Abstract
Histone mRNAs are rapidly degraded at the end of S phase or when DNA replication is inhibited. Histone mRNAs end in a conserved stem–loop rather than a poly(A) tail. Degradation of histone mRNAs requires the stem–loop sequence, which binds the stem–loop-binding protein (SLBP), active translation of the histone mRNA, and the location of the stem–loop close to the termination codon. We report that the initial step in histone mRNA degradation is the addition of uridines to the 3′ end of the histone mRNA, both after inhibition of DNA replication and at the end of S phase. Lsm1 is required for histone mRNA degradation and is present in a complex containing SLBP on the 3′ end of histone mRNA after inhibition of DNA replication. We cloned degradation intermediates that had been partially degraded from both the 5′ and the 3′ ends. RNAi experiments demonstrate that both the exosome and 5′-to-3′ decay pathway components are required for degradation, and individual histone mRNAs are then degraded simultaneously 5′ to 3′ and 3′ to 5′.
[Keywords: Histone, cell cycle, mRNA stability, decapping, exosome, oligouridylation]
Replication-dependent histone mRNA levels are tightly coupled to DNA replication. Proper control of histone protein synthesis is essential to ensure proper chromatin formation. A major step in regulation of mammalian histone mRNA is a rapid alteration in the half-life of histone mRNA in response to changes in the rate of DNA synthesis, allowing the cell to rapidly adjust the rate of histone protein synthesis to maintain balanced synthesis of DNA and histone protein. The regulation of the half-life of histone mRNA is mediated by the unique 3′ end of histone mRNA (Pandey and Marzluff 1987; Kaygun and Marzluff 2005b). The metazoan replication-dependent histone mRNAs are the only eukaryotic mRNAs that are not polyadenylated (Marzluff 2005). The stem–loop-binding protein (SLBP) binds the conserved stem–loop present at the 3′ end of all five classes of histone mRNAs (Wang et al. 1996; Martin et al. 1997). SLBP is required for histone pre-mRNA processing (Marzluff 2005) and accompanies the histone mRNA to the cytoplasm (Erkmann et al. 2005), where it is required for histone mRNA translation (Sanchez and Marzluff 2002). SLBP plays a direct role in histone mRNA stability through an interaction with Upf1 (Kaygun and Marzluff 2005a).
Inhibition of DNA replication using pharmacologic agents results in a rapid degradation of histone mRNA. Degradation requires active translation of the mRNA and the stem–loop at the 3′ end. The location of the stem–loop relative to the stop codon, 45–80 nucleotides (nt), is also essential, as increasing this distance results in a loss of regulated degradation and persistent expression (Graves et al. 1987; Kaygun and Marzluff 2005b). These characteristics suggest that a terminating ribosome must be in close approximation to the stem–loop and likely SLBP. Supporting this model, we recently found that Upf1, a factor involved in translation termination and nonsense-mediated decay (NMD), is required for histone mRNA degradation (Kaygun and Marzluff 2005a).
The initial step in degradation of most polyadenylated mRNAs in eukaryotes is shortening of the poly(A) tail by a deadenylase complex (Parker and Song 2004). Deadenylated mRNAs can subsequently return to a translationally active state on polysomes, likely by the enzymatic action of cytoplasmic poly(A) polymerases (Coller and Parker 2005) or can be degraded from either terminus using a distinct set of nucleases. Upon poly(A) shortening, the Lsm1–7 complex is thought to recruit a decapping complex (Dcp1/Dcp2) to remove the 5′ cap of the mRNA (Coller et al. 2001). The decapped mRNA can then be degraded 5′–3′ by the Xrn1 exoribonuclease or can be exposed to a large complex of 3′–5′ exonucleases known as the exosome (Mitchell et al. 1997). Several of the proteins involved in catalyzing 5′–3′ degradation are localized to discrete cytoplasmic loci termed P bodies or GW bodies (Eystathioy et al. 2003; Sheth and Parker 2003). Whether 5′–3′ degradation is restricted to P bodies is not known. Exosome components are located in both the nucleus and the cytoplasm (Lejeune et al. 2003). Whether both the 5′–3′ and 3′–5′ pathways can contribute to the degradation of a single mRNA molecule is not clear.
The detailed biochemical pathway of regulated histone mRNA degradation is not understood. Here we report a novel pathway for histone mRNA degradation that involves initial addition of uridines to the 3′ end of histone mRNA. Treatment of cells with hydroxyurea (HU) results in a dramatic increase in oligouridylated histone mRNAs, and at the end of S phase, cells also accumulate oligouridylated histone mRNAs. The 3′ oligo(U) tail likely binds Lsm1–7, initiating degradation of the histone mRNA. RNAi experiments also suggest a role for both the exosome and the decapping enzymes in degradation. In agreement with this result, we find intermediates of degradation that have been decapped and fragments of histone RNAs that have been degraded from both the 5′ and 3′ ends. In addition, we find intermediates where there has been initial partial degradation on the 3′ side of the stem–loop, and subsequent uridylation of the partially degraded stem–loop, reminiscent of the degradation of bacterial mRNAs (Kushner 2004). Two candidate terminal uridylyl transferases (TUTases) have been identified that are both required for histone mRNA degradation. These two TUTases are localized to the cytoplasm, where they likely uridylate actively translating histone mRNA substrates, modulating a dramatic decrease in their half-lives.
Results
Knockdown of 3′hExo does not affect histone mRNA degradation in vivo
Polyadenylated mRNAs are primarily degraded by a pathway that initiates deadenylation, removing most of the poly(A) tail, followed either by decapping and 5′-to-3′ degradation or degradation 3′-to-5′ by the exosome (Parker and Song 2004). Although it is known that the stem–loop at the 3′ end of histone mRNA is the cis element that regulates histone mRNA degradation (Pandey and Marzluff 1987), the biochemical details of histone mRNA degradation are not known. Early studies by Ross and coworkers (Ross and Kobs 1986; Ross et al. 1986, 1987) suggested that degradation of histone mRNA proceeded 3′ to 5′. Since we recently identified an exonuclease, 3′hExo, that can specifically degrade histone mRNA from the 3′ end in vitro (Dominski et al. 2003), and which can form a ternary complex with the stem–loop of histone mRNA and SLBP, we tested whether the 3′hExo might play a role in initiating histone mRNA degradation. Since the critical cis element for histone mRNA degradation is also at the 3′ end of the mRNA, we tested known factors in decay of polyadenylated mRNAs to see if they might also be involved in histone mRNA degradation.
For all RNAi experiments, we knocked down the targeted proteins using two sequential treatments with siRNA (Wagner and Garcia-Blanco 2002) and used two different siRNAs for each protein. Cells were treated with a control siRNA (C2) that did not knock down any of the proteins. We also used siRNAs that targeted the splicing factor polypyrimidine tract-binding (PTB) protein, as a control. The cells were treated with 5 mM HU, and the amount of histone mRNA degradation was measured over a 45-min time course.
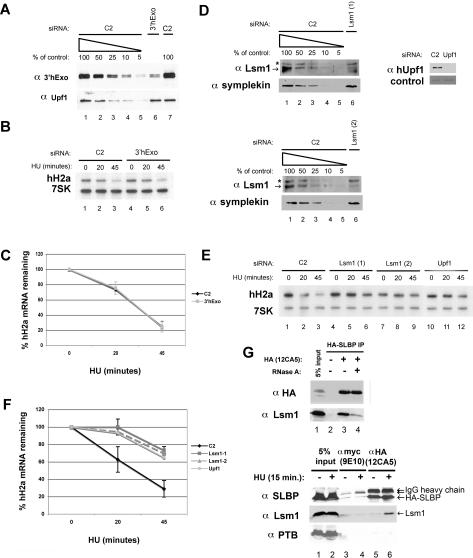
The transfected siRNA reduced 3′hExo protein levels by 80%–90% as estimated by Western blot analysis of a dilution series of control treated lysates (Fig. 1A). Total RNA from these cells was subjected to Northern blot analysis simultaneously probing for both histone H2a mRNA and 7SK RNA as a loading control. Knockdown of 3′hExo has no effect on histone H2a mRNA degradation in HeLa cells (Fig. 1B,C). There was also no change in the cell cycle distribution as determined by FACS (Supplemental Fig. S1A). The steady-state levels of histone mRNA in the exponentially growing cells (0 time point) was similar in both the C2 (control) and 3′hExo knockdowns (Fig. 1B, cf. lanes 1 and 4). Thus, knockdown of 3′hExo had no discernable effect on histone mRNA metabolism. Knocking down PTB also had no effect on histone mRNA regulation or cell growth (Supplemental Fig. S2A–C).
Figure 1.
Effect of knockdown of 3′hExo or Lsm1 on histone mRNA degradation. HeLa cells were treated with the 3′hExo (A–C) or Lsm1 (D–F) siRNAs as described in Materials and Methods, and then treated with 5 mM HU and total cell RNA prepared 0, 20, or 45 min after HU treatment. (A,D) A dilution series of the control (C2) cell lysate together with a lysate from cells treated with the indicated siRNA was analyzed by Western blotting for 3′hExo and Upf1. The asterisk indicates a cross-reacting band. (B,E) Two micrograms of total cell RNA were resolved by urea acrylamide gel electrophoresis, transferred to N+ nitrocellulose, and probed with a mixture of histone H2a mRNA and 7SK snRNA probes. (C,F) The average of three independent experiments is shown. (C) (◆) C2; (■) 3′hExo. (F) (◆) C2; (■) Lsm1-1; (▴) Lsm1-2; (●) Upf1. Standard deviations are indicated by vertical bars. (G) Lysates were prepared from S-phase cells expressing HA-SLBP 15 min after treatment with HU as described previously (Kaygun and Marzluff 2005a). (Top, lane 3) A portion of the lysate was treated with RNase A (20 μg/mL) for 15 min prior to immunoprecipitation with anti-HA antibody. The immunoprecipitates were analyzed for SLBP and Lsm1 by Western blotting. (Bottom) S-phase cells expressing HA-SLBP were treated with HU for 15 min; lysates were prepared and subjected to immunoprecipitation with anti-myc (lanes 3,4) or anti-HA (lanes 5,6) antibodies. The immunoprecipitates were analyzed for SLBP, Lsm1, and PTB as a control by Western blotting. (Lanes 1,2) The input lanes show 5% of the total lysate.
Lsm1 plays an integral role in replication-dependent histone mRNA degradation
We tested several decay factors and enzymes involved in regulating poly(A) mRNA stability by RNAi to see if any of these factors affected histone mRNA degradation. The Lsm1–7 complex is a heteroheptameric set of polypeptides that is a positive regulator of decapping in Saccharomyces cerevisiae (Tharun et al. 2000; Tharun and Parker 2001) and has also been shown to be important in regulating the stability of mRNAs containing AU-rich elements (ARE) (Mukherjee et al. 2002; Stoecklin et al. 2006). We carried out a series of RNAi experiments assessing the role of various factors using C2 siRNA as a negative control and Upf1 as a positive control for a factor that reduces histone mRNA degradation. Each experiment shown represents an example of a panel of experiments done on parallel cultures.
The down-regulation of Lsm1 protein varied from 75% to 95% among different experiments as estimated from the protein dilution series of the control (C2) sample (Fig. 1D). Each siRNA had a similar degree of knockdown, and Symplekin, a scaffold protein involved in mRNA 3′ end formation (Takagaki and Manley 2000; Kolev and Steitz 2005), served as a loading control in this experiment. We showed previously that the NMD factor hUpf1 is important in regulating the rapid decay of histone mRNA (Kaygun and Marzluff 2005a), and we used this as a positive control. Transfection of either of two siRNAs targeting Lsm1 resulted in stabilization of histone mRNA at the 20-min time point (Fig. 1E, cf. lanes 2, 5, and 8) and over a 2.5-fold stabilization at the 45-min time point (Figs. 1E [cf. lanes 3, 6, and 9], 3G [below]) relative to C2 siRNA-transfected cells. Figure 1F summarizes three independent RNAi experiments. The cell cycle distributions of the cells were not significantly changed by the Lsm1 knockdown (Supplemental Fig. S1B).
Figure 3.
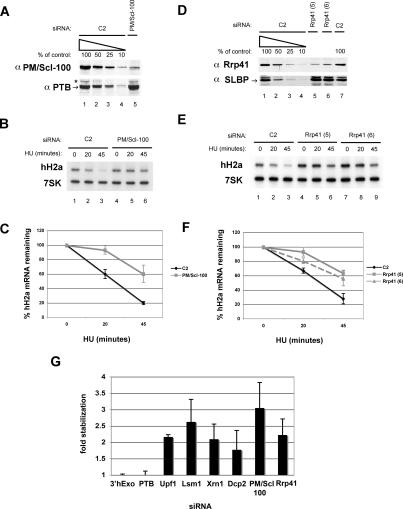
Effect of knockdown of exosome components on histone mRNA degradation. (A,D) HeLa cells were treated with the indicated siRNAs to PM/Scl-100 (A–C) or Rrp41 (D–F) and were treated with 5 mM HU, and the levels of each protein were determined by Western blotting as in Figures 1 and 2. (B,E) Two micrograms of total cell RNA from cells with PM/Scl100 knocked down (B) or Rrp41 knocked down with two siRNAs (5 and 6) to Rrp41 were resolved by urea-acrylamide gel electrophoresis and probed with a mixture of histone H2a mRNA and 7SK snRNA probes as in Figure 1. (C,F) The average of three independent knockdowns to PM/Scl-100 (C: ◆, C2; ■, PM/Scl-100) and Rrp41 [F: ◆, C2; ■, Rrp41(1); -▴-, Rrp41(2)] is shown. (G) Average fold stabilization of all experiments shown in Figures 1, 2, 3, with standard deviations indicated by vertical bars.
Given the apparent role of Lsm1 in histone mRNA decay, we asked whether we could detect Lsm1 in SLBP immunoprecipitates when histone mRNAs were being rapidly degraded. Lysates were prepared 15 min after HU treatment from S-phase cells stably expressing HA-tagged SLBP (Erkmann et al. 2005) and immunoprecipitated with anti-HA antibody. We then probed for Lsm1. Detectable amounts of Lsm1 were coimmunoprecipitated with HA-SLBP. When we treated the cell lysates with RNase A prior to immunoprecipitation, the amount of Lsm1 coimmunoprecipitated was greatly reduced (Fig. 1G, top, lanes 3,4). This result suggests that both SLBP and Lsm1 can be bound to histone mRNA but that they do not stably interact with each other, in contrast to the interaction of SLBP and Upf1, which is resistant to RNase digestion (Kaygun and Marzluff 2006). In a separate experiment, we did not detect association of Lsm1 with SLBP in S-phase cells prior to HU treatment, although it was readily detected 15 min after HU treatment (Fig. 1G, bottom, lanes 5,6). Another RNA-binding protein, PTB, was not present in any of the SLBP immunoprecipitates, demonstrating the specificity of the interaction. Thus Lsm1 only interacts with SLBP when histone mRNA is being rapidly degraded.
Down-regulation of the decapping enzyme Dcp2 and the 5′–3′ exonuclease Xrn1 reduces histone mRNA decay
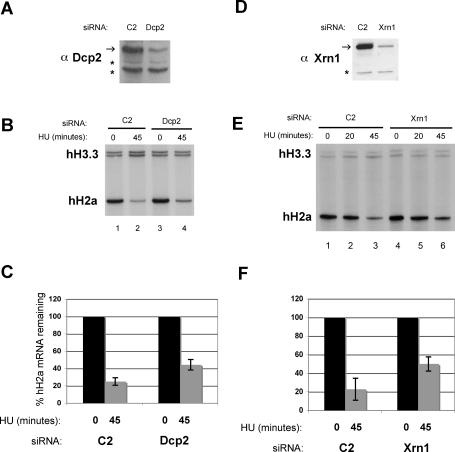
Since the Lsm1–7 complex is a positive regulator of decapping and 5′–3′ decay and had an effect on histone mRNA stability, we examined other components of the 5′–3′ mRNA decay apparatus. We knocked down the catalytic subunit of the decapping complex Dcp2 (Wang et al. 2002) and the 5′–3′ exonuclease Xrn1/Rat1 (Long and McNally 2003) using targeted siRNAs to these enzymes. Each protein was knocked down by an estimated 80%–90% (Fig. 2A,D). As an alternative method to Northern blots, we used an S1 nuclease protection assay to monitor histone H2a mRNA levels (Kaygun and Marzluff 2005a, b). HU treatment does not affect polyadenylated H3.3 mRNA levels and thus serves as a loading control. The degradation of histone mRNA was reduced by knocking down Dcp2 and Xrn1, and after 45 min of HU treatment, there was twice as much histone mRNA remaining compared with control treated cells (Figs. 2B,C,E,F, 3G).
Figure 2.
Effect of decapping complex and 5′–3′ exonuclease knockdown on histone mRNA degradation. HeLa cells were treated with the indicated siRNAs to Dcp2 (A–C) or Xrn1 (D–F) and treated with 5 mM HU as described in Figure 1. (A,D) Protein levels of knockdowns were measured by Western blot using antibodies to Dcp2 (A) and Xrn1 (D). (B,E) S1 nuclease protection assays of 5 μg of total RNA isolated from cells treated with 5 mM Hu for 0, 20, or 45 min from Dcp2 (B) or Xrn1 (E) knockdowns. Human H3.3 and H2a mRNAs were detected by mixing probes to detect the 3′ and 5′ ends of H3.3 and H2a mRNAs, respectively. (C,F) The average of three independent experiments from Dcp2 (C) and Xrn1 (F) knockdowns is shown. The vertical bars indicate the standard deviation.
The down-regulation of both Dcp2 and Xrn1 had minimal effects on the cell cycle distribution of the cells (Supplemental Fig. S1C,D). There was a small increase in the percentage of S-phase cells of the Dcp2 knockdown compared with the control (Supplemental Fig. S1C). This likely accounts for the small increase in steady-state histone H2a mRNA levels (Fig. 2B, cf. lanes 1 and 3) in the Dcp2 knockdown cells. These data lead us to conclude that components of the 5′-to-3′ degradation pathway also play a role in histone mRNA degradation.
Knocking down exosome components PM/Scl-100 and Rrp41 inhibits histone mRNA degradation
Polyadenylated mRNAs may also be degraded by the exosome following deadenylation, in addition to degradation 5′ to 3′ through the decapping pathway. It is possible that the Lsm1–7 complex could also recruit the exosome. To determine whether the exosome may also play a role in histone mRNA degradation, we knocked down two components of the exosome by RNAi. We chose PM/Scl-100 and Rrp41, a core exosome component (Liu et al. 2006). PM/Scl-100 has been shown to be present in the cytoplasm of human cells (Lejeune et al. 2003), in contrast to initial reports (Allmang et al. 1999). PM/Scl-100 has substantial homology with Escherichia coli RNase D, a 3′–5′ exonuclease, and Rrp41 has been shown to have 3′–5′ exonuclease activity in vitro (Mitchell et al. 1997).
The RNAi treatment reduced PM/Scl-100 levels by ∼80% (Fig. 3A), and two different siRNAs against Rrp41 reduced its levels by at least 80% (Fig. 3D). Knocking down either PM/Scl100 (Fig. 3B) or Rrp41 (Fig. 3E) had a significant effect on histone mRNA degradation, similar to that seen with knocking down Lsm1. The average of three independent experiments is shown in Figure 3, C and F. There was a threefold stabilization of histone mRNA 45 min after HU treatment when PM/Scl-100 is knocked down and a 2.3-fold stabilization when Rrp41 was knocked down (Fig. 3G). Knocking down the exosome components had no effect on cell growth or cell cycle distribution (Supplemental Fig. S1E,F).
Determination of the 5′ and 3′ ends of individual histone mRNA molecules in vivo
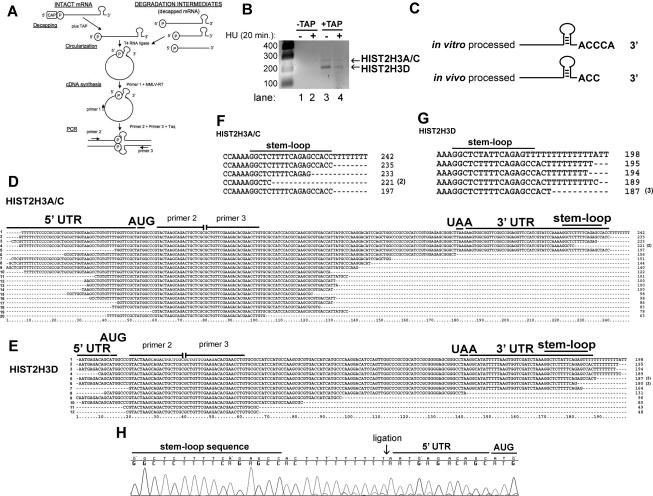
To directly identify molecules that had been partially degraded, we used a circularization RT–PCR (cRT–PCR) strategy in which the 5′ (containing a 5′ phosphate) and 3′ ends (with a 3′ OH) are ligated to each other, forming a circular RNA. One can then amplify the region containing the ligated 5′ and 3′ ends by RT–PCR. This method allows one to determine the 5′ end and the 3′ end of individual molecules. The ends of molecules that have 5′ caps can be detected by first treating the RNA with tobacco acid pyrophosphatase (TAP). This method has been used to directly analyze deadenylated mRNAs in cells (Fromont-Racine et al. 1993; Couttet et al. 1997). A schematic of this RT–PCR strategy, adapted from Couttet et al. (1997), is depicted in Figure 4A. We designed primers to target histone H3 mRNA from two identical genes (HIST2H3A and HIST2H3C) of the 12 annotated histone H3 genes in the human genome (Marzluff et al. 2002).
Figure 4.
Detection of the 5′ and 3′ ends of capped histone H3 mRNA in vivo and determination of the sequence of decapped histone H3 mRNA degradation intermediates. (A) The cRT–PCR strategy to determine the ends of intact histone mRNA and degradation intermediates is shown (adapted from Couttet et al. 1997; © 1997 National Academy of Sciences, USA). (B) cRT–PCR reactions with and without decapping from total cell RNA were separated on 1.5% agarose gels, and amplicons were detected by ethidium bromide staining. Two human histone H3 mRNAs are visualized in the +TAP lanes. (C) The 3′ end of a synthetic histone pre-mRNA substrate processed in vitro and decapped differs from histone mRNA decapped and retrieved from the cell. In vitro processed pre-mRNA and total cell RNA were decapped (+TAP), subjected to cRT–PCR, and cloned. Complete sequences of these experiments are presented in Supplemental Figure S4. (D,E) cRT–PCR was performed on total cell RNA, and the histone H3 products were amplified, cloned, and sequenced. HIST2H3A/C (D) or HIST2H3D (E) mRNAs that were not capped were cloned by cRT–PCR. Primers 2 and 3 represent sites where we targeted amplification toward the 5′ and 3′ ends. ORF sequences between these two oligonucleotides are missing. Numbers in parentheses are the number of times each clone was obtained. (F,G) The 3′ ends from the largest HIST2H3A/C (F) and HIST2H3D (G) mRNA degradation intermediates are shown. The number of times each sequence was obtained is indicated by a number in parentheses. (H) Chromatogram from a HIST2H3D mRNA degradation intermediate containing eight untemplated Us (Ts in the DNA sequence). The 3′ and 5′ ends were ligated (arrow) together.
As a control to demonstrate that we could identify the authentic 3′ ends of histone mRNAs, we analyzed the 3′ end of histone H2a mRNA formed in an in vitro processing reaction. After processing, we recovered only histone RNA that ended with the conserved ACCCA nucleotides following the base of the stem–loop, precisely where we and others (Scharl and Steitz 1994; Dominski et al. 2005) mapped the cleavage site in vitro (Supplemental Fig. S3A). These results demonstrate that we were able to recover RNA molecules that had their 3′ and 5′ ends intact, and hence nuclease activity in our reagents was minimal.
Figure 4B shows the results from a cRT–PCR experiment using RNAs from control cells and cells treated with HU for 20 min, at which time ∼40% of the histone mRNA has been degraded. Two amplicons were detected (∼190 and 235 nt) when TAP was used to decap the mRNA (Fig. 4B, lanes 3,4), and these were more abundant in RNA from untreated cells compared with HU-treated cells. No amplified products were detected by ethidium bromide staining when the RNA was not treated with TAP (Fig. 4B, lanes 1,2).
We cloned these two PCR products and sequenced multiple clones from each fragment. The 235-nt fragment was derived from the HIST2H3A, HIST2H3C genes we initially set out to amplify (Marzluff et al. 2002), while the 187-nt band was a previously unannotated histone H3 gene also found in the minor histone cluster on chromosome 1q21 (nucleotides 276,048– 275,157), which we named HIST2H3D (see below). Each of these genes is highly expressed in HeLa cells. Supplemental Figure S3, B and C, shows sequences for the 5′ and 3′ regions of these two genes. The top line (HIST2H3A/C or HIST2H3D) gives the genomic sequence of these genes, extending from the region upstream of the TATA box into the ORF, and from the stop codon past the HDE. The 5′ untranslated region (UTR) of the HIST2H3A/C mRNAs is 47 nt, with the G at the start site located 27 nt from the TATAA box, the expected distance for an RNA polymerase II transcription initiation site. This G was ligated to an ACC sequence that follows the stem–loop in 10 of the 11 circularized clones isolated. The cytoplasmic histone mRNA is thus 2 nt shorter than the mRNA formed by the in vitro processing reaction. In one clone the last 2 nt of the stem were missing, and these were followed by several nontemplated Us (Fig. 4C; Supplemental Fig. S4B, clone 2). As shown below, we believe that this clone likely represents a degradation intermediate. This clone had two additional nucleotides, a GT that could be part of the nontemplated 3′ UTR or result from slippage in the oligo(T) stretch in the 5′ UTR.
Similar results were observed with the clones from the 187-nt fragment. These clones were also essentially identical and correspond to the HIST2H3D gene. The 5′ UTR of this mRNA is extremely short, only 11 nt from the A in the ATG codon in 23 of the 28 clones, which is located 27 nt from the TATAA box. The small amount of heterogeneity at the 5′ end of the mRNA (one clone, Supplemental Fig. S3C, clone 2) starting at the C, 1 nt upstream, and another (Supplemental Fig. S3C, clone 1) at an A 3 nt upstream of the major start site, likely represents the use of closely spaced transcription start sites, all of which were ∼25 nt from the TATAA box. The 3′ end of the mRNA in 23 of the 28 clones (Supplemental Fig. S3C, clones 2 and 3) is 3 nt after the stem–loop; one ends 2 nt after the stem–loop (Supplemental Fig. S3C, clone 4), and the other four clones (Supplemental Fig. S3C, clone 1) may have a single nontemplated U (or the U may represent an alternative start site and may be the first nucleotide of the transcript). Note that this gene has an unusual sequence after the stem–loop ACTGC rather than the consensus ACCCA.
These results confirmed our ability to obtain full length 5′ and 3′ UTRs using the cRT–PCR procedure from total cellular RNA. Although the histone mRNA formed by cleavage of the histone pre-mRNA ends 5 nt after the stem–loop, the cytoplasmic histone mRNAs end only 2–3 nt after the stem–loop. Thus there must be trimming of 2 nt from the processed mRNA at some point after the processing reaction, and this may be a function of the 3′hExo, which has this activity in vitro in the presence of SLBP (Dominski et al. 2003; Yang et al. 2006).
cRT–PCR products of uncapped histone H3 mRNA identifies degradation intermediates containing oligouridine on the 3′ end of the mRNA
If decapping occurs during histone mRNA degradation, then we could potentially circularize decay intermediates of histone mRNAs, allowing us to clone single molecules of partially degraded histone mRNA. Because our primers are located near the end of each ORF, we will preferentially isolate intermediates from early in the degradation process. We cloned all the products of the circular RT–PCR reactions starting with the RNAs not treated with TAP, although we could not see any amplified products in these reactions. Unlike the clones obtained by TAP treatment, these clones were heterogeneous in size, accounting for the failure to see discrete bands. We sequenced these clones, and all of them represented fragments from either the HIST2H3A/C genes (Fig. 4D) or HIST2H3D gene (Fig. 4E).
Only four of the 37 clones we obtained were from full-length mRNAs (Fig. 4D [clone 2], E [clone 5]). The remaining 33 clones encoded 28 shorter mRNAs—with nucleotides removed from either the 5′ end (Fig. 4D; Supplemental Fig. S5A, clone 5), the 3′ end (11 clones) (Figs. 4D [clones 3, 4, and 7–9], 5E [clones 6, 9, and 10]) or both (16 clones) (Fig. 5D [clones 6 and 11–20], E [clones 7, 8, and 10–12])—and five encoded mRNAs that had nontemplated Us added to the 3′ end (Fig. 5D [clone 1 from HIST2H3A/C], E [clones 1–4 from HIST2H3D]).
Figure 5.
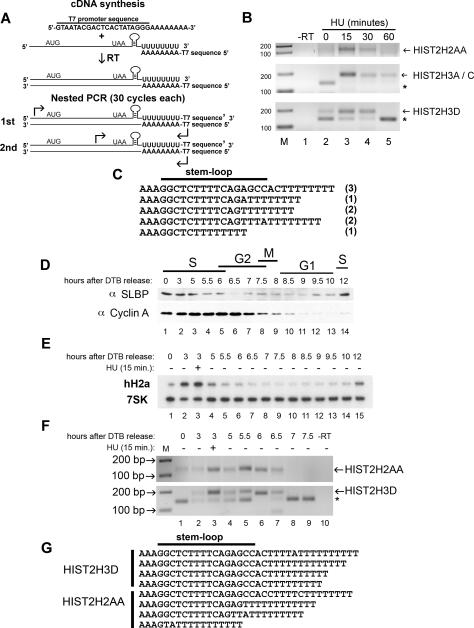
Detection of oligouridylated histone mRNAs after inhibition of DNA synthesis and at the end of S phase. (A) RT–PCR strategy to detect oligo(U)-containing histone mRNA molecules. Total RNA from HeLa cells was primed for cDNA synthesis using an oligo(dA) primer fused to a T7 sequence. Two rounds of PCR (30 cycles each) were performed using oligos targeting the 5′ UTR (first round) or the ORF (second round) and a primer complementary to the T7 sequence. (B) Oligo(dA) RT–PCR from HIST2H2AA, HIST2H3A/C, and HIST2H3D treated with HU for 0, 15, 30, and 60 min. The products were resolved by agarose gel electrophoresis and were detected by ethidium bromide staining. Asterisk indicates a nonspecific band. (C) Sequences from oligo(dA) RT–PCR reactions from the HIST2H3D mRNA when treated with HU for 15 min. The number in parentheses indicates the number of times a particular sequence was observed. (D) HeLa cells were synchronized by DTB and released into S phase, and protein samples were collected at the indicated times. Western blot analysis of SLBP and cyclin A was performed. (E) Total RNA was isolated from the same cells as in D, and H2a mRNA and 7SK RNA levels were measured by Northern blotting. (F) Oligo(dA) RT–PCR on total RNA from the middle and end of S phase into the G2 phase of the cell cycle. Total RNA from the indicated time points following release from DTB was subjected to the RT–PCR strategy depicted in A to detect HIST2H2AA and HIST2H3D mRNA and was resolved on a 2.0% agarose gel. HU was added at 3 h for 15 min in lane 3. (G) The oligo(dA) primed RT–PCR products from the 5.5- and 6.0-h time points were cloned, and several clones from the HIST2H2AA and HIST2H3D mRNAs were cloned and sequenced.
Most striking were the five mRNAs that contained eight to 10 nontemplated uridines added to the 3′ end of the mRNA, starting 3 nt after the stem–loop, the site where we mapped the 3′ end of the cytoplasmic mRNA (Supplemental Fig. S3B,C). Two of these clones contained a single A (Fig. 4E, clone 1) or C (Fig. 5B, clone 3) in the oligo(U) tract. One of these mRNAs from the HIST2H3D mRNA had lost 8 nt from the 5′ end. The mRNA from the HIST2H3A/C genes (Fig. 4D, clone 1) contains at least eight nontemplated Us at the 3′ end. Since there is also a string of five Us in the 5′ end just after the cap, it is also possible that this clone comes from a decapped mRNA that has lost a few nucleotides from the 5′ end. Figure 4, F and G, summarizes the 3′ ends of the HIST2H3A/C (Fig. 4F) and the HIST2H3D (Fig. 4G), showing the nontemplated Us as well as partial degradation of the 3′ side of the stem. An example of the sequence of a clone containing non-templated Us is shown in Figure 4H.
Histone H2a and H3 mRNAs acquire oligo(U) tails after addition of HU
In order to detect mRNAs containing oligouridylated tails added to the 3′ end of histone mRNA, we developed a nested RT–PCR strategy, depicted in Figure 5A. First we primed reverse transcription with an oligonucleotide containing a T7 promoter sequence followed by eight As to make cDNAs from all RNAs containing an oligo(U) sequence. A specific primer targeting the 5′ UTR of a histone gene and the T7 primer were used for the first round of PCR. The nested PCR reaction used a histone ORF primer with the same reverse T7 promoter primer. We targeted the same two H3 genes used in the cRT–PCR studies and in addition targeted an H2a mRNA (HIST2H2AA). We prepared total RNA at 0, 15, 30, and 60 min after initiating histone mRNA degradation by treating cells with HU to inhibit DNA synthesis. As a control, we used an oligo(dT) oligo followed by the T7 sequence (Supplemental Fig. S4B,C).
At 0 time, there is no amplicon of oligouridylated histone mRNA detected from either the HIST2H2AA mRNA or the HIST2H3A/C and HIST2H3D mRNAs (Fig. 5B, lane 2). After 15 min, a robust band is detected, which subsequently declines in intensity at 30 min and is almost entirely gone at 60 min, coinciding with the near completion of histone mRNA degradation (Fig. 5B, lanes 3–5). We cloned the amplicons from the 0- and 15-min time points. There was a striking difference in colony number on the transformed H2a plates, with only a few colonies present on the 0 min plates compared with hundreds of colonies present on the 15-min plates (Supplemental Fig. S4A), consistent with the increased amount of RT–PCR product after HU treatment. We sequenced amplicons cloned after HU treatment, and these all were derived from histone mRNAs containing oligo(U) tails at or near the 3′ end of these histone mRNAs. They contained eight Us (the number of Us in the primer) that were added either to the very terminal end (3 nt after the stem–loop) of the mRNA or somewhere within the stem–loop (Fig. 5C). The latter observation suggests that multiple rounds of oligo(U) addition may occur near the 3′ end of the mRNA, possibly because degradation may stall in the stem–loop, and the degradation must be primed again.
In the HIST2H2A/C (Fig. 5B, middle panel, lane 2) and the HIST2H3D (Fig. 5B, bottom panel, lanes 2,5) reactions, we detected a nonspecific band at the 0 time point that is absent after HU treatment for 15 or 30 min, but is present at 60 min when most of the histone mRNA has been degraded. We sequenced clones from the 0 time point of the H3 oligo(dA) RT–PCRs, and the nonspecific band was a primer dimer sequence containing the two histone primers and a small sequence of H3 ORF followed by the reverse T7 promoter primer (primer dimer of three oligos).
Addition of adenosines to the 3′ end of many RNAs is a prerequisite to degradation of RNAs by the exosome in the nucleus (LaCava et al. 2005; Vanacova et al. 2005). To test whether polyadenylation might also play a role in histone mRNA degradation, we attempted to detect poly- or oligoadenylated histone mRNAs after inhibiting DNA replication by a similar strategy used to amplify the oligouridylated histone mRNAs. As a positive control for the oligo(dT) oligo, we used SLBP knockdown cells that have been shown to result in accumulation of some polyadenylated histone mRNA as a result of misprocessing (Narita et al. 2007). We readily detected polyadenylated histone mRNAs in the SLBP knockdown cells, but did not detect any oligoadenylated histone mRNAs in control cells or cells treated with HU (Supplemental Fig. S4B,C). In addition, our unbiased sequencing of circularized histone mRNAs never yielded oligoadenylated mRNAs.
Oligouridylated histone mRNAs are present at the end of S phase but not in mid S phase
Histone mRNAs are rapidly degraded at the end of S phase. To determine if the oligouridylation is present normally at the end of S phase, we synchronized HeLa cells by double thymidine block (DTB) (Whitfield et al. 2000). As a positive control, we treated mid-S-phase cells (3 h after release from the thymidine block) for 15 min with HU. We prepared whole-cell lysates for Western analysis and total RNA for Northern blot analysis and oligo(dA) RT–PCR at various times beginning from the middle of S phase but focusing particular attention to the end of S phase. Figure 5D (top panel) is a Western blot showing that SLBP protein levels are high in S phase and decrease rapidly upon completion of DNA synthesis and increase in abundance again when cells approach the subsequent S phase (Whitfield et al. 2000). We also determined cyclin A levels, a mitotic cyclin that is degraded at anaphase as cells exit mitosis. Northern blot analysis was performed for hH2a mRNA using 7SK snRNA as a loading control (Fig. 5E). Histone H2a mRNA is rapidly degraded upon the completion of S phase, then reaccumulates upon entry into the subsequent S phase. Figure 5F shows the results from oligo(dA) RT–PCR at various times in S phase and upon the completion of DNA sythesis. At 0 and 3 h following release from DTB, there was little to no detection of oligouridylated HIST2H2AA or HIST2H3D mRNA (Fig. 5F, lanes 1,2). When HU was added for 15 min at 3 h, oligouridylated HIST2H2AA and HIST2H3D mRNA accumulated (Fig. 5F, lane 3). Starting 5 h after release into S phase, we detected oligouridylated histone mRNA (Fig. 5F, lane 4), and the amount increased at 5.5–6 h for both HIST2H2AA mRNA (Fig. 5F, lane 5) and HIST2H3D mRNA (Fig. 5F, lane 6). The levels of oligouridylated histone mRNAs declined at 6.5 h and were no longer detectable 7 h during G2 phase. We sequenced oligouridylated RT–PCR products for both HIST2H2AA and HIST2H3D from the 5.5- and 6-h time points. Consistent with the results after HU treatment (Fig. 5D), we found oligouridylated tails present at the end of S phase on both the 3′ end of the histone mRNA as well as within the stem–loop sequence. Note that the oligo(U) tails were often interrupted by another nucleotide, which is characteristic of other terminal transferases (Kwak and Wickens 2007).
Two putative cytoplasmic TUTases are required for efficient histone mRNA degradation
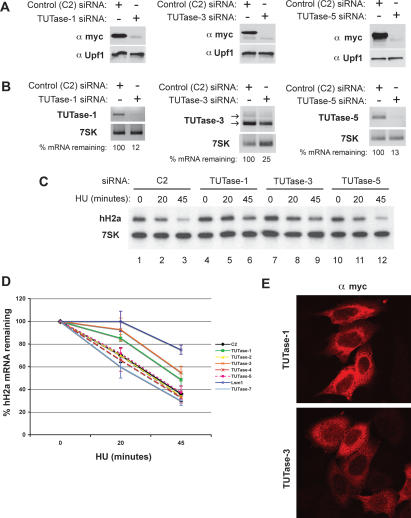
The addition of nontemplated uridines to the 3′ end of histone mRNA requires a 3′-terminal transferase activity. We searched the human genome for putative TUTase enzymes that contain the PAPD/TRF domain, a domain found in the human CID1 (Rissland et al. 2007) and U6 snRNA TUTase (Trippe et al. 2006). We identified seven putative TUTases (one of them being the U6 TUTase) and designed siRNAs targeting each one and knocked them down in HeLa cells. To demonstrate the efficiency of the siRNAs at the protein level, we coexpressed myc-tagged TUTases and found that the siRNAs efficiently depleted the exogenously expressed TUTases (Fig. 6A) by Western blotting. In addition, we determined whether the siRNAs reduced the endogenous TUTase mRNA levels by RT–PCR. We assessed the knockdown at the RNA level using 7SK snRNA as a loading control (Fig. 6B; Supplemental Fig. S6). Knocking down the U6 TUTase was lethal as previously reported (Trippe et al. 2006). Of the remaining six TUTases, knocking down two (TUTase-1 and TUTase-3) reduced the rate of histone mRNA degradation after HU treatment (Fig. 6C [lanes 4–9], D). Knocking down the other four enzymes, TUTase-2, TUTase-4, TUTase-5, and TUTase-7 (Fig. 6C [lanes 10–12], D), had no effect on histone mRNA degradation.
Figure 6.
RNAi screen to seven TUTases present in the human genome identifies two putative TUTase involved in histone mRNA degradation. The seven TUTases were knocked down using specific siRNAs as in Figure 1. (A) A representative experiment to determine the efficacy of TUTase siRNAs. Myc epitope-tagged TUTases were cloned and transfected at the time of the second hit of siRNA (see Materials and Methods). Protein lysates were harvested 48 h following the second hit and probed by Western using an anti-myc antibody. Upf1 was used as a loading control. (B) Endogenous TUTase mRNA levels were measured by RT–PCR. Two micrograms of total RNA were random primed and subjected to MMLV reverse transcriptase followed by PCR. The degree of knockdown, relative to 7SK RNA as a control, estimated by RT–PCR is indicated. (C) Knockdown of TUTase-1 and TUTase-3 stabilizes histone H2a degradation following inhibition of DNA synthesis by HU. Two micrograms of total cell RNA from cells treated with 5 mM HU were resolved by urea acrylamide gel electrophoresis and analyzed with a mixture of histone H2a mRNA and 7SK snRNA probes. (D) The rate of degradation of histone mRNA in three independent experiments where the different TUTases were knocked down. Lsm1 was included as a positive control. Standard deviations are represented by vertical bars. The U6 TUTase is essential for cell viability, and the effect on histone mRNA degradation could not be determined (see text for details). (E) TUTase-1 and TUTase-3 are localized predominantly in the cytoplasm of HeLa cells. Cells were transfected with myc-tagged clones of TUTase-1 or TUTase-3, and the tagged proteins were detected by immunofluorescence 48 h following transfection.
Since histone mRNAs are degraded in the cytoplasm, we determined the subcellular localization of the TUTase-1 and TUTase-3 by transiently transfecting HeLa cells with various doses of myc-tagged plasmids encoding these proteins, followed by confocal immunofluorescence. TUTase-1 is exclusively cytoplasmic, while TUTase-3 is predominantly cytoplasmic but is also present in the nucleus in some cells (Fig. 6E). The localization is consistent with a role for these enzymes in histone mRNA degradation.
Note that the TRAMP complex that adds adenosines to nuclear RNAs to prime their degradation are in this same family of enzymes (Vanacova et al. 2005; Rissland et al. 2007). We could not determine which of the human family members correspond to the Trf4/5 enzyme. However, since that enzyme is strictly nuclear, it is very unlikely that TUTase-1 or TUTase-3 is responsible for this activity.
Discussion
mRNA degradation is an important step in regulating gene expression, and destabilizing an mRNA is the only way to rapidly reduce mRNA levels when necessary. The pathway of mRNA degradation for most polyadenylated mRNAs has been determined over the past decade (for a recent review, see Garneau et al. 2007). The initial step is the shortening of the poly(A) tail. Once the poly(A) tail is short enough that it can no longer bind PABP, Lsm1–7 can bind the 3′ end and likely recruits Pat1, effectively inhibiting translation. The Lsm1–7 complex also stimulates decapping, allowing rapid degradation 5′ to 3′ by Xrn1, the cytoplasmic 5′-to-3′ exonuclease. Following deadenylation, the mRNA may also be degraded 3′ to 5′ by the exosome. In yeast, the 5′-to-3′ and the 3′-to-5′ degradation pathways are redundant, although the major pathway of mRNA degradation is 5′ to 3′ using the decapping machinery. Yeast mutants for members of either pathway are viable, but debilitation of both pathways results in lethality. Thus it is likely the two pathways are redundant and that most mRNAs in yeast can be degraded by either pathway (Caponigro and Parker 1996).
A third pathway of mRNA degradation is endonucleolytic cleavage, followed by 5′-to-3′ degradation of the 3′ fragment, and degradation of the 5′ fragment either by the exosome or by decapping followed by the 5′-to-3′ exonuclease. This pathway is used for degradation of mRNAs by siRNAs (Orban and Izaurralde 2005) and some endonucleases involved in degradation of specific mRNAs (Yang and Schoenberg 2004) or in NMD in Drosophila (Gatfield and Izaurralde 2004).
Histone mRNAs are the only mRNAs that are not polyadenylated, ending instead in a conserved stem–loop sequence. A major step in regulating histone mRNA levels is rapid degradation of histone mRNA, mediated by the stem–loop as the cis element that directs the degradation of histone mRNA. Despite the fact that the cis element that determines histone mRNA degradation has been known for 20 years (Pandey and Marzluff 1987), the details of the biochemical pathway for degradation of histone mRNA are not known.
In this study, we demonstrate three features of histone mRNA degradation: (1) There is oligouridylation of the 3′ end of histone mRNA when DNA replication is inhibited and at the end of S phase, and this is likely the step that initiates histone mRNA degradation. (2) Lsm1 is essential for histone mRNA degradation, suggesting that Lsm1–7 binds to the oligo(U) tail that is added to histone mRNA. (3) Both the 5′ pathway and the 3′ pathway are involved in histone mRNA degradation, and individual molecules of histone mRNA can be simultaneously degraded 5′ to 3′ and 3′ to 5′.
Both the 5′ and 3′ degradation pathways contribute to histone mRNA degradation
Previous work by Ross and coworkers (Ross and Kobs 1986; Ross et al. 1986, 1987) had implicated 3′-to-5′ degradation as the major pathway of histone mRNA degradation, largely based on in vitro studies. Despite the fact that the 3′hExo can potentially degrade the 3′ end of histone mRNA, we find no evidence for a role of this enzyme in histone mRNA degradation in mammalian cells (Fig. 1) or Drosophila (Kupsco et al. 2006). In contrast, knocking down components of both the 5′-to-3′ decay pathway or the exosome resulted in a reduced rate of degradation of histone mRNA when DNA replication was inhibited (Figs. 2, 3). Strikingly, knocking down Lsm1 had a large impact on histone mRNA degradation. The cytoplasmic complex Lsm1–7 is essential for degradation of polyadenylated mRNAs and binds to short ssRNAs at the 3′ end of deadenylated mRNAs (Tharun and Parker 2001). It functions both to block translation by recruitment of Pat1 (Coller and Parker 2005), as well as to initiate degradation of the body on the mRNA by recruitment of the decapping complex (Tharun and Parker 2001; Tharun et al. 2005), and possibly also the exosome (Lehner and Sanderson 2004). Given that histone mRNAs lack an obvious binding site for Lsm1–7, the finding that Lsm1 was critical for histone mRNA degradation was surprising.
Model of histone mRNA degradation
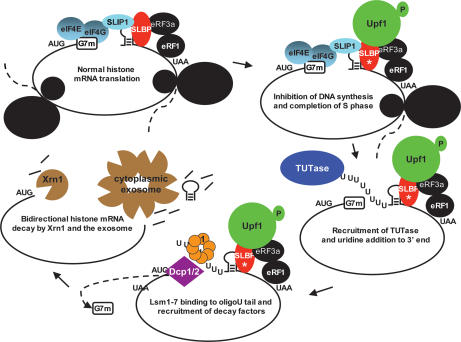
Histone mRNA degradation in the cytoplasm is triggered by inhibition of DNA replication in the nucleus. Previous work together with these results allow us to propose a model for histone mRNA degradation (Fig. 7). The stem–loop is the cis element responsible for histone mRNA degradation. It must be located an appropriate distance from the termination codon and requires ongoing translation (Graves et al. 1987; Kaygun and Marzluff 2005b). We postulated that inefficient translation termination is the critical event for triggering histone mRNA degradation (Kaygun and Marzluff 2005b), as is the case for NMD in yeast (Amrani et al. 2004). Histone mRNA degradation requires Upf1, which interacts with SLBP at the 3′ end of histone mRNA after treatment with HU (Kaygun and Marzluff 2005a). We postulate that the recruitment of Upf1 results in the recruitment of the appropriate TUTase and oligouridylation of histone mRNA. Lsm1–7 then binds to the oligo(U) tail (and may be specifically recruited by the complex at the 3′ end of the mRNA). Recent studies by Kiledjian and coworkers (Song and Kiledjian 2007) have shown that Lsm1–7 is the major complex that binds to oligo(U) in HeLa cell lysates, consistent with the proposed role of Lsm1–7 in histone mRNA degradation.
Figure 7.
Model of histone mRNA degradation. A proposed model of histone mRNA degradation showing the transition of histone mRNA from active translation as a circular mRNA, followed by the recruitment of Upf1 to the 3′ end (Kaygun and Marzluff 2006) when DNA replication is inhibited, followed by oligouridylation and degradation. SLBP is essential for translation (Sanchez and Marzluff 2002), and SLIP1 bridges SLBP to the 5′ end of histone mRNA (Cakmakci et al. 2007).
Following oligouridylation it is likely that SLBP is still bound to the histone mRNA, since SLBP and Lsm1 coimmunoprecipitate from HU-treated cells (Fig. 1G), although the association is sensitive to RNase, consistent with the Lsm1 being bound to the oligo(U) tail rather than to SLBP. Once Lsm1–7 is bound, then the decapping complex can be recruited, and histone mRNA is decapped and degradation initiated from the 5′ end. Alternatively the exosome may be recruited and the mRNA degraded from the 3′ end. Our data strongly suggest that both of these events occur on most mRNAs, resulting in simultaneous degradation of the same molecule 5′ to 3′ and 3′ to 5′. We cloned decay intermediates of single molecules by cRT–PCR and obtained histone RNAs that had both their 5′ and 3′ ends shortened, demonstrating that individual molecules of histone mRNA are degraded from both the 5′ end and the 3′ end (Fig. 4).
Initially 3′-to-5′ degradation may be relatively inefficient (possibly because of the presence of SLBP bound to the stem–loop), and we detect intermediates in which degradation has proceeded into the stem, and then the RNA is oligouridylated at that site. These molecules likely arise from stalling (and dissociation) of the exosome, requiring oligouridylation to reinitiate degradation 3′ to 5′.
Histone mRNA in the cytoplasm has been trimmed
Although the product of the in vitro processing reaction of histone mRNAs ends in 5 nt, ACCCA, the 3′ end of cytoplasmic histone mRNA ends in 2–3 nt (usually AC or ACC) after the stem–loop. Thus following processing the 3′ end must be trimmed. The properties of the 3′hExo, which can remove 2–3 nt from the stem–loop while SLBP is bound to the stem–loop (Dominski et al. 2003; Yang et al. 2006), suggests that the 3′hExo carries out this final reaction in histone mRNA maturation. The oligo(U) stretches we detected were all added to the trimmed histone mRNA.
Candidate TUTases for histone mRNA degradation
There are a group of genes initially thought to encode poly(A) polymerases that were clearly distinct from the canonical poly(A) polymerase that adds poly(A) tails to mRNAs. These enzymes, such as Cid1 in Schizosaccharomyces pombe, were thought to be poly(A) polymerases because they had some homology with the metazoan Gld2 enzymes, which are responsible for cytoplasmic polyadenylation of mRNA in metazoan oocytes (Kwak et al. 2004; Stevenson and Norbury 2006). It is now clear that at least some of these enzymes are also uridylyl transferases. S. cerevisiae lacks any proteins that are similar to Cid1 and has two family members, Trf4 and Trf5, that are clearly nuclear, while many of the Cid1-related proteins are cytoplasmic (Stevenson and Norbury 2006).
We identified seven human genes that encode putative terminal uridylyl transferases (TUTases) based on their homology with the U6-terminal transferase and the S. pombe Cid1 enzyme, and the same enzymes were identified by Kwak and Wickens (2007). Knocking down two of these enzymes, TUTase-1 and TUTase-3, but not the other five (including TUTase-7, the Cid1 homolog), resulted in a reduced rate of histone mRNA degradation (Fig. 6). These two enzymes are cytoplasmic, and we postulate that one or both of them may be required for the oligouridylation that initiates histone mRNA degradation. It is possible that one enzyme is responsible for the initial terminal addition, and the other is required for the addition to the partially degraded stem, which may be necessary to effectively complete histone mRNA degradation.
Benecke and coworkers (Trippe et al. 2006) recently isolated the enzyme that adds oligo(U) tails to U6 snRNA, an essential reaction in the maturation of U6 snRNA. TUTase-1 has been previously implicated as the mitochondrial poly(A) polymerase (Nagaike et al. 2005). However, Norbury and coworkers (Rissland et al. 2007) have shown that S. pombe Cid1 can add either uridine or adenosine to RNA in vitro and that the specificity is likely conferred by proteins associated with the TUTase, since Cid1 complexes immunoprecipitated from cells only add uridine in vitro. Thus it is possible that TUTase-1 has multiple functions in the cell.
Is oligouridylation important for other mRNA degradation pathways?
While addition of oligo(U) to a specific subset of mRNAs to initiate RNA degradation has not been previously reported in eukaryotic cells, there are examples of a potential role for oligouridylation in degradation of mRNAs following siRNA cleavage, and of terminal addition of nucleotides to prime degradation of RNAs in both the eukaryotic nucleus and in prokaryotes. Shen and Goodman (2004) reported that following miRNA cleavage of mRNAs in plants, mouse, and Epstein-Barr virus, they could detect oligouridylated mRNA fragments corresponding to the 5′ cleavage products. They suggested that these were intermediates in the degradation of the 5′ mRNA fragments produced by miRNA cleavage. In the alga Chlamydomonas reinhardtii, Ibrahim et al. (2006) suggested that the MUT68 protein, a poly(A) polymerase (CID12 ortholog in S. pombe), was responsible for oligoadenylation of RISC-induced cleavage products prior to cleavage. There is extensive evidence that degradation of improperly processed tRNAs and rRNAs are degraded in the nucleus by addition of oligo(A) by the Trf4/Trf5 complex followed by recruitment of the exosome (LaCava et al. 2005; Vanacova et al. 2005).
Bacterial mRNA degradation is initiated by addition of oligo(A) to the 3′ end of the mRNAs by a poly(A) polymerase that is homologous to Trf5. This oligo(A) tail can then bind the Hfq protein (a homolog of the eukaryotic Sm proteins) (Valentin-Hansen et al. 2004) and recruit the degradosome, a bacterial homolog of the exosome (Blum et al. 1999). In bacterial mRNA degradation, there is addition of oligonucleotides to the 3′ end of the RNA whenever the degradosome pauses or is blocked by secondary structure of the mRNA. In bacteria, this addition is catalyzed by polynucleotide phosphorylase, which adds nucleotides nonspecifically (although As predominate since ATP is the most abundant nucleotide) (Mohanty and Kushner 2000). It is thought that the single-stranded tail then serves as a platform for the degradosome to reinitiate degradation of the mRNA, and that cycles of nucleotide addition followed by exonucleolytic degradation are characteristic of bacterial mRNA degradation (Kushner 2004). Note that in addition to histone mRNAs with oligo(U) tails, we also isolated mRNAs that had oligo(U) tails added to mRNAs that had part of the stem sequence removed. It is possible that these result from partial degradation of histone mRNA, followed by stalling of the exosome, and then addition of oligo(U) to the degradation intermediate, allowing completion of mRNA degradation in a manner similar to that found in degradation of bacterial mRNAs.
A striking finding is that individual histone mRNA molecules are degraded both 5′ to 3′ and 3′ to 5′. A recent report suggests that individual ARE-containing mRNAs are also degraded from both ends (Murray and Schoenberg 2007), suggesting that this may be a common pathway for mRNA degradation in mammalian cells. Although we now know many of the requirements and the likely pathway for histone mRNA degradation, we still do not understand the molecular details of how the protein complex (including SLBP, Upf1, a uridylyl transferase, and Lsm1–7) is recruited to the 3′ end of histone mRNA to activate degradation, or the signals that activate this pathway at the end of S phase or when DNA replication is inhibited.
Materials and methods
RNAi
HeLa cells were transfected with Lipofectamine 2000 (Invitrogen) using a two-hit RNAi method (Wagner and Garcia-Blanco 2002). Forty-eight hours following the second RNAi transfection, cells were treated with 5 mM HU for 0, 20, or 45 min. Histone mRNA level was measured by Northern blotting or S1 nuclease assays. siRNAs were synthesized by Dharmacon, and the targeted sequences for the genes are listed in Supplemental Table 1.
Western blot analysis
Total cell lysates were prepared as described (Kaygun and Marzluff 2005a). Detection of endogenous proteins was made possible by generous gifts of antibodies to Lsm1/CaSm, Dcp2, Xrn1, Upf1, PM/Scl-100, Rrp41, and PTB. To determine the efficiency of TUTase knockdowns, myc-tagged transgenes were cotransfected with the second dose of siRNA. We assayed knockdown 48 h later using an anti-myc antibody (Upstate Biotechnology).
Northern blot and S1 nuclease protection assays
Total RNA was isolated using Trizol reagent (Invitrogen). For Northern blot assays, 2 μg of total RNA were separated on 6% acrylamide/7 M urea gels and transferred to Hybond N+ nitrocellulose membranes (Amersham). S1 nuclease protection assays to simultaneously measure the variant H3.3 and replication-dependent H2a mRNA were performed as described (Kaygun and Marzluff 2005a). Gels were dried and exposed to PhosphorImager cassettes, quantified using ImageQuant, and normalized to the amount of 7SK RNA.
cRT–PCR and cloning
cRT–PCR was performed similarly as described (Fromont-Racine et al. 1993; Couttet et al. 1997). Briefly, total RNA (50 μg) was treated with 25 U of DNase (Promega) and 20 U of HPRI (New England BioLabs) in a 100-μL reaction according to the manufacturer’s protocols (Promega). The RNA was then extracted with phenol-chloroform and precipitated with ethanol. For decapping, we incubated 10 μg of DNase-treated RNA, 2.5 U of TAP (Epicentre Biotechnologies), and 20 U of HPRI in a 20-μL reaction containing 1× TAP buffer (supplied by the manufacturer) for 1 h at 37°C and then raised the volume to 100 μL. The RNA was recovered as above, washed in 80% ethanol, and resuspended in 10 μL of water. Circularization of 4 μg of total decapped or nondecapped RNA was performed in a 400-μL reaction containing 20 U of T4 RNA ligase (New England BioLabs) and 20 U of HPRI (New England BioLabs) for 16 h at 16°C. The samples were then extracted and precipitated as above and were suspended in 12 μL of water.
Half of the ligation was subject to reverse transcription using MMLV RT (Invitrogen) and a human histone H3 oligo, termed primer 1 (5′-CTTCTGGTAGCGCCGGATCTC-3′), which was designed to specifically target the ORF of the HIST2H3A and HIST2H3C genes. The RT reaction was performed for 50 min at 37°C then heated for 15 min to 70°C. Subsequently, 2 μL were used in 100-μL PCR reactions using Taq polymerase. Human histone H3 primers 2 and 3 (5′-GACTTGCGAGCAGTCTGCT TAG-3′ and 5′-GCTGTTCGAAGACACGAACCTG-3, respectively) were used to target regions in the ORF that amplify toward the 5′ and 3′ UTRs. A total of 28–30 cycles were performed under the conditions of 30 sec at 95°C, 30 sec at 52°C, and 30 sec at 72°C. Twenty microliters of the reaction were analyzed on a 1.5%–3.0% agarose gel, and the identity of the PCR bands was confirmed by sequencing TA-cloned (Invitrogen) products.
RT–PCR of oligouridylated histone mRNA
Synchronized HeLa cells were treated with 5 mM HU for 0, 15, 30, or 60 min. Four micrograms of total cell RNA (treated with DNase) and a T7-oligo(A) primer (5′-GTAATACGACTCACTA TAGGGAAAAAAAA-3′) were used to prime cDNA synthesis in a RT reaction as above. The first round of PCR used a forward histone primer targeting the 5′ UTR of H2a/a, H3/2a/2c, or H3/1d (5′-GACTACTATCGCTGTCATGTCTG-3′, 5′-GGTAA GCCCTGTGTTTTGGTTCGC-3′, and 5′-GAGACAGCATGG CCCGTACTAAG-3′, respectively) and a reverse T7 primer. We performed 30 cycles on a 50-μL reaction containing a GC-rich Pfu DNA polymerase (Invitrogen), 1× Buffer A supplied by the manufacturer, and a similar cycling strategy as for the cRT–PCR (30 sec at 95°C, 30 sec at 52°C, and 30 sec at 72°C). The second round of PCR used 1 μL of product from the first round and identical cycling strategies (30 cycles) except that the reaction contained 5% DMSO and used Taq DNA polymerase. The primers in this reaction used primers to target sequences near the C terminus of the respective histone ORFs (H2a/a, 5′-GA ACTGAACAAGCCTGCTGGGCAAAG-3′ or 5′-CCTGCAGC TAGCGATCCG-3′; H3/2a/2c and H3/2d used human histone H3 primer 3 shown above). PCR products were separated on 2% agarose gels, were visualized by ethidium bromide staining, and were cloned and sequenced as described above.
Cell synchronization
HeLa cells were synchronized by DTB as described previously (Whitfield et al. 2000).
RT–PCR of TUTase mRNA
TUTases were down-regulated by siRNA as described above. Total RNA was isolated by Trizol reagent (Sigma), and 2 μg of RNA were reverse transcribed with MMLV-RT (Invitrogen) and random primers. RT reactions were subjected to PCR with the oligos described in Supplemental Table 2.
Cloning of TUTases
TUTases were cloned by PCR from cDNA clones or by RT–PCR from total HeLa RNA and cloned into pCDNA4-myc3 or pCDNA3-Flag vectors. Accession numbers of the TUTases are the following: TUTase-1 (BAB13981), TUTase-2 (BC04758), TUTase-3 (Q8NDF8), TUTase-4 (XP_038288), TUTase-5 (AB005754), the U6 TUTase (BC110910), and TUTase-7 (BC032456).
Immunofluorescence and confocal microscopy
HeLa cells plated on coverslips were transiently transfected with 0.5 or 1.5 μg of pCDNA4-myc3-Flag plasmids encoding the ORF of TUTase-1 and TUTase-3 for 48 h. Cells were prepared for immunofluorescence as described (Wagner and Marzluff 2006), and the N-terminal myc-epitope tag was detected with a monoclonal mouse anti-myc antibody (Upstate Biotechnology).
Acknowledgments
We thank Eric Wagner for many helpful discussions and critical reading of the manuscript and Zbig Dominski for the in vitro processed RNA. We thank Bob Duronio and members of the Marzluff laboratory for helpful discussions and critical reading of the manuscript. Special thanks to Kelly Sullivan for RNA from SLBP knockdown cells. We also acknowledge the generous gifts of antibodies used in this study: Dennis Watson (Medical University of South Carolina) for chicken anti-Lsm1/CaSm; Mike Kiledjian (Rutgers University) for rabbit anti-Dcp2; Jens Lykke-Andersen (University of Colorado, Boulder) for rabbit anti-Xrn1; Chris Smith (Cambridge University) for rabbit anti-Upf1 antibody; Ger Pruijn (University of Nijmegen) for rabbit anti-PM/Scl-100 and Rrp41 antibodies; and Mariano Garcia-Blanco (Duke University) for rabbit anti-PTB antibody. This work was supported by NIH grant GM29832 to W.F.M. T.M. was partially supported by NIH grant T32 CA71341.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1622708
References
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes & Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Blum E., Carpousis A.J., Higgins C.F. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- Cakmakci N.G., Lerner R.S., Wagner E.J., Zheng L.-X., Marzluff W.F. SLIP1, a factor required for activation of histone mRNA translation by the stem–loop binding protein. Mol. Cell. Biol. 2007 doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J.M., Tucker M., Sheth U., Valencia-Sanchez M.A., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P., Fromont-Racine M., Steel D., Pictet R., Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang X., Kaygun H., Marzluff W.F. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Yang X.C., Marzluff W.F. The polyadenylation factor CPSF-73 is involved in histone pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Erkmann J.A., Wagner E.J., Dong J., Zhang Y.P., Kutay U., Marzluff W.F. Nuclear import of the stem–loop binding protein and localization during the cell cycle. Mol. Biol. Cell. 2005;16:2960–2971. doi: 10.1091/mbc.E04-11-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Jakymiw A., Chan E.K., Seraphin B., Cougot N., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Bertrand E., Pictet R., Grange T. A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res. 1993;21:1683–1684. doi: 10.1093/nar/21.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gatfield D., Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- Graves R.A., Pandey N.B., Chodchoy N., Marzluff W.F. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Ibrahim F., Rohr J., Jeong W.J., Hesson J., Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005a;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 2005b;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. Upf1 functions in regulation of mammalian histone mRNA decay. In: Maquat L.E., editor. Nonsense-mediated mRNA decay. Landes Bioscience; Georgetown, TX: 2006. pp. 237–252. [Google Scholar]
- Kolev N.G., Steitz J.A. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes & Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupsco J.M., Wu M.-J., Marzluff W.F., Thapar R., Duronio R.J. Genetic and biochemical chracterization of Drosophila snipper: A promiscuous member of the metazoan 3′hExo/ERI-1 family of 3′ to 5′ exonucleases. RNA. 2006;12:2103–2117. doi: 10.1261/rna.186706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S.R. mRNA decay in prokaryotes and eukaryotes: Different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- Kwak J.E., Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.E., Wang L., Ballantyne S., Kimble J., Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lehner B., Sanderson C.M. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Li X., Maquat L.E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Long R.M., McNally M.T. mRNA decay: X (XRN1) marks the spot. Mol. Cell. 2003;11:1126–1128. doi: 10.1016/s1097-2765(03)00198-9. [DOI] [PubMed] [Google Scholar]
- Martin F., Schaller A., Eglite S., Schümperli D., Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F. Metazoan replication dependent histone mRNAs: A unique class of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Marzluff W.F., Gongidi P., Woods K.R., Jin J.P., Maltais L. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Polynucleotide phosphorylase functions both as a 3′ → 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Gao M., O’Connor J.P., Raijmakers R., Pruijn G., Lutz C.S., Wilusz J.R. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E.L., Schoenberg D.R. A + U-rich instability elements differentially activate 5′–3′ and 3′–5′ mRNA decay. Mol. Cell. Biol. 2007;27:2791–2799. doi: 10.1128/MCB.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T., Suzuki T., Katoh T., Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Orban T.I., Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N.B., Marzluff W.F. The stem–loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Rissland O.S., Mikulaslova A., Norbury C.J. Efficient RNA polyuridylation by non-canonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Kobs G. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3′ terminus and proceeds 3′ to 5′. J. Mol. Biol. 1986;188:579–593. doi: 10.1016/s0022-2836(86)80008-0. [DOI] [PubMed] [Google Scholar]
- Ross J., Peltz S.W., Kobs G., Brewer G. Histone mRNA degradation in vivo: The first detectable step occurs at or near the 3′ terminus. Mol. Cell. Biol. 1986;6:4362–4371. doi: 10.1128/mcb.6.12.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Kobs G., Brewer G., Peltz S.W. Properties of the exonuclease activity that degrades H4 histone mRNA. J. Biol. Chem. 1987;262:9374–9381. [PubMed] [Google Scholar]
- Sanchez R., Marzluff W.F. The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl E.C., Steitz J.A. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 1994;13:2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Goodman H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.G., Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson A.L., Norbury C.J. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P. ARE-mRNA degradation requires the 5′–3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y., Manley J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun S., He W.H., Mayes A.E., Lennertz P., Beggs J.D., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S., Muhlrad D., Chowdhury A., Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R., Guschina E., Hossbach M., Urlaub H., Luhrmann R., Benecke B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P., Eriksen M., Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Marzluff W.F. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S-phase. Mol. Cell. Biol. 2006;26:6702–6712. doi: 10.1128/MCB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-F., Whitfield M.L., Ingledue T.I., Dominski Z., Marzluff W.F. The protein which binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Wang Z., Jiao X., Carr-Schmid A., Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M.L., Zheng L.-X., Baldwin A., Ohta T., Hurt M.M., Marzluff W.F. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Schoenberg D.R. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol. Cell. 2004;14:435–445. doi: 10.1016/j.molcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Yang X.C., Purdy M., Marzluff W.F., Dominski Z. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J. Biol. Chem. 2006;281:30447–30454. doi: 10.1074/jbc.M602947200. [DOI] [PubMed] [Google Scholar]