hairpin RNA was robustly detected with both miR-iab-4-5p and miR-iab-4-3p probes, but not by miR-iab-8-5p or miR-iab-8-3p probes. Conversely, synthetic mir-iab-8 hairpin RNA was detected by miR-iab-8-5p and miR-iab-8-3p probes, but not by miR-iab-4-5p or miR-iab-4-3p probes. These control experiments demonstrated the specificity of these probes in Northern analysis.
We determined the 5′ ends of iab-8 miRNAs using adaptor-mediated PCR and found that while the 5′ end of miR-iab-8-5p is related to that of miR-iab-4-5p, the former contains a strict 2-nt “UU” extension (Fig. 1A). Because of their seed offset, the target site preferences of these miRNAs are expected to be mostly nonoverlapping. However, certain appropriate sequences could, in principle, accommodate the seeds of both miRNAs. Subsequently, iab-4/iab-8 miRNA clones were obtained via 454 pyrosequencing (GEO series GSE7448), which verified the ends of the iab-8 small RNAs (Ruby et al. 2007). The characteristic 3′ overhangs to the antisense miRNA duplex served as further evidence that mir-iab-8 transits the canonical miRNA biogenesis pathway (Ruby et al. 2007).
The pyrosequencing data suggested that the mature miRNAs derived from mir-iab-8 are less abundant than those from mir-iab-4, which is consistent with the much more restricted spatial and temporal expression of the antisense transcript (Fig. 1C–F; Bae et al. 2002). On the other hand, the read numbers suggested comparable levels of miR-iab-8-5p and miR-iab-8-3p RNAs (Ruby et al. 2007), a conclusion that was at odds with our Northern data. It is worth noting that the original cloning data of Aravin et al. (2003) identified one clone each of miR-iab-4-5p and miR-iab-4-3p, an equivalency of expression that was neither borne out by larger clone sets (Ruby et al. 2007), nor by our Northern analysis (Fig. 1G). Therefore, low numbers of small RNA clones likely do not permit accurate quantitation of small RNA levels.
Sense and antisense iab-4 miRNAs exhibit separable target specificities
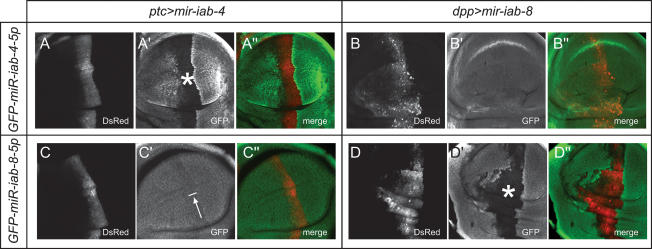
We next asked whether sense and antisense iab-4 miRNAs are active regulatory species in the animal. We used an in vivo assay in wing imaginal discs (Stark et al. 2003) to test whether mir-iab-4 or mir-iab-8 expression constructs could repress GFP “sensor” transgenes. Based on our observation that the left arm, “5p,” species from both hairpins predominate over their right arm, “3p” counterparts, we constructed tub-GFP sensors bearing antisense matches to miR-iab-4-5p or miR-iab-8-5p. We observed robust repression of the miR-iab-4-5p sensor by mir-iab-4 (Fig. 2A), and of the miR-iab-8-5p sensor by mir-iab-8 (Fig. 2D). Conversely, we observed little repression of the miR-iab-8-5p sensor by ectopic mir-iab-4, and no activity of mir-iab-8 on the miR-iab-4-5p sensor (Fig. 2B,C). Therefore, miRNAs derived from mir-iab-4 and mir-iab-8 are functional regulatory species that exhibit distinct target capabilities in vivo, which is consistent with their distinct seed regions (Fig. 1A).
Figure 2.
Sense and antisense iab-4 miRNAs are functional inhibitory RNAs that exhibit target selectivity. Shown are pouch regions of wing imaginal discs that carry the indicated tub-GFP sensors and express either UAS-DsRed-mir-iab-4 under ptc-Gal4 control (A,C) or UAS-DsRed-mir-iab-8 under dpp-Gal4 control (B,D). (A′,D′) Evidence for strong target inhibition is seen in the GFP-negative regions designated with asterisks; arrow in C′ indicates weak target inhibition.
iab-4-5p and iab-8-5p exhibit distinct BX-C gene target relationships
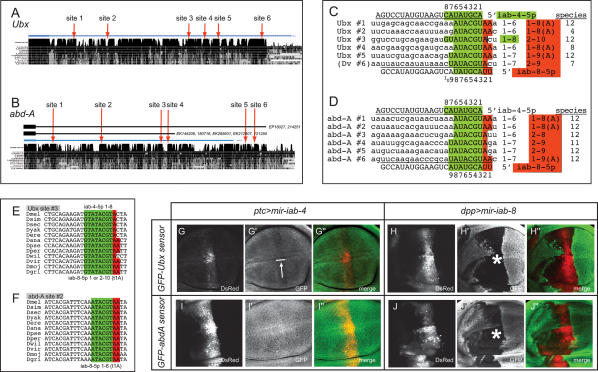
Our previous work established Ubx as a direct target of miR-iab-4-5p (Ronshaugen et al. 2005). However, only one (#3) of the many candidate sites in the Ubx 3′ untranslated region (UTR) is a canonical “2–8” seed-match site (Fig. 3A,C,E; Supplemental Fig. S1); that is, a site with Watson-Crick complementarity to positions 2–8 of a miRNA (Lai 2002; Lewis et al. 2005). Since most of the other sites are well conserved, we earlier hypothesized that Ubx might be configured to respond to a multiplicity of suboptimal miRNA-binding sites. Surprisingly then, six of these sites proved to be “2–8” seed match targets for miR-iab-8-5p (Fig. 3A,C,E). Four of these sites also have adenosines against position 1 of the miRNA (t1A features), which correlate with increased targeting efficacy (Lewis et al. 2005; Grimson et al. 2007; Nielsen et al. 2007); none of the candidate miR-iab-4-5p sites has t1A features. Therefore, Ubx appears to be a relatively modest target of miR-iab-4-5p, but a very strong target of miR-iab-8-5p.
Figure 3.
Ubx and abd-A are strong, direct targets of miR-iab-8-5p. (A,B) Alignments of the Ubx and abd-A 3′ UTRs across 12 Drosophilid genomes (obtained from the University of California at Santa Cruz Genome Browser, http://genome.ucsc.edu). The distal sites in abd-A are contained within a novel, extended isoform whose structure is supported by the indicated ESTs. (C,D) Canonical binding sites for sense and antisense iab-4 miRNAs in Ubx and abd-A. All sequences are from Drosophila melanogaster except for Ubx site #6; which is canonical only in nonmelanogaster group species (D. virilis [Dv] sequence is shown as a representative). Contiguous Watson-Crick seed-matches to miR-iab-4-5p are highlighted in green, while nucleotides that match the 5′ extension of miR-iab-8-5p are in red. The number of seed-matched nucleotides, t1A features (“A”), and depth of species conservation are tabulated to the right of each alignment. Seed-match data are highlighted green or red if predicted as a functional target of miR-iab-4-5p or miR-iab-8-5p, respectively; note that Ubx site #3 is a canonical binding site for both miRNAs. Additional conservation data are reported in Supplemental Figures S1–S3. (E,F) Examples of highly conserved miR-iab-8-5p/miR-iab-4-5p-binding sites in the 3′ UTRs of Ubx and abd-A. (G–J) Wing disc assays of the ability of sense and antisense iab-4 miRNAs to regulate Ubx or abd-A 3′ UTR sensors. Asterisks indicate strong regulation, while the arrow indicates weak regulation.
Equally striking is the degree to which miR-iab-8-5p targets abd-A (Fig. 3B,D,F; Supplemental Fig. S2). The abd-A 3′ UTR contains four “2–8” seed match sites for miR-iab-8-5p, with two additional sites distal to the annotated transcript; three of these sites have t1A features. We suspected that these downstream sites are genuine features of the abd-A transcription unit. Indeed, EST searches revealed both the canonical transcript and a novel abd-A transcript that utilizes a conserved, downstream poly(A) signal (Supplemental Fig. S2), and thus includes all six sites (Fig. 3B). None of these sites is a “2–8” seed match to miR-iab-4-5p, since all are unpaired at positions 7 and/or 8 of miR-iab-4-5p, and all of them also lack t1A pairing (Fig. 3D). The selection of Ubx and abd-A to have multiple “2–8” seed + t1A sites specifically for miR-iab-8-5p strongly suggests that they are preferred targets of the antisense miRNA mir-iab-8.
The conservation patterns of these extensive target site arrays are notable. Ubx sites #1, #3, and #5 have been perfectly conserved among all 12 sequenced Drosophilids, whereas the other three sites are found only in more limited species subsets (Supplemental Fig. S1); in fact, site #6 is a “2–8” seed-match site only in nonmelanogaster group species. In contrast, with the sole exception of site #4 in Drosophila virilis (which has a single C → T substitution introducing a G:U seed pair), all six miR-iab-8-5p sites in abd-A orthologs have been perfectly conserved among the 12 sequenced Drosophilids (Supplemental Fig. S2). The invariant nature of the large array of miR-iab-8-5p sites in Drosophilid abd-A genes contrasts with the fact that several sites in Ubx orthologs are actively evolving. We note that previous genome-wide searches revealed only one other Drosophila gene 3′ UTR with six highly conserved seed-match sites for an individual miRNA (nerfin, with six sites for miR-279/miR-286) (Grun et al. 2005; Stark et al. 2005). Thus, these Hox targets of miR-iab-8-5p are remarkable with respect to the complete landscape of Drosophila miRNA targets.
In vivo validation of Ubx and abd-A as mir-iab-8 targets
We next tested the capacity of sense and antisense iab-4 miRNAs to regulate tub-GFP-Ubx 3′UTR or tub-GFP-abd-A 3′UTR sensors in wing imaginal discs. These tests revealed a clear distinction between their regulatory activities. While ectopic mir-iab-4 induced only mild suppression of Ubx (Fig. 3G), mir-iab-8 strongly suppressed both Ubx and abd-A sensors (Fig. 3H,J). The degree of target inhibition correlated well with the qualities of the predicted target sites, yielding strong support for our notions regarding miRNA site functionality. For example, Ubx contains one well-conserved “1–8” site for miR-iab-4-5p (Fig. 3E; Ubx site #3, which is also a site for miR-iab-8-5p), and could be mildly repressed by mir-iab-4. On the other hand, even though abd-A contains six conserved sites that could pair with nucleotides 1–6 or 1–7 with iab-4-5p, we did not detect any inhibition of abd-A by ectopic mir-iab-4 (Fig. 3I).
The mild-to-nonexistent repression of these Hox targets by mir-iab-4 was not a consequence of an ineffectual transgene, since the same transgene could strongly repress its cognate perfect sensor (Fig. 2A). Conversely, these differences were not due to the chance isolation of an abnormally powerful mir-iab-8 transgene, since five independent genomic insertions of this construct showed similar activities. Instead, these data indicate that canonical “2–8” seed pairing is normally prerequisite for conventional miRNA target recognition, and cannot be compensated for simply by a large number of suboptimal target sites. Indeed, these data question whether “1–7” seed matches lacking t1A features even qualify as potential miRNA target sites.
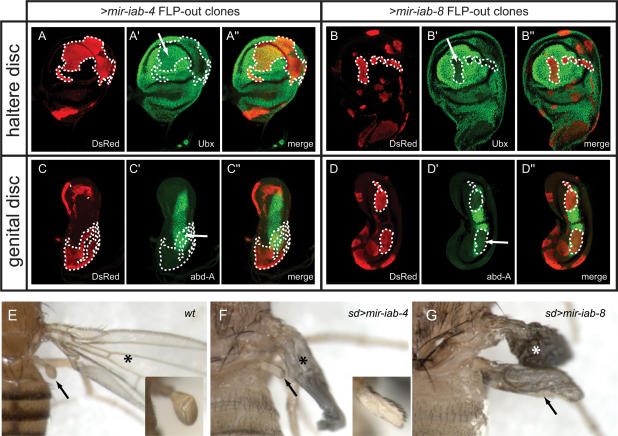
We next asked if mir-iab-4 or mir-iab-8 could affect the accumulation of endogenous target proteins. We used the FLP-out technique to generate clones of cells that express DsRed and either iab-4 or iab-8 miRNAs. We then examined the behavior of endogenous Ubx and abd-A proteins in miRNA-expressing clones in haltere discs and genital discs, respectively, using the adjacent nonclonal tissue as an internal staining control. We observed that mir-iab-4 weakly represses Ubx (Fig. 4A) as shown previously (Ronshaugen et al. 2005), while its misexpression had no effect on endogenous abd-A (Fig. 4C). In contrast, ectopic mir-iab-8 nearly eliminated the accumulation of endogenous Ubx and abd-A proteins in a cell-autonomous fashion (Fig. 4B,D). These data provide evidence for strong regulatory capability of Ubx and abd-A by miR-iab-8-5p, as supported by stringent conservation of their extensive target site arrays among the Drosophilids. In addition, the fact that the endogenous transcripts responded identically to the GFP sensors further suggests that targets lacking “2–8” seed pairing and/or t1A features are unlikely to be of functional significance.
Figure 4.
Ectopic mir-iab-8 inhibits endogenous Ubx and abd-A proteins and induces homeotic phenotypes. (A–D) FLP-out clones expressing sense or antisense iab-4 miRNAs were assayed for their effect on Ubx or abd-A. A–D show DsRed/miRNA-expressing clones (in red), A′–D′ depict endogenous Hox proteins (in green), and A″–D″ display merged images; selected clone boundaries that overlap domains of Hox protein expression are marked with dotted lines. Ectopic mir-iab-4 reduces Ubx (A) but has no effect on abd-A (C), while ectopic mir-iab-8 nearly eliminates both Hox proteins (B,D). (E–G) Haltere-to-wing respecification induced by sense and antisense iab-4 miRNAs, characteristic of Ubx loss of function. (E) Wild-type thorax. The wing is marked with an asterisk and the haltere is marked with an arrow; inset shows a higher-magnification view of the haltere. (F) sd-Gal4/X; UAS-DsRed-mir-iab-4/+ animal exhibits mild haltere-to-wing transformation including extensive specification of anterior wing margin bristles (see inset); the endogenous wing has not yet inflated. (G) sd-Gal4/X; UAS-DsRed-mir-iab-8/+ pharate lethal animal exhibits a nearly complete transformation of haltere toward wing identity; the morphology of both wings is abnormal.
Misexpression of mir-iab-8 induces a strong Ubx phenocopy
With these target data in hand, we investigated the phenotypic effects of ectopic mir-iab-8. We previously showed that misexpression of mir-iab-4 in developing halteres using sd-Gal4 subverts halteres into adopting partial wing identity (Ronshaugen et al. 2005). This is similar to the phenotype exhibited by a mild Ubx allelic combination, and is most clearly indicated by the specification of an anterior wing margin-like structure on the haltere decorated with sensory bristles (Fig. 4E,F). Misexpression of mir-iab-8 using sd-Gal4 induced larval and pupal lethality. However, by reducing the activity of Gal4 we obtained some pharate lethal females that exhibited nearly complete haltere-wing transformations (Fig. 4G). These substantial phenocopies of Lewis’ (Lewis 1978) classic four-winged flies obtained with strong Ubx mutant alleles are a palpable exhibition of the powerful regulatory relationship between miR-iab-8-5p and Ubx.
We also searched for potential transformations in abdominal segments characteristic of abd-A loss of function. DsRed/mir-iab-4+ clones were recovered in the adult abdomen, and these exhibited no overt phenotype (data not shown). In contrast, we were unable to recover clones of DsRed/mir-iab-8+ cells in the adult abdomen. Histoblast nest cells (which give rise to abdominal structures) lacking both Ubx and abd-A assume thoracic identity, and are unable to differentiate within the abdominal environment (Casanova et al. 1987). Therefore, the dual targeting of these BX-C genes by mir-iab-8 constrains the types of visible homeotic transformations that it can induce.
Endogenous sense and antisense processing of Drosophila mir-307 and mammalian mir-338
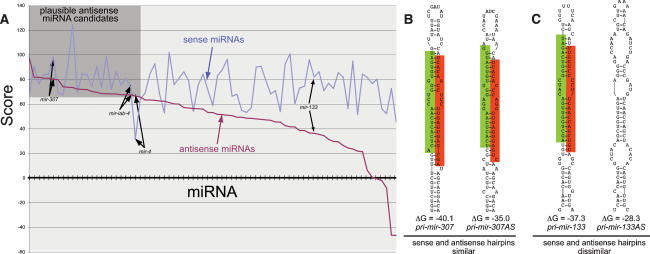
We wished to generalize the principle of endogenous antisense miRNAs. Hairpins composed solely of Watson-Crick pairs have identical secondary structures on either strand. On the other hand, since miRNA hairpins typically exhibit G:U basepairs and/or unpaired nucleotides, their corresponding sense and antisense structures are usually different (e.g., mir-133) (Fig. 5C), with the antisense version usually presenting a less compelling structure (Fig. 5A). Nevertheless, analysis of the 76 Drosophila miRBase genes on both strands suggested that more than one-quarter are plausible as miRNA genes as antisense transcripts (Fig. 5A), with mir-iab-4 in the upper quartile.
Figure 5.
Comparison of miRNA precursors as sense and antisense transcripts. (A) Sense and antisense miRNA hairpins of known Drosophila miRNAs were assessed according to their free energy and structural parameters (Lai et al. 2003). The X-axis denotes each miRNA hairpin folded and scored on its sense strand (blue) and antisense strand (purple); the Y-axis denotes the score of the miRNA candidate. Most antisense strands are poorer candidates than their sense counterparts (e.g., mir-133); however, nearly one-third of them (gray box) achieve a score that is plausible as a miRNA candidate (e.g., mir-307). In a few cases, such as mir-4, the antisense strand is more compelling than the sense strand. The full analysis is available in Supplemental Figure S4. (B) Example of a miRNA locus that adopts comparable structures on both strands. (C) Example of a miRNA locus whose antisense structure is dissimilar to its sense counterpart.
Another distinguishing feature of mir-iab-4 is that both sides of its hairpin are under tremendous evolutionary constraint. In fact, both arms have been perfectly preserved among flies, mosquitoes, bees, and beetles, species that have been separated for >400 million years (Ronshaugen et al. 2005). We believe this to be a likely consequence of the critical functions of sense and antisense miRNAs produced from both arms of this locus.
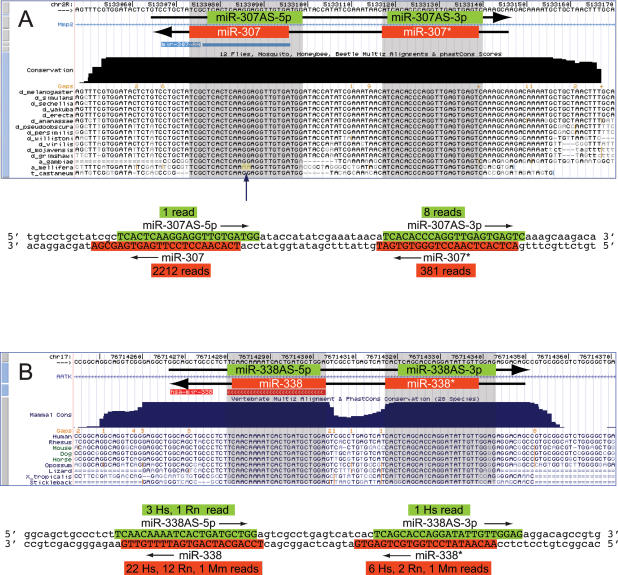
We noticed through this analysis that the mir-307 hairpin was the sixth highest scoring antisense candidate (Fig. 5; Supplemental Fig. S4) and has been strictly conserved on both arms during insect evolution (Lai et al. 2003). In fact, while mature miR-307 (which derives from the “3p” hairpin arm) contains a 2-nt divergence in Apis (although it is completely conserved in the more distant Tribolium species), miR-307* is 100% conserved among these widely divergent insects (Fig. 6A). Might the particular constraint on miR-307* actually reflect an antisense miRNA? Indeed, analysis of small RNAs cloned from female Drosophila heads revealed both mature miRNA and miRNA* species for antisense (AS) mir-307, with miR-307AS-3p as the predominant species. miR-307 and miR-307AS-3p have different seeds, and consequently have distinct regulatory capacity. Although miR-307AS-3p is rarer than miR-307, its higher evolutionary constraint indicates that its trans-regulatory function is under stringent selection. It is conceivable that, as is the case with mir-iab-4 and mir-iab-8, mir-307AS is transcriptionally active in a much more spatially and/or temporally restricted domain than is mir-307.
Figure 6.
Drosophila mir-307 and mammalian mir-338 produce antisense miRNAs. (A) Multiple alignment of the insect mir-307 locus (obtained from the University of California at Santa Cruz Genome Center, http://genome.ucsc.edu). Previously cloned miR-307 derives from the bottom strand (miR-307, “3p” arm). Both arms of the hairpin have been highly conserved during insect evolution (blue-shaded boxes), but miR-307 has diverged slightly in Apis (yellow, arrow). Below this is the DNA sequence of the mir-307 hairpin, color-coded for cloned sense (in red) and antisense (in green) miRNAs. (B) Multiple alignment of the vertebrate mir-338 locus. Previously identified miR-338 derives from the bottom strand. Both arms of the hairpin are strongly conserved in placental mammals. Below this is the DNA sequence of the mir-338 hairpin, color-coded for cloned sense (in red) and antisense (in green) miRNAs.
The head small RNAs also revealed candidate antisense miRNAs for several other loci. The most compelling loci were mir-13b-2AS, which was the seventh highest-scoring antisense candidate, and mir-305AS, for which two clones were obtained. Finally, we note that Ruby et al. (2007) also reported antisense clones for mir-307, mir-305, mir-124. Thus, there is candidate evidence for at least five processed antisense miRNA loci in Drosophila.
Does the phenomenon of antisense miRNAs apply to other species? We analyzed a recently published mammalian small RNA data set (Landgraf et al. 2007) and identified several candidate antisense miRNAs. Of these, the most compelling example was mir-338, since the antisense strand of human mir-338 generated both cloned miRNA and miRNA* species (Fig. 6B). The modest number of antisense clones did not permit confident assignment of the miRNA* strand, and comparison of the relative free energies at the ends of its small RNA duplex did not suggest a particular strand asymmetry (Supplemental Fig. S5). However, we found that miR-338AS-5p was also cloned from rat. This indicates that the production of antisense miRNAs is a conserved feature of the mammalian mir-338 locus, and plausibly suggests miR-338AS-5p as the antisense miRNA of preferred endogenous usage. Consistent with this notion, the DNA sequence that jointly encodes miR-338 and miR-338AS-5p is more highly conserved than the sequence that encodes their partner small RNAs (Fig. 6B).
In summary, these additional examples generalize the principle of antisense miRNAs in both invertebrates and vertebrates. In the three most confident cases—for which both antisense miRNA and miRNA* were cloned—the register of hairpin processing on the sense and antisense strands was distinct (Figs. 1, 6). This results in antisense miRNA seeds that are radically different from their sense counterparts. Therefore, antisense miRNA transcription and processing can serve to diversify the activities of miRNA genes in both Drosophila and mammals.
Discussion
Our studies of BX-C miRNAs reveal two principal insights into miRNA regulatory biology. First, we identified a new Hox cluster miRNA, mir-iab-8, and showed using gain-of-function assays that it can strongly inhibit Ubx and abd-A and generate homeotic phenotypic transformations. Indeed, the Hox-regulatory properties of mir-iab-8 are far more potent than those of mir-iab-4 (Ronshaugen et al. 2005), and correlate directly with the properties of its target sites in their 3′ UTRs (Figs. 3, 4; Supplemental Fig. S1, S2). Curiously, both BX-C miRNAs obey organizational and functional rules previously defined for the protein-encoding members of the BX-C (Lewis 1978). These regulatory RNAs exhibit colinearity, in that transcription of pri-mir-iab-8 initiates more distally on the chromosome and is expressed more posteriorly in the embryo relative to pri-mir-iab-4 (Fig. 1A, C–F). They also exhibit posterior prevalence, in that both sense and antisense iab-4 miRNAs directly repress multiple homeotic genes located more anteriorly in the Hox cluster (Figs. 1, 3, 4). In fact, we identified the next most-anterior Hox gene Antp as a third likely endogenous iab-miRNA target that contains highly conserved target sites with t1A features for miR-iab-8-5p (Fig. 1A; Supplemental Fig. S3). On the other hand, Abd-B contains no conserved sites for either iab-4 or iab-8 miRNAs in its long (>2 kb) 3′ UTR. Therefore, BX-C miRNAs and homeobox genes are governed by the same regulatory logic.
It is worth recalling that saturation mutagenesis screens of the BX-C revealed only three loci that are required for viability and exhibit homeotic defects, corresponding to the homeobox genes Ubx, abd-A, and Abd-B (Sanchez-Herrero et al. 1985; Tiong et al. 1985). On the other hand, pioneering studies by Lewis (1978) that also considered rearrangements and dominant alleles suggested the existence of at least eight homeotic “factors” in this region of the genome. Although many of these are now recognized as cis-regulatory elements that regulate Hox gene transcription (Maeda and Karch 2006), our work with BX-C miRNAs reveals two bona fide Hox regulators that are capable of inducing severe dominant homeotic transformations. The endogenous requirement for iab-4/iab-8 miRNAs appears to be subtle, possibly due to compensatory transcriptionally based regulatory mechanisms. Nevertheless, loss of function analysis in an accompanying manuscript corroborates that these miRNAs are required for normal expression of Hox targets in the nervous system and for normal development (Bender 2008). These data emphasize that loss-of-function and gain-of-function genetics are complementary approaches to uncover important regulatory molecules.
Second, we uncovered antisense transcription and processing as a mechanism to generate new functional miRNAs. Bioinformatic analysis suggests that a large fraction of miRNA loci are theoretically competent to produce antisense miRNAs (Fig. 5; Supplemental Fig. S4). Extant cloning efforts suggest that few miRNA loci actively produce large quantities of antisense miRNAs. Nevertheless, the sequencing effort reported here and in a contemporary analysis (Ruby et al. 2007) revealed additional instances of putative antisense miRNAs. Although none of these was cloned more than twice, genetics demonstrates that rare miRNAs (e.g., lsy-6, expressed by a handful of neurons in a whole animal) can be critical components of regulatory networks and can have potent biological activities (Johnston and Hobert 2003). Therefore, assessment of the biological relevance of the other antisense miRNA candidates awaits further study.
In the case of the iab-4 locus, the regulatory diversity afforded by sense and antisense transcription of a single miRNA hairpin is manifested by altering the seed regions of their respective miRNA products and by deploying the sense and antisense pri-miRNA transcripts in distinct spatial domains. It might in fact be deleterious for a given locus to be simultaneously transcribed on both strands—either because of transcriptional interference from colliding polymerase complexes, or because of the possibility to inadvertently generate dsRNA. Further analysis is needed to test the notion that it is favorable for sense/antisense miRNA pairs not to be expressed in the same cells. Overall, though, as animal genomes are quite extensively transcribed (Manak et al. 2006; Kapranov et al. 2007), and many miRNA genes adopt extensive hairpins on both strands (Fig. 5), the potential for endogenous antisense processing of miRNA hairpins is theoretically quite broad. We propose that antisense transcription of other miRNA loci might generate novel small RNAs whose potentially beneficial regulatory activities are available for selection and stabilization by natural selection (Chen and Rajewsky 2007). Our identification of several confident examples of antisense miRNAs, whose processing and/or targets have been conserved among diverse species, provides compelling support for this hypothesis.
Materials and methods
Northern analysis
Endogenous total RNAs were isolated from staged Canton S embryos using Trizol (Life Technologies). Synthetic pre-mir-iab-4 and pre-mir-iab-8 templates were generated by first annealing a long pre-miRNA oligo with a T7 promoter oligo and extending with Taq polymerase. Oligos are as follows, with T7 sequences in lowercase: T7-promoter, taatacgactcactatag; T7-pre-mir-iab-4, GTTACGTATACTGAAGGTATACCGGATAG GATACACTCAGGATACATTCAGTATACGTCCctatagtgagtc gtatta; T7-pre-mir-iab-8, TAAACGTATACTGAATGTATCCT GAGTGTATCCTATCCGGTATACCTTCAGTATACGTAAC Cctatagtgagtcgtatta.
These templates were then transcribed using T7 RNA polymerase (Ambion). RNA samples (20 μg of total embryonic RNA or specified amounts of synthetic RNA per lane) were run on 12% acrylamide denaturing gels and then electrophoretically transferred to Genescreen Plus (Perkin Elmer) membranes. After transfer, they were cross-linked with 120 mJ of UV. LNA or DNA probes were labeled using T4 polynucleotide kinase (New England Biolabs) and γ-32P ATP and hybridized to the membranes at 45°C in hybridization buffer (50% Formamide, 5× SSPE, 5× Denhardt’s, 0.5% SDS, 20 μg/mL sheared fish DNA) overnight. Membranes were washed at 45°C, twice with 2× SSC, 0.1% SDS, and three times with 0.2× SSC, 0.1% SDS, followed by PhosphorImager analysis (Fuji, BAS2500).
The following probes were used: miR-iab-4-5p, TCAGGATA CATTCAGTATACGT (LNA, Exiqon); miR-iab-4-3p, TTACG TATACTGAAGGTATACCG (LNA, Exiqon); miR-iab-8-5p, CGGTATACCTTCAGTATACGTAA (LNA, Exiqon); miR-iab-8-3p, TAAACGTATACTGAATGTATCCT (LNA, Exiqon); 2S rRNA, (TACAACCCTCAACCATATGTAGTCCAAGCA) (DNA, IDT).
Drosophila strains
dpp-Gal4, ptc-Gal4, and sd-Gal4 driver strains were obtained from the Bloomington Stock Center. tub-GFP-Ubx 3′ UTR, UAS-DsRed-mir-iab-4 were described previously. The mir-iab-8 expression construct was created by inserting the reverse complement of a previously described mir-iab-4 genomic fragment (Ronshaugen et al. 2005) into the 3′ UTR of UAS-DsRed. Perfect sensors for iab-4-5p and iab-8-5p were generated by cloning the following oligos into the XhoI and XbaI sites of tub-GFP (Stark et al. 2003): miR-iab4-5 sensor, 5′-CTAGAcTCAG GATACATTCAGTATACGTtcaacaatcaccTCAGGATACATT CAGTATACGTtC-3′ and 3′-TgAGTCCTATGTAAGTCAT ATGCAagttgttagtggAGTCCTATGTAAGTCATATGCAaGAG CT-5′; miR-iab8-5 sensor, 5′-CTAGATCCGGTATACCT TCAGTATACGTAACACGAGcaacaatcacTCCGGTATACCT TCAGTATACGTAACACGAGcC-3′ and 3′-TAGGCCATA TGGAAGTCATATGCATTGTGCTCgttgttagtgAGGCCATAT GGAAGTCATATGCATTGTGCTCgGAGCT-5′.
We generated the GFP-abd-A 3′ UTR sensor by amplifying a 2.3-kb fragment containing the entire 3′ UTR from w1118 genomic DNA and cloned it into the NotI and XbaI sites of tub-GFP: abdA_RA 5′Not, ggggcggccgcTAAgctgacaaccacccatc; and abdA_RA 3′Xba, ggggtctagaGTATATGCGGGTTCTTGAAC.
Transgenic strains were generated by BestGene, Inc., and at least three independent insertions were isolated for each construct.
In situ hybridization
For in situ hybridizations, we used PCR to generate two different amplicons of the iab-4 region, incorporating flanking T7 and T3 promoters. These templates were used to make sense and antisense pairs of digoxigenin-labeled probes, which were hybridized to embryos using a standard method (Kosman and Small 1997). Both probe sets gave identical staining patterns; the data shown in the paper used the “4–5” probe set below.
This primer set generated an ∼1-kb fragment centered on the iab-4 hairpin: iab4T3, AATTAACCCTCACTAAAGGGAGA accaccgaacacaagacaca; iab4T7, TAATACGACTCACTATAGG GAGAgcggtttcatgacacagaga. The second primer set was “4–5,” described by Bae et al. (2002): 4–5 s, ACCACAAGAAGGAG CAGTCG; and 4–5 as, GCACTCTCACCTACACGAATGC.
Small RNA libraries
We prepared total RNA from adult female Canton S Drosophila heads using Trizol, and cloned the ∼18- to 26-nt fraction using a previously published method (Lau et al. 2001). The library DNA was then adapted with primers that permitted them to be loaded onto Solexa flow cells. One sequencing lane was analyzed, and the mapped miRNA data were deposited in GEO (record GPL6072).
Sensor assays
We crossed X-Gal4; tub-GFP-Y trans-heterozygous females to UAS-DsRed-mir-Z males, where “X” is either dpp-Gal4 or ptc-Gal4, “Y” is either a perfect sensor (bearing tandem antisense matches to miR-iab-4-5p or miR-iab-8-5p) or a 3′ UTR sensor (containing the 3′ UTR of abd-A or Ubx), and “Z” is either a pri-miRNA fragment of mir-iab-4 or mir-iab-8. We then selected GFP+, DsRed+ larvae for dissection. It was necessary to express mir-iab-4 using ptc-Gal4 and mir-iab-8 using dpp-Gal4 because reciprocal combinations were lethal before the third larval instar.
Clonal expression of miRNAs
To analyze the effect of ectopic miRNAs on Hox protein accumulation, we heat-shocked y w hsFlp/+; tub-α1 > FRT Gal80, y+ FRT > Gal4/+; UAS-DsRed-miRNA/+ first instar larvae for 30 min at 38°C. Flp-mediated excision of the FRT-Gal80 cassette induced Gal4 and miRNA expression in DsRed-marked cell clones. Fluorescent antibody staining of third instar imaginal discs was then performed as described previously (Lai and Rubin 2001). We used the following antibodies: rabbit α-GFP (1:1250; Molecular Probes), mouse α-Ubx (1:10; from Nipam Patel), mouse α-Abd-A (1:10; from Ian Duncan), and Alexa-coupled secondary antibodies (1:400; Molecular Probes).
Acknowledgments
Bergin Tam and Nancy Arango helped to clone miRNA constructs, and Emily Hodges performed Solexa sequencing. Julius Brennecke, Stephen Cohen, Nipam Patel, and Ian Duncan generously provided antibodies and plasmids. K.O. was supported by grants from the Japan Society for the Promotion of Science and Charles H. Revson Foundation. J.W.H. was supported by the Mildred and Emil Holland Scholarship, the Margaret and Herman Sokol Fellowship and NIH MSTP grant GM07739. E.B. was supported by a VIDI grant (NWO). G.J.H. is an investigator of the Howard Hughes Medical Institute. E.C.L. was supported by grants from the Leukemia and Lymphoma Foundation, the Burroughs Wellcome Foundation, the V Foundation for Cancer Research, the Sidney Kimmel Foundation for Cancer Research and the National Institutes of Health (GM083300).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1615208
References
- Aravin A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Bae E., Calhoun V.C., Levine M., Lewis E.B., Drewell R.A. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W. MicroRNAs in the Drosophila bithorax complex. Genes & Dev. 2008 doi: 10.1101/gad.1614208. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J., Sanchez-Herrero E., Busturia A., Morata G. Double and triple mutant combinations of bithorax complex of Drosophila. EMBO J. 1987;6:3103–3109. doi: 10.1002/j.1460-2075.1987.tb02619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Zaratzian A., Sakonju S. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 1990;87:3259–3263. doi: 10.1073/pnas.87.9.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell R.A., Bae E., Burr J., Lewis E.B. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 2002;99:16853–16858. doi: 10.1073/pnas.222671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Wang Y.-L., Langenberger D., Gunsalus K.C., Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLOS Compt. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kosman D., Small S. Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development. 1997;124:1343–1354. doi: 10.1242/dev.124.7.1343. [DOI] [PubMed] [Google Scholar]
- Lai E.C. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai E.C. microRNAs: Runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Rubin G.M. neuralized functions cell-autonomously to regulate a subset of Notch-dependent processes during adult Drosophila development. Dev. Biol. 2001;231:217–233. doi: 10.1006/dbio.2000.0124. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Tomancak P., Williams R.W., Rubin G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N., Lim L., Weinstein E., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Leaman D., Chen P.Y., Fak J., Yalcin A., Pearce M., Unnerstall U., Marks D.S., Sander C., Tuschl T., Gaul U. Antisense-mediated depletion reveals essential and specific functions of MicroRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lewis E.B. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lipshitz H.D., Peattie D.A., Hogness D.S. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes & Dev. 1987;1:307–322. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- Maeda R.K., Karch F. The ABC of the BX-C: The bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M., Biemar F., Piel J., Levine M., Lai E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes & Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Herrero E., Vernos I., Marco R., Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R.B., Cohen S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A., Bushati N., Jan C., Kheradpour P., Hodges E., Brennecke J., Bartel D.P., Cohen S.M., Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes & Dev. 2008 doi: 10.1101/gad.1613108. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiong S., Bone L.M., Whittle J.R. Recessive lethal mutations within the bithorax-complex in Drosophila. Mol. Gen. Genet. 1985;200:335–342. doi: 10.1007/BF00425445. [DOI] [PubMed] [Google Scholar]