Abstract
Transcriptional initiation is a key step in the control of mRNA synthesis and is intimately related to chromatin structure and histone modification. Here, we show that the ubiquitylation of H2A (ubH2A) correlates with silent chromatin and regulates transcriptional initiation. The levels of ubH2A vary during hepatocyte regeneration, and based on microarray expression data from regenerating liver, we identified USP21, a ubiquitin-specific protease that catalyzes the hydrolysis of ubH2A. When chromatin is assembled in vitro, ubH2A, but not H2A, specifically represses the di- and trimethylation of H3K4. USP21 relieves this ubH2A-specific repression. In addition, in vitro transcription analysis revealed that ubH2A represses transcriptional initiation, but not transcriptional elongation, by inhibiting H3K4 methylation. Notably, ubH2A-mediated repression was not observed when H3 Lys 4 was changed to arginine. Furthermore, overexpression of USP21 in the liver up-regulates a gene that is normally down-regulated during hepatocyte regeneration. Our studies revealed a novel mode of trans-histone cross-talk, in which H2A ubiquitylation controls the di- and trimethylation of H3K4, resulting in regulation of transcriptional initiation.
[Keywords: Nucleosome, ubiquitylation, histone code, transcription, USP21, H2A]
Eukaryotic gene expression is regulated by the cellular network of cis-acting elements and trans-acting factors. Transcriptional initiation of protein-encoding genes represents a major control point for gene expression in eukaryotes, which is mediated by RNA polymerase II and a surprisingly complex array of general transcription factors (GTFs) that initiate transcription and are highly conserved from yeast to humans (Hochheimer and Tjian 2003) In the eukaryotic nucleus, genomic DNA interacts with numerous proteins to form chromatin. Packaging of the template into chromatin appears to affect all stages of transcription, including initiation and elongation (for reviews, see Kadonaga 1998; Orphanides and Reinberg 2002; Li et al. 2007).
The nucleosome is the fundamental unit of chromatin and is composed of 147 base pairs (bp) of DNA wrapped 1.65 turns around the histone octamer of the four core histones (H2A, H2B, H3, and H4) (Luger et al. 1997). Nucleosomes act as general repressors of multiple stages of transcription, including initiation, promoter clearance, elongation, and termination. According to recent concepts of the histone code, post-translational modifications of the histone tail represent a major mechanism by which cells control the structure and function of chromatin (Strahl and Allis 2000; Fischle et al. 2003; Kouzarides 2007). Diverse histone modifications—such as acetylation, methylation, and polyadenylation, in addition to ubiquitylation—exist, many of which have been linked to the regulation of cellular activities such as transcription, repair, and replication. Although the precise mechanisms by which histone modifications contribute to the transcription process are not fully understood, increasing evidence suggests that they work together in the form of a histone code to regulate the recruitment of chromatin-modulating factors (Berger 2002; Mellor 2005).
On the other hand, three decades have passed since the discovery that the core histone H2A is monoubiquitylated, which changes during hepatocyte regeneration. Histone H2A was the first protein identified as being ubiquitylated (Goldknopf et al. 1975), and the ubiquitylation site has been mapped to the highly conserved residue Lys 119 (Goldknopf and Busch 1977). Since then, a variety of cellular processes including protein degradation, stress response, cell cycle regulation, protein trafficking, endocytosis signaling, and transcriptional regulation have been linked to mono- or polyubiquitylation of proteins (Pickart 2004). However, the role of the monoubiquitylation of H2A (ubH2A) remains not fully understood.
Since liver retains the capacity to regenerate in response to changes in mass or function in both humans and animals, it has been used in many studies, including those analyzing H2A ubiquitylation. These studies used a liver regeneration model after partial hepatectomy that was introduced seven decades ago (Higgins and Anderson 1931). Following a two-thirds hepatectomy, normally quiescent hepatocytes undergo one or two rounds of replication to restore the liver mass by a process of compensatory hyperplasia. A large number of genes comprises the gene expression network that is associated with liver regeneration (for reviews, see Michalopoulos and DeFrances 1997; Costa et al. 2003; Mangnall et al. 2003; Fausto et al. 2006).
These genes include many growth factors and cytokines, such as hepatocyte growth factor, epidermal growth factor, and “immediate early genes” such as c-Fos and c-Jun (for reviews, see Taub 1996; Michalopoulos and DeFrances 1997). In addition to growth factors and immediate early genes, global programs are also important for the homeostatic and proliferative responses in regenerating liver. These programs include hormone biosynthesis, lipid and amino acid metabolisms, and nucleotide biosynthesis. The transcription network revealed by microarray expression profiling during hepatocyte regeneration indicated that Igfbp1, LCN2, and Saa2 increase but, in contrast, Car3, Serpina6 (also called CBG; corticosteroid-binding globulin), and Cyp2f2 are manifestly decreased ∼12 h after hepatectomy (Togo et al. 2004; White et al. 2005). These genes are also important for hepatic regeneration, in terms of homeostasis and hepatic metabolism.
Some early studies suggested the involvement of H2A ubiquitylation in gene activation (for review, see Jason et al. 2002); however, recent studies have indicated a role for H2A ubiquitylation in gene repression (for reviews, see Zhang 2003; Shilatifard 2006). An H2A ubiquitin ligase was identified, and its complex has been suggested to play a role in Polycomb group (PcG) protein-mediated gene silencing by H2A ubiquitylation (Wang et al. 2004). Using a different approach, it was found that ubH2A and a subgroup of PcG proteins were located on the inactive X chromosome, but were abolished from the inactive X chromosome in a mouse line with deleted copies of Polycomb repressor complex protein Ring1B and its closely related homolog Ring1A (de Napoles et al. 2004). The finding that H2A ubiquitylation is required for PcG protein-regulated gene regulation suggests that it may be part of the mechanism underlying PcG protein-regulated gene silencing.
Here, we show that in mouse regenerating liver chromatin, ubH2A is enriched in the promoter region of repressed genes and in silenced chromatin. Based on the microarray profile during dynamic changes in histone H2A ubiquitylation after hepatectomy, we found that USP21, the presence of which is related to hepatic regeneration, hydrolyzes an isopeptide bond in ubH2A. We also show that ubH2A represses transcription initiation but not elongation by histone code cross-talk in vitro, and that this repression is canceled by USP21 in vitro and in vivo.
Results
Histone H2A ubiquitylation correlates with repressed genes during hepatocyte regeneration
Since the state of histone H2A ubiquitylation changes robustly in concert with hepatic regeneration and its concomitant gene regulation network, regenerating liver chromatin provides a superb model to study gene regulation by ubH2A (Michalopoulos and DeFrances 1997; Costa et al. 2003; Mangnall et al. 2003; Fausto et al. 2006). Within this network, the monoubiquitylation of core histone H2A dynamically changes, with the bulk of this modification decreasing during hepatocyte regeneration (Fig. 1A,B). However, it is noteworthy that some foci of ubH2A persisted during hepatocyte regeneration (Fig. 1B, panel f).
Figure 1.
Histone H2A ubiquitylation plays a role in gene silencing during hepatocyte regeneration. (A) Experimental scheme. Hepatectomized liver was used for quiescent (0 h) specimens, whereas residual mouse liver was removed and analyzed as regenerating liver at the indicated times after hepatectomy. (B) ubH2A was detected by anti-ubH2A antibodies with quiescent and regenerating hepatocytes, which were obtained by hepatectomy and residual liver resected 12 h after hepatectomy, respectively. Although most of the nuclear ubH2A disappeared 12 h after hepatectomy, a significant amount of ubH2A still persisted in the regenerating nuclei. (C,D) RT–PCR analysis of six representative genes, with altered expression during hepatocyte regeneration, based on expression microarray data together with GAPDH that exists in multiple loci in the mammalian genome. cDNAs for RT–PCR were obtained from mouse livers at the indicated times after hepatectomy, and the primers for RT–PCR are described in Supplemental Table 1. (E–G) ChIP analysis was carried out using quiescent and regenerating hepatocytes at the indicated time points. Samples were homogenized and immunoprecipitated using 25 μg of rabbit anti-ubH2A (Supplemental Fig. 1), 25 μg of anti-H3 dimethyl K4 (Supplemental Fig. 2), 5 μL of anti-H3 trimethyl K4 serum (Upstate Biotechnology #07-473), and 25 μg of anti-H3 trimethyl K9 (Supplemental Fig. 3) antibodies. The promoter regions of LCN2 and Serpina6 were analyzed by ChIP assays together with GAPDH that exists in multiple loci as a control. Nonspecific IgG was used as a control for immunoprecipitation. Primers for PCR are described in Supplemental Table 1.
To explore the role of histone H2A monoubiquitylation, we examined the regulation of hepatic regeneration-related genes whose expression changes after several hours, coinciding with changes in H2A ubiquitylation. Six-week-old to 8-wk-old BALB/c mice were subjected to partial hepatectomy, as previously described (Higgins and Anderson 1931). Microarray-based expression data illuminated transcriptional regulation changes that play important roles in hepatocyte metabolism and in expression of genes that trigger liver regeneration after partial hepatectomy. The expression patterns of these genes were examined by RT–PCR with mRNA from residual liver at different time points after partial hepatectomy. Genes such as Igfbp1, LCN2, and Saa2 are up-regulated, whereas Car3, Serpina6, and Cyp2f2 are down-regulated (Fig. 1C,D; Togo et al. 2004; White et al. 2005).
Although histone H2A was the first protein identified as being ubiquitylated (Goldknopf et al. 1975), at the conserved Lys 119 (Goldknopf and Busch 1977), the role of H2A monoubiquitylation during hepatocyte regeneration is not fully understood. To find out the role of these, histone modification antibodies were generated against H3 dimethylK4, H3 trimethylK9, ubiquitylated H2A (ubH2A), and Serpina6 and are characterized in Supplemental Figures 1–4. Chromatin immunoprecipitation (ChIP) assays were performed with the promoter regions of LCN2 and Serpina6, which are up- and down-regulated after partial hepatectomy, respectively, along with GAPDH existing in multiple loci in mammals as a control (Fig. 1E–G). We showed that H2A ubiqutylation and H3K9 trimethylation decreases and H3K4 di- and trimethylation increases in an up-regulated gene. In contrast however, we showed that H2A ubiqutylation and H3K9 trimethylation increases and H3K4 di- and trimethylation decreases in down-regulated genes.
The result that H2A ubiquitylation increases on a down-regulated promoter indicates that specifically ubiquitylated H2A on the gene promoter participates in gene silencing even if the bulk of ubiquitylated H2A decreases during hepatocyte regeneration. The observation that mRNA from Serpina6 is still detected 6 h after hepatectomy even if the nucleosome around its promoter regions has been modified by H2A ubiquitylation, H3K9 methylation, and H3K4 hypomethylation at the same time is explained by the time taken for the decay of the mRNA.
GAPDH is encoded in multiple loci, and most of them have the same coding sequences. Thus, PCR primers for ChIP assays were designed to detect common genomic DNA within one nucleosome from the core promoter with multiple loci (Supplemental Table 1). Using common primers, we could characterize modifications of the nucleosome assembled on core promoters of multiple GAPDH gene loci as a control. Thus, our results suggested that some loci are active, as revealed by H3K4 methylation, whereas other loci are inactive, as revealed by H2A ubiquitylation and H3K9 trimethylation (Fig. 1G).
We also examined the distribution of ubH2A in chromatin. Samples of bulk chromatin, from residual regenerating liver at different time points after hepatectomy and quiescent hepatocytes, were analyzed by immunoprecipitation with well-characterized antibodies recognizing dimethyl H3K4 or trimethyl H3K9, followed by a Western blot analysis with the ubH2A-specific monoclonal antibody, E6C5. The amount of ubH2A in bulk chromatin from the hepatocyte nucleus decreased after partial hepatectomy (Supplemental Fig. 5A). Immunoprecipitation of bulk chromatin with anti-H3 dimethyl K4 antibodies resulted in coimmunoprecipitation of ubH2A in a pattern that reflected the ubH2A content in it (Supplemental Fig. 5B). From quiescent hepatocytes with relatively abundant ubH2A, a part of ubH2A was precipitated with anti-H3 dimethyl K4 antibodies. This result suggests the possibility that these two histone modifications coincide in the quiescent hepatocyte, which is consistent with the recent concept that the inner core of the core nucleosome is very stable, whereas histone H2A–H2B of nucleosomes exchange dynamically (Kimura and Cook 2001).
In sharp contrast, immunoprecipitation of bulk chromatin with anti-trimethyl H3K9 antibodies yielded a pattern of ubH2A coimmunoprecipitation that is the opposite of that of the bulk chromatin; that is, the coimmunoprecipitated ubH2A increased after partial hepatectomy (Supplemental Fig. 5C). Hence, there is a strong correlation between the levels of ubH2A and trimethyl H3K9, which are associated with transcriptional repression. These findings further support a role for ubH2A in silent chromatin.
USP21 hydrolyzes an isopeptide bond of ubiquitylated H2A
We next sought to identify the enzyme responsible for the deubiquitylation of ubH2A. Based on the expression microarray data after hepatectomy, we found two ubiquitin-specific proteases, designated as USP21 and USP4 (Supplemental Fig. 6A,B). We synthesized His-tagged USP21 and USP4 in Escherischia coli and purified them (Fig. 2A). USP21 but not USP4 catalyzed the hydrolysis of mouse liver chromatin ubH2A (Fig. 2B). USP21 is a well-conserved cysteine protease among species (Supplemental Fig. 7). Although truncated USP21 was reported to be a nonspecific deubiquitylase by Gong et al. (2000), full-length USP21 hydrolyzed nucleosomal ubH2A, but not free ubH2A (Fig. 2C), suggesting that the N terminus of USP21 might be important for nucleosomal ubH2A specificity.
Figure 2.
Recombinant USP21 specifically hydrolyzes an isopeptide bond of ubiquitylated H2A in nucleosome form. (A) Expression of USP21 and USP4 in E. coli. (B) Recombinant USP21 (0.013–0.1 pmol) and the same amount of USP4 were incubated with native liver chromatin, prepared by glycerol gradient centrifugation after micrococcal nuclease digestion. After a 2-h incubation, the components were analyzed by Western blotting with a 1:250 dilution of mouse anti-ubH2A (Upstate Biotechnology #06-678) as primary antibody, 2 ng/μL of rabbit anti-mouse (Zymed #61-6400) as secondary antibody, and 125I protein A for detection. As negative control, buffer was used. Amidoblack staining of the immunoblot filter is shown to rule out the degradation of the core histones by the USPs. (C) Recombinant USP21 (0.16 pmol) was incubated with native liver chromatin and an equal amount of free core histones for the indicated times in minutes.
USP21 is conserved among species, as shown in Supplemental Figure 7. Triplet amino acids essential for cysteine protease, which were identified based on homology, are Cys 221, His 518, and aspartate 534, respectively. Mutants of these three were expressed and tested for activity. USP21wt catalyzes hydrolysis of ubiqutylated H2A, resulting in free ubiquitin as shown in lanes 2 and 3 of Supplemental Figure 8B. The three USP21 mutants, USP21CA (Cys 221 to alanine), USP21HA (His 518 to alanine), and USP21DN (aspartate 534 to asparagine) lacked activity as shown in Supplemental Figure 8C.
To confirm substrate specificity of USP21, we investigated whether there was USP21 substrate in the nuclear extract using Western blotting with anti-ubiquitin antibodies, as shown in Supplemental Figure 9. We incubated for only 10 min in the presence of recombinant USP21 to exclude the effect of native deubiquitylase activity. Ubiquitylated H2A is found to be the major substrate of USP21.
Ubiquitylation of H2A represses the di- and trimethylation of H3K4 but not that of H3K9
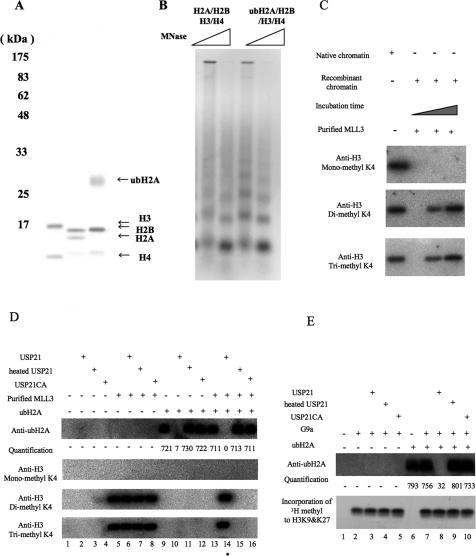
To characterize the effect of ubiquitylation in vitro, we purified H3–H4 tetramers and H2A–H2B and ubH2A–H2B dimers from mouse liver nuclei. The purified core histones are shown in Figure 3A. With all four core histones, the chromatin was assembled using NAP-1 and ACF (Fig. 3B; Ito et al. 1997). There were no significant differences in the nucleosome repeat length or the efficiency of histone deposition with either H2A–H2B dimers or ubiquitylated H2A–H2B dimers.
Figure 3.
Chromatin assembled with ubH2A directly inhibits H3K4 di- and trimethylation by MLL. (A) Coomassie staining of purified H3–H4 tetramer, H2A–H2B, and ubiquitylated H2A–H2B. Separation of ubH2A–H2B from H2A–H2B was performed with rabbit anti-ubH2A antibody purification and elution with bifurcated peptides. Both H2A–H2B and ubH2A–H2B were then further purified by SP Sepharose chromatography. (B) Chromatin assembly with native purified histones by NAP-1/ACF. The samples were partially digested with various concentrations of micrococcal nuclease and then deproteinized. For each set of reaction conditions, the different lanes represent increasing concentrations, from left to right, of micrococcal nuclease used to digest the chromatin. The resulting DNA fragments were subjected to agarose gel electrophoresis and visualized by staining with ethidium bromide. (C) Purified MLL3 was tested for its activity. Antibodies used to detect H3K4 mono- and trimethylation by Western blotting were Upstate Biotechnology #07-436 and Upstate Biotechnology #07-473, respectively. Antibodies used to detect H3K4 dimethylation by Western blotting are characterized in Supplemental Figure 1. As a positive control, native chromatin was used. For H3K4 methylation, a chromatin template containing 300 ng of all four core histones was incubated with purified mouse MLL in TKM buffer (50 mM Tris-Cl at pH 8.0, 20 mM KCl, 1 mM MgCl2) in the presence of 200 μM of SAM. (D,E) A chromatin template was made with the pGIE4 plasmid and purified H3–H4 in addition to H2A–H2B or ubiquitylated H2A–H2B, as indicated. To determine whether ubH2A is hydrolyzed by 0.16 pmol of USP21 or heat-inactivated USP21, Western blotting was performed with anti-ubH2A antibodies. (D) For H3K4 methylation, a chromatin template containing 300 ng of all four core histones was incubated with purified mouse MLL, USP21, heated USP21, and USP21CA, as indicated. H3K4 methylation was detected by Western blotting with anti-H3 mono-, di-, and trimethyl K4 antibodies. (E) For the methylation of H3K9&K27, a chromatin template containing 300 ng of all four core histones was incubated with purified mouse MLL, USP21, heated USP21, and USP21CA, as indicated. Then the template was subsequently incubated with 50 ng of G9a in TKM buffer in the presence of 10 kBq of 3H SAM. To detect the incorporation of 3H methyl groups, fluorography was performed.
Core histone modification is known to allow cross-talk between other modifications, such as H2B ubiquitylation and H3K4 or H3K36 methylation (Dover et al. 2002; Sun and Allis 2002; Henry et al. 2003). Since ubH2A is closely correlated with H3K9 trimethylation, but not H3K4 methylation during hepatocyte regeneration as shown in Supplemental Figure 1, we investigated whether ubH2A affects H3K9 methylation and H3K4 methylation. For these studies, we used chromatin assembled onto the pGIE4 plasmid, which contains Gal4 recognition sites and theAdE4 core promoter (Pazin et al. 1994).
To test the effect of H3K4 methylation using the reconstituted pure system in vitro, we purified native H3K4 methylase from mouse livers and identified MLL3 (Tan and Chow 2001; Goo et al. 2003) as the major detectable H3K4 methylase (data not shown). MLL3 introduced di- and trimethylation of H3K4, but not monomethylation detected by Western blotting with anti-H3 mono-, di-, and trimethyl K4 antibodies (Fig. 3C). Using purified components in an in vitro system, H3K4 was not methylated in the absence of MLL3 (Fig. 3D, lanes 1–4,9–12). MLL3 introduced di- and trimethylation of H3K4 but not monomethylation of H3K4 in the nucleosome with nonubiquitylated H2A, even in the presence of USP21, heat-inactivated USP21, or USP21CA (Fig. 3D, lanes 5–8). However, the ubiquitylation of H2A inhibited H3K4 methylation (Fig. 3D, lane 13), and USP21, but not heat-inactivated USP21 or USP21CA, relieved this inhibition by deubiquitylating ubH2A (Fig. 3D, lanes 14–16). These results thus reveal a trans-histone interaction in which ubiquitylation of H2A blocks di- and trimethylation of H3K4, and are consistent with the role of ubH2A in transcriptional repression.
Next, we investigated whether ubH2A could affect core histone methylation by G9a. G9a has been reported to induce the methylation of H3K9 and H3K27 using salt-dialyzed recombinant chromatin (Tachibana et al. 2002). Neither H2A ubiquitylation (Fig. 3E, lanes 2,7) nor additional USP21, heat-inactivated USP21, and USP21CA affected H3K9 or H3K27 methylation (Fig. 3E, lanes 3–5,8–10). We also tested the effect of H2A ubiquitylation on histone acetylation by p300 (Ito et al. 2000), a well-characterized coactivator. The ubiquitylation of H2A did not appear to affect the acetylation at H2A K5 or at H3 K14 by p300 (data not shown). It is notable that ubH2A specifically inhibits MLL3 but not G9A or p300.
Ubiquitylation of H2A represses transcriptional initiation but not elongation by inhibiting H3K4 di- and trimethylation
To clarify the effect of ubiquitylation on gene expression, we examined whether ubH2A affects in vitro transcription. For these experiments, we found that a chemical compound termed KYT-36, which was developed as a cysteine protease inhibitor (Kadowaki et al. 2004), inhibits deubiquitylation but does not affect transcription. KYT-36 can inhibit both recombinant USP21 and the intrinsic ubiquitin-specific protease activity present in Drosophila nuclear extract preparations, used as sources of GTFs (Supplemental Fig. 10). For in vitro transcription, the chromatin was assembled with either ubH2A–H2B or H2A–H2B.
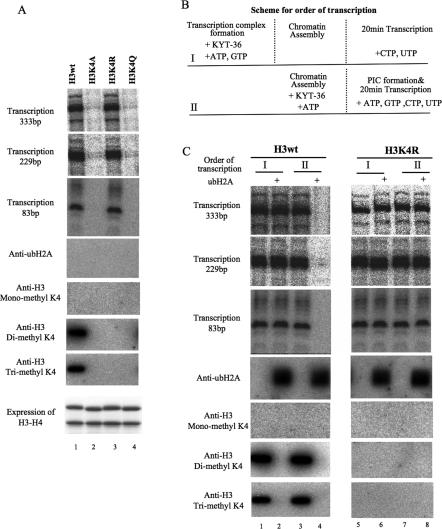
To investigate the importance of trans-histone cross-talk between H3K4 methylation and ubH2A, we introduced mutations into H3K4, resulting in H3K4A, H3K4R, and H3K4Q, which were assembled into chromatin by assembly factors as shown in Supplemental Figure 11. We initially tested the effects of the mutations on transcriptional activity in vitro. These experiments revealed that a significant amount of transcriptional initiation and elongation were detected from chromatin containing H3K4wt or H3K4R, but not from the chromatin containing H3K4A or H3K4Q (Fig. 4A). The K4 residue in wild-type H3 is di- and trimethylated during transcription (Fig. 4A, lane 1) by the pre-existing methylase in Drosophila nuclear extract used for transcription.
Figure 4.
ubH2A represses transcriptional initiation but not elongation, and the H3K4R mutation cancels this repression in vitro. (A) H3wt, cloned in pET11a (gift from James T. Kadonaga), was mutated into alanine(H3K4A), arginine(H3K4R), and glutamine(H3K4Q). The H3–H4 tetramer was expressed and purified, as shown in the lowest panel, basically as described previously (Aihara et al. 2004). Chromatin was assembled using recombinant H3–H4 and native H2A–H2B, and was subjected to GAL4-VP16-mediated transcription. Simultaneously, the chromatin was examined by Western blotting using anti-H3 mono-, di-, and trimethyl K4 antibodies. At the bottom, purified recombinant H3–H4 is shown. For primer extension to test transcription including elongation, oligonucleotide primers were designed to detect different length of transcripts (Supplemental Table 1). (B) Scheme of the order of transcription. For order I, the transcription complex was formed in the presence of GAL4-VP16, nuclear extract, and with limited nucleotides before chromatin assembly and subsequent transcription. For order II, the chromatin was assembled and then GAL4-VP16-mediated transcription was performed. (C) In vitro transcription was performed with chromatin made with H3wt or H3K4R by a different order of transcription, as illustrated in the scheme. Transcription was detected by primer extension as shown in A, and the template was examined by Western blotting with anti-ubH2A and anti-H3 mono-, di-, and trimethyl K4 antibodies after transcription.
We then used the mutant H3 histones to explore the relationship between H3K4 methylation and H2A ubiquitylation, as well as to determine which transcription step is repressed by ubiquitylation. In these experiments, we altered the order of chromatin assembly and transcription complex assembly, as illustrated in Figure 4B. Chromatin was assembled with recombinant wild-type H3 or H3K4R and native purified H2A or ubH2A. In one set of reactions (Fig. 4B, I), we first assembled transcription complexes (with ATP and GTP), and then assembled the chromatin prior to the addition of the remaining rNTPs that are needed for transcription. In the second set of reactions (Fig. 4B, II), we first assembled the chromatin and then carried out the transcription reactions. These experiments revealed that ubH2A inhibits the steps leading to transcription initiation (Fig. 4C, lanes 3,4), but not transcription from a pre-established transcription complex (Fig. 4C, lanes 1,2) or transcript elongation (Fig. 4C, lanes 1,2,5–8). The presence of ubH2A also inhibited H3K4 di- and trimethylation. In addition, transcriptional repression by ubH2A was observed with wild-type H3 but not with H3K4R (Fig. 4C, lanes 4,8). Therefore, these results show that transcriptional repression by ubH2A occurs at the stage of transcription initiation and specifically involves the inhibition of H3K4 di- and trimethylation through trans-histone cross-talk.
USP21 regulates transcription in vivo
We introduced USP21wt and USP21CA (catalytic domain mutant; cysteine to alanine) fused with DsRed fluorescence into liver and explored the effect of deubiquitylation in vivo, together with a fluorescent protein as a control (Fig. 5A,B). The introduced USP21wt decreased the amount of ubH2A, in both quiescent and regenerating hepatocytes (Fig. 5C, rows II,III, arrow), but the introduction of USP21CA had no detectable effect (Fig. 5C, rows IV,V, arrow).
Figure 5.
USP21 regulates transcription in vivo. (A) Scheme for DsRed-USP fusion or control constructs that were introduced into liver by in vivo electroporation. (B) The fluorescent protein construct was introduced into liver 48 h before hepatectomy. Samples removed by hepatectomy were designated as quiescent hepatocytes, and the residual liver removed 12 h after hepatectomy was designated as regenerating hepatocytes. (C) Quiescent and regenerating hepatocytes that expressed fluorescent protein are shown, together with DNA staining and immunostaining by anti-ubH2A antibodies, and these three panels are merged, as shown in the rows designated I–V. The arrow indicates a cell with ubH2A decreased by the introduction of USP21wt in rows II and III, and without a significant change in ubH2A by the introduction of USP21CA in rows IV and V. “Q” and “R” indicate quiescent and regenerating hepatocytes, respectively. (D) Quiescent and regenerating hepatocytes that expressed fluorescent protein are shown, together with DNA staining and immunostaining by anti-Serpina6 antibodies, and three panels are merged, as shown in the rows designated I–V. The arrowhead indicates a cell with increased Serpina6 expression by the introduction of USP21wt in row III and with decreased Serpina6 expression by the introduction of USP21CA in row IV. (E,G) USP21 decreased H2A ubiquitylation in both quiescent and regenerating hepatocytes. (F) USP21CA down-regulates Serpina6 in quiescent hepatocytes. (H) USP21wt up-regulates Serpina6 in regenerating liver. (E–H) Fifty positive nuclei, together with at least two associated controls, were counted for each experiment, and an error bar is indicated in each graph.
To explore the regulation of gene expression affected by ubiquitylation and deubiquitylation during hepatocyte regeneration, we focused on Serpina6, since its down-regulation correlated with increased H2A ubiquitylation (Fig. 1F) and decreased histone H3K4 methylation (data not shown). The introduction of USP21wt into liver up-regulated Serpina6 in regenerating liver, which was expected to be down-regulated after hepatocyte regeneration (Fig. 5D, row III, arrowhead). Conversely, the introduction of USP21CA into liver down-regulated Serpina6 in quiescent liver, even though it is usually up-regulated before hepatocyte regeneration (Fig. 5D, row IV, arrowhead).
The intensities of Serpina6 and ubH2A were measured and calculated using paired staining with and without fluorescent protein, using the same tissue section as shown in Supplemental Figure 12. This analysis clearly indicated that the introduction of USP21wt into liver decreased ubH2A and up-regulated Serpina6 during hepatocyte regeneration (Fig. 5E–H). These observations support the model that ubH2A at Lys 119 regulates transcription during hepatocyte regeneration. The introduction of USP21CA mutant down-regulated Serpina6 in quiescent liver. Although a significant increase in ubH2A was not clearly observed, probably due to H2A dynamics, it might be difficult to detect by immunostaining.
Discussion
Although the ubiquitylation of H2A was first described three decades ago, very little is presently known about the physiological role of this modification. We now proved that histone H2A ubiquitylation affects transcriptional initiation, but not elongation, through its impact on H3K4 di- and trimethylation. It is notable that H3K4 di- and trimethylation is essential for transcriptional initiation, as shown by the different orders of transcription and chromatin assembly.
As we showed a direct connection between H2A ubiquitylation and H3K4 di- and trimethylation, in addition to the important role of H3K4 methylation for transcript initiation, it is likely that deubiquitylation by USP21 regulates transcriptional initiation partly by affecting H3K4 di- and trimethylation. In addition, H3K4 di- and trimethylation can be functionally replaced by changing Lys 4 to arginine, as chromatin containing H3K4R is also an active template (Fig. 4A, lane 3). Since the pKR values of lysine and arginine are 10.5 and 12.5, respectively, arginine might mimic methylated lysine in terms of its positive charge. These findings suggest that a positive charge on the side chain is important for transcriptional initiation and that H3K4 methylation, affected by ubH2A, is indispensable for transcriptional initiation. Thus, our biochemical results suggest that USP21 deubiquitylates H2A, which then allows H3K4 methylation and transcriptional activation. We found a novel connection between ubH2A, H3K4 methylation, and transcriptional initiation, as depicted in Figure 6.
Figure 6.
Model for transcriptional initiation from a chromatin template. Deubiquitylation of histone H2A by USP21 activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. The designations “ub” and “me” indicate histone H2A ubiquitylation and H3K4 di- and trimethylation, respectively.
Concerning the connection between H2A deubiquitylation and H3K4 methylation, ubH2A may change the substrate specificity of MLL3 and other H3K4 methylases that normally methylate H3K4 in the nucleus without ubH2A. According to the crystal structure of the nucleosome, the C terminus of H2A and the N terminus of H3 form the entrance for the DNA (Luger et al. 1997). Thus, according to the three-dimensional structure, they are close together, and the substrate specificity of the methylase could be altered by the H2A ubiquitylation. How this H2A ubiquitylation affects only MLL3, but not G9A, needs to be explored in the future, in the context of the nucleosome structure.
In addition, our results investigating overexpression of USP21 and USP21CA in vivo supports the biochemical data that USP21 regulates transcription. Although USP21 is induced and Serpina6 is repressed after hepatectomy, overexpression of USP21 might activate Serpina6 that is normally repressed during hepatocyte regeneration. It is also suggested that overexpression of USP21CA behaved like a dominant-negative and repressed Serpina6 that is normally expressed before hepatectomy. Thus, our in vivo experiments with overexpression of USP21 and USP21CA strengthen our in vitro results.
Recent genomic-scale analyses of histone modifications indicated that H3K4 methylation, including di- and trimethylation, is associated with active genes (Bernstein et al. 2005). Several factors have been proposed to recognize methylated H3K4 tails via the PHD domain and chromodomain, thus stimulating transcription (for review, see Bannister and Kouzarides 2005). These factors bind to methylated H3K4 tails and could be part of a histone modification regulatory pathway involving the deubiquitylation of H2A, as a prerequisite for H3K4 methylation, with subsequent transcriptional activation. Such factors include CHD1 (Pray-Grant et al. 2005; Sims et al. 2005), Isw1 (Santos-Rosa et al. 2003), WDR5 (Wysocka et al. 2005), and a PHD finger of NURF (Li et al. 2006; Wysocka et al. 2006). Using an in vitro transcription system in which GTFs are assembled on the core promoter to form transcription complexes, it would be important to investigate what is lacking in these complexes when H3K4 Lys 4 has been mutated to alanine or glutamine. These studies will shed more light on the mechanistic aspects of transcriptional initiation in the nucleosome structure.
In addition to the trans-histone cross-talk between H2A ubiquitylation and H3K4 methylation shown in this manuscript, it has been found that histone H2B ubiquitylation controls the outcome of methylation at H3K4 in Saccharomyces cerevisiae. Thus, H2B ubiquitylation may also act as a switch for gene expression through trans-histone cross-talk (Briggs et al. 2002; Sun and Allis 2002). Futhermore, the H2B ubiquitylation mediated by Rad6 and H2B deubiquitylation, caused by SAGA-associated Ubp8, defines the regions of active chromatin by affecting the methylation of histone H3 Lys 4 and Lys 79 (Henry et al. 2003; Kao et al. 2004). Although these data are from yeast, it is possible that the deubiquitylation of H2A and the ubiquitylation of H2B regulate gene expression from one promoter in concert.
Recently, there have been other reports of H2Aub-specific deubiquitylases. Other H2Aub-specific deubiquitylases play a role during development and as a coactivator of nuclear receptors (Joo et al. 2007; Zhu et al. 2007). USP21 is up-regulated during hepatic regeneration and expressed in proliferating cultured cells. USP21 also associates with RNA polII in hepatocytes (T. Nakagawa and T. Ito, unpubl.). Thus, it is likely that USP21 plays a role in transcription during mammalian hepatic regeneration as well as in cell proliferation.
In perspective, it also should be pointed out that these trans-histone cross-talks might depend on a specific promoter, and H2A ubiquitylation and H2B ubiquitylation might act separately to generate diversity in gene expression. These points need to be investigated further, considering the different cis-elements and the multiple trans-histone cross-talk networks.
Materials and methods
ChIP
Hepatocytes were recovered at the indicated time point. After homogenizing and washing with PBS, 20 mg of nucleus were subjected to DNA histone cross-linking by adding 18.5% formaldehyde to a final concentration of 1%, incubating for 20 min, aspirating the formaldehyde, and washing the nucleus with PBS. After washing with solution I (10 mM HEPES at pH 7.5, 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100) and solution II (10 mM HEPES at pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA), the supernatant was removed, and 500 μL of solution III (150 mM NaCl, 25 mM Tris at pH 7.5, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% Deoxycholate) were added. Subsequently, samples were sonicated extensively (20%∼30% output, 12 sec × 15 times) and spun at 14,000 rpm for 10 min at 4°C. The supernatant was used for ChIP, using 25 μg of anti-ubH2A (Supplemental Fig. 1), 25 μg of anti-H3 dimethyl K4 (Supplemental Fig. 2), 5 μL of anti-H3 trimethyl K4 serum (Upstate Biotechnology #07-473), and 25 μg of anti-H3 trimethyl K9 (Supplemental Fig. 3) antibodies, together with 25 μg of rabbit IgG (Sigma #I-5006) as a control. Resulting immunoprecipitated DNA was analyzed with PCR for quantification. The linear range of detection for the various PCR products was determined by varying the cycle number from 19 to 31 in a three-cycle increment using the gene-specific primers (Supplemental Table 1). Twenty-eight cycles for serpina6 and LCN2 and 22 cycles for GAPDH were performed for 30 sec at 94°C, 1 min at 55°C, and 1 min at 72°C. PCR products were separated on an agarose gel and stained with ethidium bromide. The resulting gels were exposed to UV illumination, and densitometoric analyses were conducted on visible bands using the FluorChem Imaging Systems (Alpha Innotech Corp.) (Raju and Bird 2003; Park et al. 2005).
Purification of core histones
For the purification of the ubiquitylated H2A–H2B dimer, mouse liver was homogenized and digested with micrococcal nuclease. This preparation was fractionated on a 15%–40% (w/v) glycerol gradient, and the peak fraction was applied to a hydroxyapatite column. Core histones were eluted with a linear salt concentration gradient and divided into H3–H4 tetramer and H2A–H2B dimers. The H2A–H2B fraction contained the H2A–H2B dimer and the ubiquitylated H2A–H2B dimer. The ubiquitylated H2A–H2B dimer was purified using the anti-ubiquitylated H2A antibodies characterized in Supplemental Figure 3. These antibodies could immunodeplete the ubiquitylated H2A–H2B dimer from the bulk H2A–H2B dimer. The ubiquitylated H2A–H2B dimer could be immunoeluted from an affinity column using a bifurcated peptide. The purified H2A–H2B and ubiquitylated H2A–H2B dimers were further purified by SP Sepharose column chromatography.
Chromatin assembly reactions
Chromatin assembly reactions were performed essentially as described previously using supercoiled plasmid DNA (Bulger et al. 1995; Ito et al. 1997). A standard reaction contained plasmid DNA (0.4 mg), purified core histones from Drosophila embryos (0.33 mg), purified recombinant dNAP-1, purified recombinant ACF, ATP (3 mM), and an ATP-regenerating system (30 mM phosphocreatine and 1 mg/mL creatine phosphokinase) in HEG buffer (25 mM Hepes [K+] at pH 7.6, 0.1 mM EDTA, 10% [vol/vol] glycerol) containing 50 mM KCl and 5 mM MgCl2.
Reconstitution of nucleosomes by salt dialysis
Typically, chromatin was reconstituted with 100 μg of supercoiled pGIE0 plasmid DNA, which has GAL4-binding sites before the AdE4 promoter, and 100 μg of purified Drosophila core histones at a 1 M NaCl concentration, using salt dialysis techniques (Camerini-Otero et al. 1976; Germond et al. 1976). The final dialysis buffer contained 20 mM Tris-HCl (pH 7.2), 0.2 mM EDTA, and 50 mM NaCl. The most highly reconstituted chromatin was then enriched by 15%–40% (w/v) glycerol gradient sedimentation (60,000 rpm, 6 h, 4°C; Beckman SW60 rotor). The gradient fractions that contained the fastest migrating chromatin (which had the highest density of nucleosomes) were pooled, dialyzed against HEG buffer containing 50 mM KCl, and stored at 4°C.
In vitro transcription
For in vitro transcription with the order of preinitiation complex formation before assembly, purified Gal4-VP16 (50 nM final concentration), nuclear extract, 3 mM ATP, and 500 nM CTP were added to 75 ng of pGIE0 plasmid DNA prior to chromatin assembly by assembly factors and, subsequently, 500 nM concentrations of UTP and GTP were added to start transcription. For in vitro transcription with the order of preinitiation complex formation after assembly, 75 ng of pGIE0 plasmid DNA were assembled into chromatin by assembly factors, and the resulting chromatin samples were then subjected to in vitro transcription by adding nuclear extract and 500 nM of each NTP. Transcripts were detected by primer extension and subsequent 8% denaturing gel electrophoresis. To detect transcriptional elongation, different primers—E1, E2, and E3—that are located 83 bp, 229 bp, and 333 bp from the initiation site, respectively, were used for a primer extension reaction (Supplemental Table 1).
In vivo electroporation
Each mouse was anesthetized with an intraperitoneal injection of Nembutal (30 mg/kg body weight), and the liver was exposed by a mid-abdominal incision. A 300-μL (240 μg) aliquot of DNA solution was injected into both the left and caudate lobes of the liver, under compression of the portal vein. Electric pulses were delivered using an electric pulse generator (Electroporator CUY-21, Tokiwa Science). Each lobe was held between a pair of electrodes, and square electric pulses were applied seven times with a voltage of 40 V and a 100-msec pulse length, with intervals of 700 msec, during each injection.
Immunostaining and quantification
At the indicated time after injection of the construct, the liver was resected and placed in OCT compound, wrapped in foil, and frozen in a dry ice/ethanol bath. Five-micron sections were prepared and subjected to immunostaining with 50 ng/μL purified anti-ubH2A (Supplemental Fig. 3) or a 1:20 dilution of anti-Serpina6 serum (Supplemental Fig. 4) as primary antibody, and 10 ng/μL Alexa 633-labeled goat anti-rabbit (Invitrogen #A21070) as secondary antibody, together with 0.1 μM YOYO-1 (Invitrogen #Y3601) for DNA staining to observe the DsRed fluorescent protein. For quantification, the intensity and counts of the nuclei where the DsRed fluorescent protein was incorporated were measured, and the ratio was calculated using the intensity and the counts of the same sized area of the same tissue section, where the DsRed fluorescent protein was not incorporated, as a control.
Acknowledgments
We thank James T. Kadonaga for helpful discussions and advice. We are grateful to W. Lee Kraus for the p300-expressing baculovirus, and to Yoichi Shinkai for the G9a plasmid. We also thank Masaya Oki, Daishi Inoue, Kumi Yoshimoto, and Hikari Ikeda for helpful discussions and technical support.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1609708
References
- Aihara H., Nakagawa T., Yasui K., Ohta T., Hirose S., Dhomae N., Takio K., Kaneko M., Takeshima Y., Muramatsu M., et al. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes & Dev. 2004;18:877–888. doi: 10.1101/gad.1184604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Briggs S.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Bulger M., Ito T., Kamakaka R.T., Kadonaga J.T. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc. Natl. Acad. Sci. 1995;92:11726–11730. doi: 10.1073/pnas.92.25.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R.D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: Evidence for an arginine-rich histone kernel. Cell. 1976;8:333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Costa R.H., Kalinichenko V.V., Holterman A.X., Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J.E., Wakao R., Tang Y.A., Endoh M., Appanah R., Nesterova T.B., Silva J., Otte A.P., Vidal M., et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Fausto N., Campbell J.S., Riehle K.J. Liver regeneration. Hepatology. 2006;43 (Suppl. 1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Allis C.D. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Germond J.E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976;3:3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I.L., Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I.L., Taylor C.W., Baum R.M., Yeoman L.C., Olson M.O., Prestayko A.W., Busch H. Isolation and characterization of protein A24, a ‘histone-like’ non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- Gong L., Kamitani T., Millas S., Yeh E.T. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem. 2000;275:14212–14216. doi: 10.1074/jbc.275.19.14212. [DOI] [PubMed] [Google Scholar]
- Goo Y.H., Sohn Y.C., Kim D.H., Kim S.W., Kang M.J., Jung D.J., Kwak E., Barlev N.A., Berger S.L., Chow V.T., et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., Berger S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins G.M., Anderson R.M. Experimental pathology of the liver. Arch. Pathol. 1931;12:186–202. [Google Scholar]
- Hochheimer A., Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes & Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger M., Pazin M.J., Kobayashi R., Kadonaga J.T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Ito T., Ikehara T., Nakagawa T., Kraus W.L., Muramatsu M. p300-mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone. Genes & Dev. 2000;14:1899–1907. [PMC free article] [PubMed] [Google Scholar]
- Jason L.J., Moore S.C., Lewis J.D., Lindsey G., Ausio J. Histone ubiquitination: A tagging tail unfolds? Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- Joo H.Y., Zhai L., Yang C., Nie S., Erdjument-Bromage H., Tempst P., Chang C., Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Kadowaki T., Baba A., Abe N., Takii R., Hashimoto M., Tsukuba T., Okazaki S., Suda Y., Asao T., Yamamoto K. Suppression of pathogenicity of Porphyromonas gingivalis by newly developed gingipain inhibitors. Mol. Pharmacol. 2004;66:1599–1606. doi: 10.1124/mol.104.004366. [DOI] [PubMed] [Google Scholar]
- Kao C.F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M.A. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes & Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Cook P.R. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Li H., Ilin S., Wang W., Duncan E.M., Wysocka J., Allis C.D., Patel D.J. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mangnall D., Bird N.C., Majeed A.W. The molecular physiology of liver regeneration following partial hepatectomy. Liver Int. 2003;23:124–138. doi: 10.1034/j.1600-0676.2003.00812.x. [DOI] [PubMed] [Google Scholar]
- Mellor J. The dynamics of chromatin remodeling at promoters. Mol. Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Park S.O., Zheng Z., Oppenheimer D.G., Hauser B.A. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development. 2005;132:841–849. doi: 10.1242/dev.01654. [DOI] [PubMed] [Google Scholar]
- Pazin M.J., Kamakaka R.T., Kadonaga J.T. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Pray-Grant M.G., Daniel J.A., Schieltz D., Yates J.R., Grant P.A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Raju J., Bird R.P. Energy restriction reduces the number of advanced aberrant crypt foci and attenuates the expression of colonic transforming growth factor β and cyclooxygenase isoforms in Zucker obese (fa/fa) rats. Cancer Res. 2003;63:6595–6601. [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bernstein B.E., Karabetsou N., Morillon A., Weise C., Schreiber S.L., Mellor J., Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Sims R.J., Chen C.F., Santos-Rosa H., Kouzarides T., Patel S.S., Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun Z.W., Allis C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.C., Chow V.T. Novel human HALR (MLL3) gene encodes a protein homologous to ALR and to ALL-1 involved in leukemia, and maps to chromosome 7q36 associated with leukemia and developmental defects. Cancer Detect. Prev. 2001;25:454–469. [PubMed] [Google Scholar]
- Taub R. Liver regeneration 4: Transcriptional control of liver regeneration. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- Togo S., Makino H., Kobayashi T., Morita T., Shimizu T., Kubota T., Ichikawa Y., Ishikawa T., Okazaki Y., Hayashizaki Y., et al. Mechanism of liver regeneration after partial hepatectomy using mouse cDNA microarray. J. Hepatol. 2004;40:464–471. doi: 10.1016/j.jhep.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- White P., Brestelli J.E., Kaestner K.H., Greenbaum L.E. Identification of transcriptional networks during liver regeneration. J. Biol. Chem. 2005;280:3715–3722. doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes & Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Zhu P., Zhou W., Wang J., Puc J., Ohgi K.A., Erdjument-Bromage H., Tempst P., Glass C.K., Rosenfeld M.G. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]