Abstract
In the budding yeast Saccharomyces cerevisiae, the protein phosphatase Cdc14 triggers exit from mitosis by promoting the inactivation of cyclin-dependent kinases (CDKs). Cdc14’s activity is controlled by Cfi1/Net1, which holds and inhibits the phosphatase in the nucleolus from G1 until metaphase. During anaphase, two regulatory networks, the Cdc14 Early Anaphase Release (FEAR) network and the Mitotic Exit Network (MEN), promote the dissociation of Cdc14 from its inhibitor, allowing the phosphatase to reach its targets throughout the cell. The molecular circuits that trigger the return of Cdc14 into the nucleolus after the completion of exit from mitosis are not known. Here we show that activation of a ubiquitin ligase known as the Anaphase-Promoting Complex or Cyclosome (APC/C) bound to the specificity factor Cdh1 triggers the degradation of the Polo kinase Cdc5, a key factor in releasing Cdc14 from its inhibitor in the nucleolus.
[Keywords: Cdc14, Cdc5, cell cycle, mitosis]
Progression through mitosis is governed by ubiquitin-dependent protein degradation (for review, see Harper et al. 2002). Chromosome segregation at the metaphase–anaphase transition is initiated by the targeting of Securin (Pds1 in budding yeast) for degradation by a ubiquitin ligase termed the Anaphase-Promoting Complex or Cyclosome (APC/C) associated with its specificity factor Cdc20. This process liberates Separase (Esp1 in budding yeast), a protease that severs the linkages that hold sister chromatids together (for review, see Nasmyth 2001). The anaphase–G1 transition, also known as exit from mitosis, requires APC/C activity as well. In this cell cycle stage, APC/C-Cdc20 and APC/C-Cdh1 trigger the degradation of mitotic cyclins (Clb cyclins in yeast) (Peters 2002), thereby promoting cyclin-dependent kinase (CDK) inactivation, which in turn triggers exit from mitosis.
Clb–CDK inactivation is not only brought about by cyclin degradation but also by binding of the CDK inhibitor Sic1 to the Clb–CDK complex (for review, see Stegmeier and Amon 2004). Both events, cyclin degradation and Sic1 accumulation, are triggered by the protein phosphatase Cdc14 dephosphorylating CDK substrates (Jaspersen et al. 1998; Visintin et al. 1998; Zachariae et al. 1998). Cdc14’s activity is controlled by Cfi1/Net1, which sequesters and inhibits the protein phosphatase in the nucleolus from G1 until metaphase. At the onset of anaphase, Cdc14 dissociates from its inhibitor through the action of the Cdc14 Early Anaphase Release (FEAR) network (for review, see Stegmeier and Amon 2004). During later stages of anaphase, another signaling network, the Mitotic Exit Network (MEN), further promotes Cdc14 dissociation from its inhibitor (for review, see Stegmeier and Amon 2004).
The FEAR network initiates the release of Cdc14 from the nucleolus at the metaphase–anaphase transition. The network includes the protease Esp1 (for review, see Nasmyth et al. 2001), the Esp1 substrate and binding protein Slk19, and the nucleolar proteins Spo12 and Fob1, as well as the Polo kinase Cdc5 (Stegmeier et al. 2002, 2004). The FEAR network promotes the phosphorylation of Cfi1/Net1 by Clb–CDKs (Azzam et al. 2004) and appears to bring about this anaphase-specific phosphorylation by down-regulating the protein phosphatase 2A (PP2A) (Queralt et al. 2006).
The MEN is essential for exit from mitosis. In the absence of MEN function, Cdc14 is transiently released from the nucleolus during early anaphase by the FEAR network but returns into the nucleolus during late stages of anaphase, and cells fail to exit from mitosis (for review, see Stegmeier and Amon 2004). The signaling pathway resembles a Ras-like signaling cascade and is composed of a GTPase Tem1, a GTPase-activating protein complex Bub2–Bfa1, a putative Guanine Nucleotide Exchange Factor (GEF) Lte1, and two protein kinases Cdc15 and Dbf2, as well as the Dbf2-associated factor Mob1 (for review, see Bardin and Amon 2001). The scaffold protein Nud1 anchors the GTPase and its downstream protein kinases to the outer plaque of the spindle pole body destined to move into the daughter cell (Gruneberg et al. 2000). There, the pathway is thought to be activated when the SPB moves into the bud during anaphase.
The Polo kinase Cdc5 is a component of the FEAR network and a key regulator of the MEN (Hu et al. 2001; Hu and Elledge 2002; Pereira et al. 2002; Stegmeier et al. 2002; Geymonat et al. 2003). In cells lacking the protein kinase, Cdc14 is not released from the nucleolus at all. Because Cdc5 is required for the release of Cdc14 from the nucleolus during early anaphase, the protein kinase has been classified as a FEAR network component. Cdc5 also contributes to Cdc14 release from the nucleolus by phosphorylating the Bub2–Bfa1 complex, thereby inhibiting the GAP. Owing to Cdc5’s role in both the FEAR network and the MEN, it is not surprising that overproduction of the protein kinase can promote Cdc14 release from the nucleolus in cell cycle stages other than anaphase (Visintin et al. 2003).
The mechanisms that bring about the release of Cdc14 from the nucleolus have been largely elucidated. Cdc14 released by the FEAR network stimulates MEN activity, and Cdc14 released by the MEN further activates the MEN. This feed-forward mechanism coupled to a positive feedback loop results in the rapid release of Cdc14 from the nucleolus. How this activation loop is disrupted once exit from mitosis has been completed to cause Cdc14 to return into the nucleolus is not understood. Here we show that Cdc14 itself is responsible for its inactivation. By activating the APC/C-Cdh1, Cdc14 induces the degradation of Cdc5, thereby silencing both the FEAR and the MEN.
Results
Protein synthesis is not required for the return of Cdc14 into the nucleolus
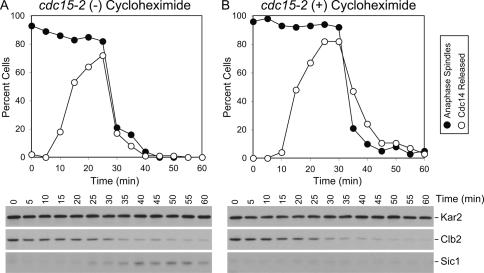
To determine how Cdc14 returns into the nucleolus once it has promoted mitotic exit, we first determined whether this process requires protein synthesis. Cells carrying a temperature-sensitive allele in the MEN component CDC15 arrest in anaphase with Cdc14 sequestered in the nucleolus (Shou et al. 1999; Visintin et al. 1999). Upon return to the permissive temperature, cdc15-2 mutants rapidly release Cdc14 from the nucleolus (Fig. 1A; Supplemental Fig. 1 provides examples of Cdc14 localization). This release coincides with mitotic CDK inactivation and hence exit from mitosis as judged by the degradation of the mitotic cyclin Clb2, the accumulation of the CDK inhibitor Sic1, and the disassembly of anaphase spindles (Fig. 1A). Concomitant with anaphase spindle disassembly, Cdc14 reaccumulates in the nucleolus. The presence of the protein synthesis inhibitor cycloheximide did not significantly alter the kinetics of Cdc14 release or resequestration into the nucleolus. Clb2 degradation and mitotic spindle disassembly also occurred without a delay (Fig. 1B). However, Sic1 did not accumulate as cells entered G1, indicating that cycloheximide treatment did efficiently inhibit protein synthesis. Our results show that following release from a cdc15-2 block, neither the release nor the return of Cdc14 into the nucleolus requires protein synthesis. Rather, post-translational events control the localization of the phosphatase. Our findings further exclude the possibility that the targeting of Cdc14 into the nucleolus after the completion of exit from mitosis is brought about by the degradation of released Cdc14 and simultaneous accumulation of de novo synthesized protein in the nucleolus.
Figure 1.
De novo protein synthesis is not required for Cdc14 return into the nucleolus. cdc15-2 cells carrying a CDC14-3HA fusion (A1674) were arrested in YEPD at the restrictive temperature (37°C) for 2.5 h. Cells were then released at the permissive temperature (25°C) into YEPD in the absence (A) or presence (B) of 1 mg/mL cyclohexymide. Samples were taken at the indicated times to determine the percentage of cells with anaphase spindles (closed circles) and of cells with Cdc14 released from the nucleolus (open circles). Mitotic CDK inactivation was assayed by analyzing Clb2 protein levels and accumulation of Sic1. Note that the absence of Sic1 in the cycloheximide-treated culture indicates that translation was effectively inhibited. Kar2 was used as an internal loading control in Western blots.
APC/C-Cdh1 and Bub2–Bfa1 are required for the return of Cdc14 into the nucleolus after exit from mitosis
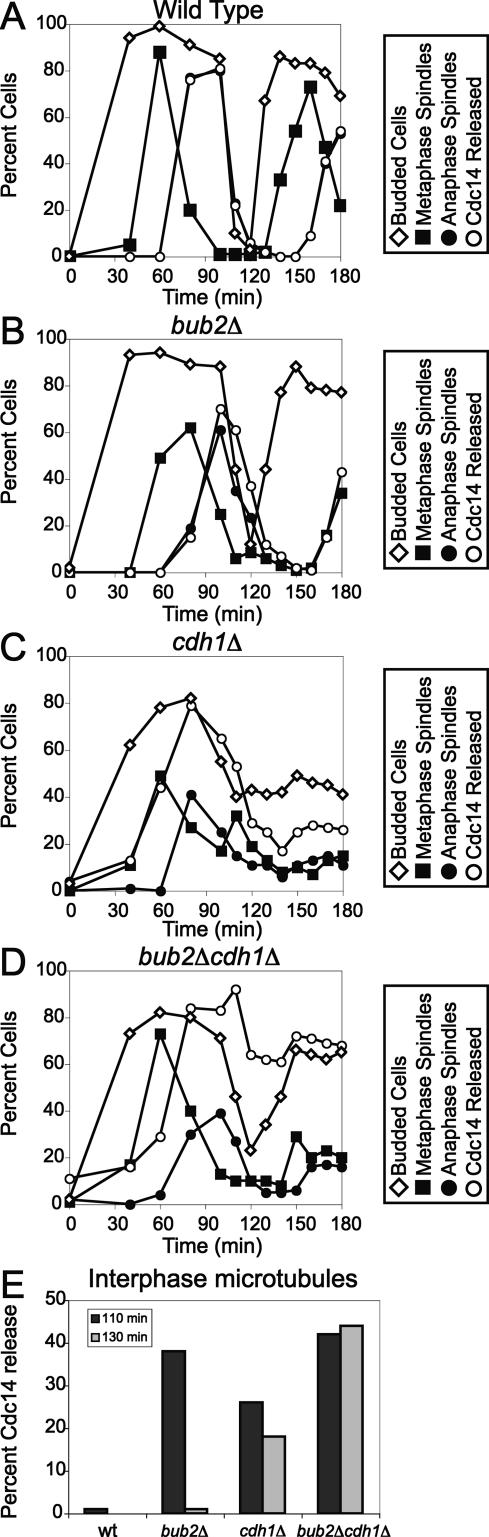
APC/C-Cdh1-dependent protein degradation could be a post-translational event important for the return of Cdc14 into the nucleolus because several factors implicated in promoting the release of Cdc14 from the nucleolus (Spo12, Clb–CDKs, and Cdc5) are targets of the APC/C-Cdh1 (for review, see Peters 2002). To test this hypothesis, we analyzed the localization of Cdc14 in wild-type and cdh1Δ cells. Upon release from an α-factor pheromone-induced G1 arrest, Cdc14 was released from the nucleolus during anaphase in wild-type cells (Fig. 2A). Although cells lacking CDH1 are somewhat resistant to α-factor (Schwab et al. 1997), they can nevertheless be arrested using high concentrations of pheromone and prolonged incubation times (Fig. 2C, percent budded cells). In cells lacking CDH1, Cdc14 release from the nucleolus occurred prematurely during metaphase, and the protein was slow to return into the nucleolus after exit from mitosis occurred (Fig. 2C).
Figure 2.
APC/C-Cdh1-dependent proteolysis is required for the return of Cdc14 in the nucleolus. (A–D) Wild-type (A2747), bub2Δ (A5954), cdh1Δ (A5952), and bub2Δ cdh1Δ (A5950) cells carrying a CDC14-3HA fusion were arrested in G1 in YEPD medium with α-factor pheromone (5 μg/mL). When the arrest was complete (after 3 h), cells were released into medium lacking pheromone. The percentage of budded cells (open diamonds), cells with metaphase spindles (closed squares), and anaphase spindles (closed circles), as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles), was determined at the indicated times. In E, cells with interphase microtubules were selected at the indicated time points to determine the percentage of cells with Cdc14 released from the nucleolus (n = 100).
APC/C-Cdh1-dependent protein degradation plays an important role in promoting the timely return of Cdc14 into the nucleolus, but it is not essential, as Cdc14 does return into the nucleolus after a delay (Fig. 2C). Thus, other mechanisms must contribute to this process. Silencing of the MEN by the GTPase-interacting protein Amn1 (Wang et al. 2003) and by the GAP complex Bub2–Bfa1 has been implicated in the return of Cdc14 into the nucleolus after mitotic exit is complete (Hu et al. 2001; Visintin and Amon 2001; Pereira et al. 2002; Visintin et al. 2003; Wang et al. 2003). Indeed, deletion of BUB2 slightly delayed the return of Cdc14 into the nucleolus after exit from mitosis as judged by a delay in Cdc14 reaccumulation in the nucleolus (Fig. 2B; Visintin and Amon 2001; Visintin et al. 2003). To determine whether the roles of APC/C-Cdh1 and BUB2 in bringing about the return of Cdc14 into the nucleolus were additive, we examined the effects of deleting both CDH1 and BUB2 on exit from mitosis. Cdc14 was released from the nucleolus prematurely, and nucleolar reaccumulation was delayed in the double mutant (Fig. 2D).
To further demonstrate that cells lacking APC/C-Cdh1 or APC/C-Cdh1 and Bub2 are defective in the timely return of Cdc14 into the nucleolus, we examined Cdc14 localization in cells that had just completed exit from mitosis. Such cells were identified as cells with an interphase microtubule array in time points collected 110 and 130 min after release from the G1 block. Cdc14 was found released from the nucleolus in a significant fraction of such cells (Fig. 2E). Our results indicate that APC/C-Cdh1 and the MEN GAP complex Bub2–Bfa1 both contribute to the timely return of Cdc14 into the nucleolus. The fact that after prolonged arrest in G1 with pheromone, Cdc14 is found in the nucleolus of cdh1Δ bub2Δ cells (Fig. 2D, 0 time point) also indicates that mechanisms in addition to silencing of the MEN and APC/C-Cdh1-mediated protein degradation exist that promote the sequestration of Cdc14 in the nucleolus once exit from mitosis has been completed.
CDK inactivation does not promote the return of Cdc14 into the nucleolus
The APC/C-Cdh1 substrates Cdc5, Spo12, and Clb–CDKs have been implicated in promoting the release of Cdc14 from the nucleolus (for review, see Stegmeier and Amon 2004). First, we tested whether Clb cyclin degradation by the APC/C-Cdh1 was important for the return of Cdc14 into the nucleolus. If Clb2 degradation by the APC/C-Cdh1 was important for the resequestration of Cdc14 into the nucleolus after exit from mitosis, deletion of the major mitotic cyclin CLB2 should ameliorate the Cdc14 localization defect of cdh1Δ bub2Δ double mutants. Deletion of CLB2 did not allow Cdc14 to return into the nucleolus in cdh1Δ bub2Δ mutants (Supplemental Fig. 2), suggesting that Clb2 degradation was not important for the return of Cdc14 into the nucleolus.
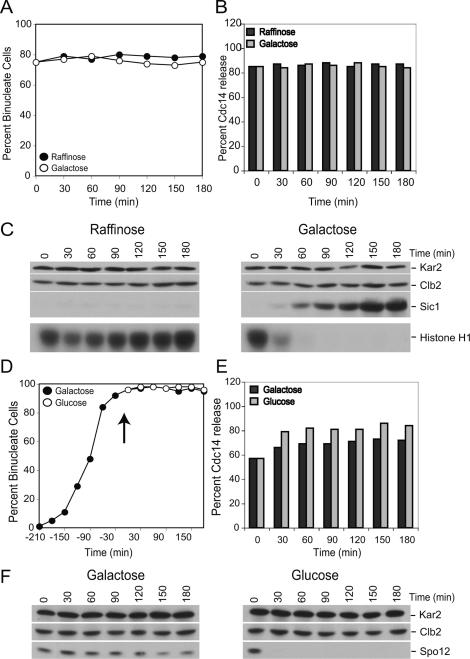
In the absence of CLB2, the other Clb cyclins could maintain Cdc14 in its released state. We therefore also examined the consequences of inactivating all Clb cyclins on Cdc14 localization. To this end, we first arrested cells in a stage of the cell cycle when Cdc14 was released from the nucleolus and then inactivated Clb–CDKs. Cells lacking PDS1 and depleted for Cdc20 (by repressing transcription of CDC20 from the methionine-repressible MET3 promoter; MET-CDC20 pds1Δ) arrest in late anaphase with Cdc14 released from the nucleolus due to a failure to degrade Clb5 (Shirayama et al. 1999). To inactivate all Clb–CDKs, we overexpressed the Clb–CDK inhibitor SIC1 from the galactose-inducible GAL1-10 promoter. MET-CDC20 pds1Δ cdh1Δ cells were arrested in anaphase by growing cells in the presence of methionine (Fig. 3A). Clb2–CDK activity was efficiently inhibited upon overexpression of SIC1 (Fig. 3C), but Cdc14 did not return into the nucleolus (Fig. 3B). Our results indicate that APC/C-Cdh1 does not promote the return of Cdc14 into the nucleolus by inactivating Clb–CDKs.
Figure 3.
Clb–CDK or Spo12 inactivation is not required for the return of Cdc14 into the nucleolus. (A–C) pMET3-CDC20 pds1Δ cdh1Δ pGAL-SIC1 (A13767) cells were grown in medium lacking methionine supplemented with 2% raffinose (−MetR). Cells were transferred in YEP supplemented with 2% raffinose (YEPR) medium containing 8 mM methionine to arrest pMET3-CDC20 pds1Δ cdh1Δ cells in anaphase. Six hours after the transfer, when >70% of cells were budded and had segregated DNA masses (binucleate cells), the culture was split in two. One-half was maintained in the same medium, whereas 2% galactose was added to the other half to induce SIC1 expression. Samples were taken at the indicated times to analyze the percentage of binucleate cells (A) and Clb2 and Sic1 protein levels and Clb2-CDK kinase activity (C). In B, anaphase cells were selected at the indicated time points to determine the percentage of cells that had Cdc14 released from the nucleolus (n = 100 per time point). (D–F) pMET3-CDC20 pds1Δ pGAL-URL-3HA-SPO12 (A13304) cells were grown in medium lacking methionine supplemented with 2% raffinose and 0.5% galactose (−MetR0.5%G). Cells were arrested in early S phase with hydroxyurea (HU; 10 mg/mL) for 3 h followed by release into −MetR0.5%G medium containing 8 mM methionine to arrest pMET3-CDC20 pds1Δ cells in anaphase. Three-and-a-half hours after the release, when ∼90% of cells were budded and had segregated DNA masses (binucleate cells), the culture was split in two. One-half was maintained in the same medium, whereas 2% glucose was added to the other half to deplete Spo12. Samples were taken at the indicated times to analyze the percentage of binucleate cells (D) and Spo12 and Clb2 protein levels (F). In E, anaphase cells were selected at the indicated time points to determine the percentage of cells that had Cdc14 released from the nucleolus (n = 100 per time point).
Inactivation of SPO12 does not promote the return of Cdc14 into the nucleolus
Spo12 is degraded during exit from mitosis by the APC/C-Cdh1 (Shah et al. 2001). To determine whether it is Spo12 degradation that facilitates the return of Cdc14 into the nucleolus after the completion of exit from mitosis, we depleted Spo12 in Cdc20-depleted pds1Δ cells, when Cdc14 is released from the nucleolus. To deplete cells of Spo12, we fused the ORF of SPO12 to a ubiquitin–arginine–lacZ fusion (URL–SPO12) that causes the protein to be degraded by the N-end rule pathway (Bachmair et al. 1986). When the fusion was placed under the control of the GAL1-10 promoter, Spo12 was efficiently depleted from cells in the presence of glucose (Fig. 3F). However, depletion of Spo12 did not promote the return of Cdc14 into the nucleolus (Fig. 3D,E), indicating that factors other than Spo12 are the essential targets of the APC/C-Cdh1 in bringing about the return of Cdc14 into the nucleolus.
Cdc5 functions after metaphase onset to bring about the release of Cdc14 from the nucleolus in a MEN-dependent and -independent manner
The Polo kinase Cdc5 is an excellent candidate for being the APC/C-Cdh1 substrate whose degradation is necessary for the reaccumulation of Cdc14 in the nucleolus. Cdc5 affects Cdc14 localization in multiple ways. Cdc5 partially inactivates the MEN GAP Bub2–Bfa1 during anaphase, thereby activating the MEN (Hu et al. 2001). Cdc5 was also proposed to be a component of the FEAR network because Cdc5 was required for the release of Cdc14 from the nucleolus during early anaphase (Pereira et al. 2002; Stegmeier et al. 2002; Yoshida and Toh-e 2002). This conclusion was, however, called into question (Queralt et al. 2006). Instead it was proposed that the defect of cdc5 mutants in releasing Cdc14 from the nucleolus during early anaphase was due to a yet-to-be-defined premetaphase function of Cdc5, because the dissociation of Cdc14 from the nucleolus during early anaphase was not affected in cdc5-4 cells released from a metaphase block (Queralt et al. 2006). Re-examination of all CDC5 alleles revealed that the cdc5-4 allele is a partial-loss-of-function allele. cdc5-4 cells do not arrest in anaphase but are only delayed in this cell cycle stage (Supplemental Fig. 3A–D) and, in fact, form colonies at 37°C (Supplemental Fig. 3E,F).
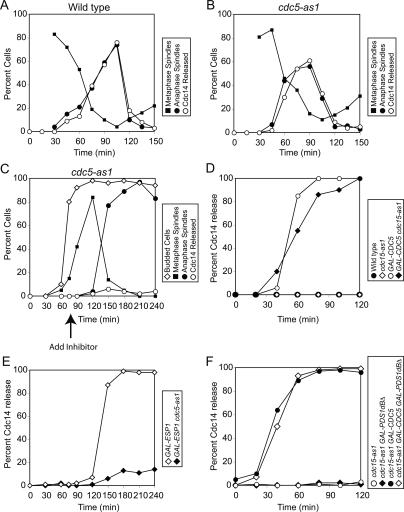
To determine whether CDC5 was required prior to metaphase to bring about the release of Cdc14 from the nucleolus, we used an allele of CDC5 that can be inhibited using an ATP analog (cdc5-as1) (Zhang et al. 2005). cdc5-as1 cells were arrested in G1 using α-factor and were released from the block into nocodazole-containing medium, which causes cells to arrest in metaphase. The release medium also contained the cdc5-as1 inhibitor CMK to inactivate the protein kinase during these early stages of the cell cycle. When cells had arrested in metaphase, they were released from the block in the absence of inhibitor, and cell cycle progression was monitored. cdc5-as1 cells released Cdc14 from the nucleolus and exited from mitosis with similar kinetics as wild-type cells (Fig. 4A,B). In contrast, inhibition of the cdc5-as1 allele either at the time of release from the pheromone-induced G1 arrest (Supplemental Fig. 4) or at the time when cells entered metaphase (Fig. 4C) effectively inhibited the release of Cdc14 from the nucleolus and exit from mitosis. Our execution point studies show that CDC5 is not required prior to metaphase to bring about the release of Cdc14 from the nucleolus and exit from mitosis. The essential function of CDC5 in promoting these events is confined to anaphase and perhaps metaphase.
Figure 4.
Cdc5’s role in promoting Cdc14 release is restricted to metaphase and anaphase. (A,B) Wild-type (A1411; A) and cdc5-as1 (cdc5L158G; Ry1260; B) cells carrying a CDC14-3HA fusion were arrested in G1 in YEPD with α-factor pheromone (5 μg/mL). After 3 h, cells were released into medium lacking pheromone but containing nocodazole (15 μg/mL) and the CMK inhibitor (5 μM). When the arrest was complete, cells were released into YEPD supplemented with 1% DMSO. The percentage of cells with metaphase spindles (closed squares) and anaphase spindles (closed circles), as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles), was determined at the indicated times. Note: Spindle morphology was not analyzed 0 and 15 min after release from the nocodazole block because the spindle had not yet reformed. (C) cdc5-as1 GAL-ESP1 CDC14-3HA (Ry1287) cells were arrested in G1 in YEP + 2% Raffinose (YEPR) with α-factor pheromone (5 μg/mL). Two-and-a-half hours into the arrest, 2% galactose (YEPR + G) was added to induce GAL-ESP1 expression. When the arrest was complete (3 h total), cells were released into YEPR + G lacking the pheromone. Seventy-five minutes after the release, when ∼80% of cells had formed a bud and were about to enter metaphase, CMK inhibitor (5 μM) was added. The percentage of budded cells (open diamonds), cells with metaphase spindles (closed squares) and anaphase spindles (closed circles), and the percentage of cells with Cdc14-HA released from the nucleolus (open circles) was determined at the indicated times. (D) Wild-type (A1411; closed circles), cdc15-as1 (A11617; open diamonds), GAL-CDC5 (A3317; open circles), and cdc15-as1 GAL-CDC5 (Ry1245; closed diamonds) cells carrying CDC14-3HA were arrested in G1 in YEP supplemented with 2% Raffinose (YEPR) with α-factor pheromone (5 μg/ mL). After 3 h, cells were released into medium lacking pheromone but supplemented with hydroxyurea (10 mg/mL) and the cdc15-as1 inhibitor (5 μM). After 2.5 h, when cells had arrested in S phase, 2% galactose was added to induce the expression of CDC5 from the inducible GAL1-10 promoter. The percentage of cells with Cdc14-HA released from the nucleolus was determined at the indicated times. (E) GAL-ESP1 (A4150) and cdc5-as1 GAL-ESP1 (Ry1287) cells carrying a CDC14-3HA fusion were treated as described in Figure 6C, except that cells were released into YEPR + G lacking pheromone but supplemented with 15 μg/mL nocodazole. (F) cdc15-as1 (Ry1386; open circles), cdc15-as1 GAL-PDS1dBΔ (Ry1385; closed diamonds), cdc15-as1 GAL-CDC5 (Ry1384; closed circles), and cdc15-as1 GAL-PDS1dBΔ GAL-CDC5 (Ry1383; open diamonds) cells were arrested with hydroxyurea (10 mg/mL). When the arrest was complete, the cdc15-as1 inhibitor (5 μM) was added. One hour after the inhibitor addition, 2% galactose was added to induce the expression of CDC5 and PDS1dBΔ from the inducible GAL1-10 promoter. The percentage of cells with Cdc14-HA released from the nucleolus was determined at the indicated times.
Does Cdc5 promote the release of Cdc14 from the nucleolus solely through the MEN, or does it function in additional ways to activate the phosphatase? To address this question, we examined whether the ectopic release of Cdc14 from the nucleolus brought about by high levels of Cdc5 required MEN function. If Cdc5 functioned solely through the MEN to bring about the release of Cdc14 from the nucleolus, the Cdc14 release caused by overproduced Cdc5 should require MEN activity. This was not the case. Overexpression of CDC5 in S-phase-arrested cells caused the release of Cdc14 from the nucleolus (Fig. 4D; Visintin et al. 2003). Cells carrying an ATP-analog-sensitive allele of CDC15 (cdc15-as1) (Bishop et al. 2001) arrest within one cell cycle in anaphase, indicating that treatment of cells with the cdc15-as1 inhibitor (PP1 analog 9) effectively inhibits Cdc15-as1 function (Supplemental Fig. 5). Inactivation of the cdc15-as1 allele in S-phase-arrested cells did not interfere with the release of Cdc14 induced by overproduced Cdc5 (Fig. 4D). This observation, together with the finding that Cdc5 does not regulate the MEN downstream from CDC15 (a hyperactive allele of Cdc15 can promote the release of Cdc14 from the nucleolus in cells depleted for Cdc5 function [Visintin et al. 2003]), demonstrates that Cdc5 promotes the release of Cdc14 from the nucleolus also in a MEN-independent manner that, according to our execution point studies, occurs during anaphase and perhaps metaphase.
CDC5 functions downstream from or in parallel to ESP1 to promote Cdc14 release from the nucleolus
Having established that Cdc5 brings about the release of Cdc14 from the nucleolus also in a MEN-independent manner, we re-examined the relationship between Cdc5 and the FEAR network. Previous studies came to the conclusion that Cdc5 functions downstream from or in parallel to all other FEAR network components because overproduced Cdc5 was able to suppress the Cdc14 release defect of slk19Δ, spo12Δ bns1Δ, and esp1-1 mutants (Visintin et al. 2003). esp1-1 mutants, like deletions of FEAR network components, only cause a delay in exit from mitosis. However, it was recently reported that ESP1 is essential for exit from mitosis (Queralt et al. 2006), raising the possibility that the epistasis analysis using the esp1-1 allele was misleading due to the hypomorphic nature of the mutation.
During our attempt to re-examine the relationship between ESP1 and CDC5, we found, to our surprise, that none of the ESP1 alleles, not even the esp1-2 allele made unstable by fusing it to the degron fusion (esp1-2td) (Queralt et al. 2006), arrested in anaphase. Upon release from a pheromone-induced G1 arrest, cells carrying the esp1-2td allele reproducibly (n = 5) entered mitosis with a delay, as judged by the delay in bud formation and metaphase spindle assembly (Supplemental Fig. 6A,B). To compare the kinetics of exit from mitosis between the esp1-1 and the esp1-2td allele, we therefore adjusted the graphs so that the onset of budding overlapped (Supplemental Fig. 6C). After accounting for the differences in timing of cell cycle entry, it was evident that Cdc14 release from the nucleolus and exit from mitosis occurred with similar kinetics in esp1-1 mad 1Δ, esp1-2td, and esp1-2td mad 1Δ cells as judged by the ability of dumbbell cells to form a bud (rebudding) (Supplemental Fig. 6D,E). Rebudding occurs because cytokinesis is incomplete in cells lacking ESP1 (Stegmeier et al. 2002). Mother and daughter cells therefore remain connected, and the daughter cell, which receives the nucleus (Ross and Cohen-Fix 2004), forms a bud upon entry into the subsequent cell cycle. Cells overexpressing a nondegradable form of the Esp1 inhibitor Pds1 (GAL-PDS1dbΔ) also released Cdc14 from the nucleolus and exited mitosis (as judged by rebudding) with similar kinetics as cells carrying the esp1-1 allele (Supplemental Fig. 6F–H). Exit from mitosis occurred even in cells, which, in addition to carrying an esp1-1 allele, expressed a nondegradable form of the Esp1 inhibitor Pds1 (Supplemental Fig. 7). Thus, none of the methods aimed at inactivating Esp1 function developed to date prevent exit from mitosis. Why the esp1-2td allele causes an anaphase arrest in the study published by Queralt et al. (2006) but not in our hands is at present unclear. Irrespective of the reason(s), our results suggest that the epistasis results obtained with the esp1-1 allele likely reflect the behavior of a null or close-to-null mutation of ESP1 with respect to the protein’s mitotic exit function.
Next we reinvestigated the relationship between CDC5 and ESP1 by extending our epistasis analysis between gain- and loss-of-function alleles of the two genes. Overexpression of ESP1 induces the release of Cdc14 from the nucleolus in cells arrested in metaphase using the microtubule-depolymerizing drug nocodazole (Fig. 4E; Sullivan and Uhlmann 2003; Visintin et al. 2003; Queralt et al. 2006). Inhibition of the analog-sensitive cdc5-as1 allele greatly reduced the ability of GAL-ESP1 cells to induce the release of Cdc14 from the nucleolus (Fig. 4E). Furthermore, we found that high levels of Cdc5 were able to induce Cdc14 release from the nucleolus even in the absence of both MEN and FEAR network function. Upon overexpression of CDC5, Cdc14 release from the nucleolus occurred in cells in which the MEN was inactivated using the cdc15-as1 allele and in which the FEAR network component Esp1 was inactivated by overexpression of a stabilized version of its inhibitor Pds1 (GAL-PDS1dbΔ) (Fig. 4F). Together with previous findings (Visintin et al. 2003), our data indicate that CDC5 functions downstream from or in parallel to ESP1.
The depletion of Cdc5 is sufficient to promote the return of Cdc14 into the nucleolus
Having demonstrated that Cdc5 functions during anaphase in a MEN-dependent and FEAR network-dependent manner to release Cdc14 from the nucleolus, we next wished to determine whether Cdc5 was continuously required during anaphase to maintain Cdc14 in its released state. Because the cdc5-as1 allele causes lethality in cells lacking Cdc20 and Pds1 (data not shown), we depleted Cdc5 by fusing the ORF of CDC5 to a ubiquitin–arginine–lacZ fusion (URL–CDC5) (Bachmair et al. 1986) and placed the fusion under the control of the GAL1-10 promoter. Depletion of an unstable form of Cdc5 (pGAL-URL-3HA-CDC5) by the addition of glucose in Cdc20-depleted pds1Δ cells caused Cdc14 to return into the nucleolus (Fig. 5A–C), indicating that CDC5 was continuously required for maintaining Cdc14 in the released state during anaphase.
Figure 5.
CDC5 is required for maintaining Cdc14 in its release state in cdh1Δ cells during anaphase. (A–C) pMET3-CDC20 pds1Δ pGAL-URL-3HA-CDC5 (A12396) cells were grown in medium lacking methionine supplemented with 2% raffinose and 0.5% galactose (−MetR0.5%G). Cells were arrested in early S phase with hydroxyurea (HU; 10 mg/mL) for 3 h followed by release into −MetR0.5%G medium containing 8 mM methionine to arrest pMET3-CDC20 pds1Δ cells in anaphase. Three hours after the release, when ∼70% of cells were budded and had segregated DNA masses (binucleate cells), the culture was split in two. One-half was maintained in the same medium, whereas 2% glucose was added to the other half to deplete Cdc5. Samples were taken at the indicated times to analyze the percentage of binucleate cells (A) and Cdc5 and Clb2 protein levels (C). In B, anaphase cells were selected at the indicated time points to determine the percentage of cells that had Cdc14 released from the nucleolus (n = 100 per time point). (D–F) pMET3-CDC20 pds1Δ pGAL-URL-3HA-CDC5 cdh1Δ (A12595) cells were treated as described in A–C with the exception that it took 3.5 h to arrest cells in the cdc20Δ pds1Δ block instead of 3 h. Samples were taken at the indicated times to analyze the percentage of binucleate cells (D) and Cdc5 and Clb2 protein levels (F). In E, anaphase cells were selected at the indicated time points to determine the percentage of cells that had Cdc14 released from the nucleolus (n = 100 per time point). (G–I) pMET3-CDC20 pds1Δ pGAL-URL-3HA-CDC5 pGAL-URL-3HA-SPO12 (A13804) cells were treated as described in A–C. Samples were taken at the indicated times to analyze the percentage of binucleate cells (G) and Cdc5, Spo12 and Clb2 protein levels (I). In H, anaphase cells were selected at the indicated time points to determine the percentage of cells that had Cdc14 released from the nucleolus (n = 100 per time point).
To determine whether Cdc5 was the critical target of the APC/C-Cdh1 that needed to be degraded for Cdc14 to return to the nucleolus, we depleted Cdc5 in anaphase arrested MET-CDC20 pds1Δ cdh1Δ cells. Upon depletion of Cdc5, Cdc14 returned into the nucleolus in this strain (Fig. 5D–F). Depletion of Spo12 did not significantly accelerate the kinetics with which Cdc14 was resequestered into the nucleolus in Cdc5-depleted cells (Fig. 5G–I). Our results indicate that the APC/C-Cdh1 promotes the return of Cdc14 into the nucleolus by degrading Cdc5.
Expression of a proteolysis-resistant CDC5 allele delays the return of Cdc14 into the nucleolus
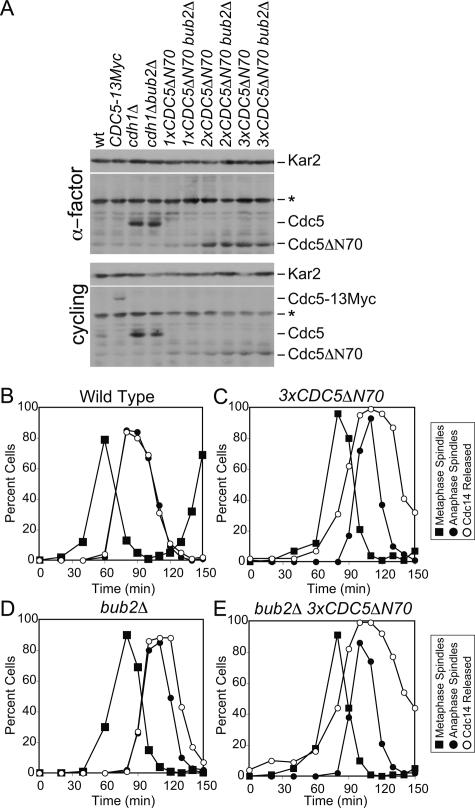
APC/C substrates carry recognition motifs known as destruction boxes and KEN-boxes (for review, see Harper et al. 2002). Cdc5 contains two destruction boxes in its N terminus, which have been shown to be essential for the protein’s degradation during exit from mitosis (Charles et al. 1998; Shirayama et al. 1998). To determine whether Cdc5 degradation was indeed important for the timely return of Cdc14 into the nucleolus, we examined the consequences of expressing a Cdc5 mutant lacking the first 70 amino acids of the protein and hence both of Cdc5’s destruction boxes (CDC5ΔN70) (Shirayama et al. 1998). Even three copies of CDC5ΔN70 did not produce as much Cdc5 as seen in cdh1Δ mutants (Fig. 6A). This is likely due to the CDC5ΔN70 not being completely stabilized and being expressed at lower levels than wild-type Cdc5 caused by the expression from a truncated promoter. However, although the Cdc5ΔN70 mutant protein was expressed to only ∼50% of the Cdc5 levels seen in cdh1Δ bub2Δ cells (Fig. 6A), cells carrying the construct exhibited dramatic defects in Cdc14 localization (Fig. 6C; Supplemental Fig. 8). Cdc14 was released from the nucleolus prematurely and returned into the nucleolus with a delay (Fig. 6C). Deletion of BUB2 slightly enhanced the effects of expressing a stabilized version of Cdc5, with respect to Cdc14 localization (Fig. 6D,E). This is not surprising as Cdc5 only partially inactivates Bub2–Bfa1 (Hu et al. 2001). Expression of CDC5ΔN70 as the sole source of CDC5 also affected cell cycle progression. Consistent with a delay in Cdc14’s return into the nucleolus, CDC5ΔN70 and CDC5ΔN70 bub2Δ cells exited mitosis prematurely, as judged by the kinetics with which anaphase spindle disassembly occurred. (Note that cells expressing CDC5ΔN70 enter metaphase with a 20-min delay, yet anaphase spindle disassembly occurred virtually at the same time as in wild-type cells.) Our results indicate that the degradation of Cdc5 by the APC/C-Cdh1 plays an important role in promoting the return of Cdc14 into the nucleolus.
Figure 6.
Cdc5 degradation is important for the timely return of Cdc14 into the nucleolus. (A) Wild-type (A2587), CDC5-13MYC (A2976), cdh1Δ (A5952), cdh1Δ bub2Δ (A5950), 1xCDC5ΔN70 (A10203), 1xCDC5ΔN70 bub2Δ (A10202), 2xCDC5ΔN70 (A10206), 2xCDC5ΔN70 bub2Δ (A10356), 3xCDC5ΔN70 (A10853), and 3xCDC5ΔN70 bub2Δ (A10856) were either arrested in G1 in YEPD medium with α-factor (5 μg/mL) or grown to exponential phase. Cdc5 protein levels were assessed by Western blot. Asterisk (*) marks a cross-reacting band on Western blots. The number of CDC5ΔN70 integrants was assessed by Southern blot analysis. (B–E) Wild-type cells (A2747; B), cells carrying three copies of CDC5ΔN70 construct (3xCDC5ΔN7; Α10853; C), bub2Δ cells (A5954; D) and bub2Δ 3xCDC5ΔN70 cells (Α10856; Ε) carrying a CDC14-3HA fusion were arrested with α-factor (5 μg/mL). After 3 h, cells were released into medium lacking pheromone, and the percentage of cells with metaphase spindles (closed squares), anaphase spindles (closed circles), and with Cdc14 released from the nucleolus (open circles) was determined at the indicated times.
Discussion
Here we investigate how the protein phosphatase Cdc14 returns into the nucleolus after having promoted exit from mitosis. Our data indicate that the phosphatase itself initiates this event. Cdc14 activates the MEN inhibitor Bub2–Bfa1 (Pereira et al. 2002). In addition, we find that Cdc14 activates the ubiquitin ligase APC/C-Cdh1. Among the Cdc14 activators degraded by the APC/C-Cdh1, we identified the Polo kinase Cdc5 as the critical target. Cdc5, which we confirmed to function during anaphase as a FEAR network component and a MEN activator, is continuously required during anaphase to maintain Cdc14 in the released state. Furthermore, targeted degradation of Cdc5 is sufficient to promote the return of Cdc14 into the nucleolus in cdh1Δ cells. Finally, we show that cells expressing a version of Cdc5 that is resistant to APC/C-Cdh1-mediated degradation are delayed in resequestering Cdc14 after exit from mitosis has been completed. Together these data indicate that Cdc14 initiates its return into the nucleolus. Through this negative feedback mechanism, cells ensure that this potent antagonist of CDKs is active for only a short period of time during exit from mitosis.
Cdc14 can promote exit from mitosis in the absence of CDK inactivation
The final step in CDK inactivation that triggers exit from mitosis is brought about by two redundant mechanisms: degradation of Clb cyclins by the APC/C-Cdh1 and binding of the Clb–CDK inhibitor Sic1 to the kinase complex. Our finding that the return of Cdc14 into the nucleolus was delayed in cdh1Δ cells provides an explanation for the previously puzzling observation that cells lacking CDH1 and SIC1, despite failing to inactivate Clb2–CDK kinase activity at the end of mitosis, do not arrest in anaphase but only exhibit a delay of 45 min in mitotic exit (Supplemental Fig. 9; Wasch and Cross 2002). In cells lacking CDH1, the period during which Cdc14 is active is prolonged. This is sufficient to antagonize the effects of CDKs and to allow for the dephosphorylation of CDK substrates to bring about mitotic spindle disassembly, cytokinesis, and origin licensing. However, it is important to note that Cdc14 is not capable of bringing about exit from mitosis in the complete absence of Clb–CDK inactivation. When degradation of Clb cyclins by APC/C-Cdc20 is prevented, Cdc14 despite becoming eventually fully released from the nucleolus cannot promote exit from mitosis (Shirayama et al. 1999). Thus, whereas prior to the metaphase–anaphase transition Clb–CDK inactivation is absolutely essential for exit from mitosis to occur, thereafter cells enter a state in which either CDK inactivation or activation of the Cdc14 phosphatase can bring about exit from mitosis.
Re-evaluating Cdc5’s role in exit from mitosis
Cdc5 promotes mitotic exit in multiple ways. Best understood is the mechanism whereby the protein kinase stimulates the MEN. Cdc5 partially inactivates the MEN GAP Bub2–Bfa1 during anaphase (Hu et al. 2001). It is possible that Cdc5 regulates the MEN in additional ways, but it is clear that the protein kinase is not essential for MEN activity once the MEN kinase Cdc15 is active. In cells expressing a truncated, hyperactive version of CDC15 (amino acids 1–750), Cdc14 release from the nucleolus occurred even when Cdc5 function was eliminated by depleting the protein (Visintin et al. 2003).
In addition to a MEN-dependent role in mitotic exit, our data support a MEN-independent role for Cdc5 in promoting the release of Cdc14 from the nucleolus during anaphase. (1) The cdc5-4 allele, which was used to show that CDC5 was dispensable for Cdc14 release from the nucleolus during early anaphase, was only a partial loss-of-function allele, indicating that Cdc5 was not completely inactive in this experiment. (2) Execution point studies using the cdc5-as1 allele show that CDC5 is not required prior to metaphase to bring about exit from mitosis, but that it is needed during anaphase (and perhaps metaphase) to perform its essential mitotic exit-promoting function. (3) In the experiment that implicated CDC28 rather than CDC5 in promoting Cdc14 release from the nucleolus during early anaphase (Queralt et al. 2006), Cdc5 protein was rapidly lost from cells, presumably due to activation of the APC/C-Cdh1 (Supplemental Fig. 10). (4) Overexpression of CDC5 can promote the release of Cdc14 from the nucleolus in the absence of MEN and ESP1 function (Fig. 4F). (5) The release of Cdc14 brought about by high levels of ESP1 requires CDC5 function. Together these results show that CDC5 functions during anaphase downstream from or in parallel to ESP1 to bring about the release of Cdc14 from the nucleolus. We favor the idea that CDC5 functions downstream from ESP1 rather than in parallel to it because Cdc5 is still capable of promoting the release of Cdc14 from the nucleolus even in the complete absence of all the known pathways that regulate this event.
The silencing of the mitotic exit machinery
Our data and that of others show that silencing of the MEN through inhibition of the GTPase Tem1 by Amn1 and Bub2–Bfa1 helps to return Cdc14 into the nucleolus (Pereira et al. 2001; Visintin and Amon 2001; Wang et al. 2003). However, the contribution of GAP activation appears minor compared with that of the APC/C-Cdh1. Our data further indicate that deletion of CDH1 not only delays the return of Cdc14 into the nucleolus after the completion of exit from mitosis, but it also causes the phosphatase to be released from the nucleolus prematurely (as does expression of a stabilized version of Cdc5). We propose that this premature release is due to high levels of Cdc5 during metaphase caused by a failure to degrade Cdc5 protein during the preceding exit from mitosis and onset of de novo synthesized during entry into mitosis.
While degradation of Spo12 could contribute to the inactivation of the pathways that promote Cdc14 release from the nucleolus, Cdc5 degradation is primarily responsible. Elimination of Cdc5 by means other than APC/C-Cdh1-mediated degradation is sufficient to promote the return of Cdc14 into the nucleolus in cells lacking CDH1. Furthermore, expression of a stabilized version of Cdc5 leads to a delay in the resequestration of the phosphatase. The fact that cells expressing a stabilized version of Cdc5 exhibit a less severe Cdc14 localization defect than cells lacking CDH1 is likely to be due in part to Cdc5ΔN70 protein levels being lower than Cdc5 levels in cdh1Δ bub2Δ cells. However, it is also possible that yet-to-be-identified proteins promoting the release of Cdc14 from the nucleolus during anaphase are APC/C-Cdh1 substrates. What is also clear is that Bub2–Bfa1 and APC/C-Cdh1 activity are not the only mechanisms that bring about the return of Cdc14 into the nucleolus. After prolonged arrest in G1, cells lacking BUB2 and CDH1 eventually resequester Cdc14 into the nucleolus. The factors that bring about this delayed return remain to be identified.
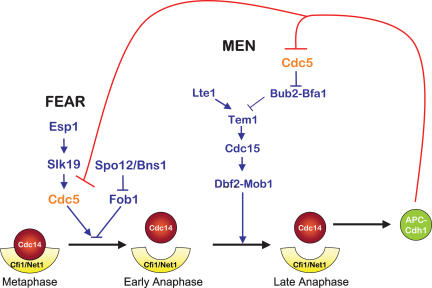
Based on previous work and our present study, we propose the following model for how Cdc14 activity is restricted to a small window during exit from mitosis (Fig. 7). At the end of mitosis, Cdc14 released by the FEAR network stimulates MEN activity. This feed-forward mechanism is coupled to a positive feedback loop, where Cdc14 released by the MEN later in anaphase further activates the MEN. Once the phosphatase is fully activated by the MEN, it dephosphorylates Cdh1 to bring about activation of APC/C-Cdh1. This not only rapidly inactivates Clb–CDKs and promotes exit from mitosis, it also initiates the return of the phosphatase into the nucleolus, where it binds its inhibitor. Among the APC/C-Cdh1 targets, Cdc5 appears to be critical. Because Cdc5 is required for the release of Cdc14 from the nucleolus as a component of the FEAR network and an activator of the MEN, the down-regulation of the protein kinase effectively silences both pathways. By using a negative feedback loop wherein Cdc14 released by Cdc5 triggers the degradation of the protein kinase, cells ensure that the machinery promoting exit from mitosis is restricted to a narrow window of the cell cycle.
Figure 7.
A model for how APC/C-Cdh1 promotes the return of Cdc14 into the nucleolus. See text for details.
Materials and methods
Yeast strains and growth conditions
All strains were derivatives of strain W303 (A2587). The pGAL-URL-3HA-CDC5 and pGAL-URL-3HA-SPO12 fusions were constructed by using the PCR-based method described in Longtine et al. (1998). The URL sequence (Johnson et al. 1992) was cloned in front of the 3HA epitope in the pFA6a-kanMX6-pGAL-3HA (Longtine et al. 1998). The resulting cassette was used as template with primer F4 and R3 (Longtine et al. 1998). CDC5ΔN70 was cloned under the control of its own promoter using the following strategy. About 500 base pairs (bp) of the promoter were amplified using the following primers: 5′-GGA ATTCTCGAGGACCTGGGATGATCTTTG-3′ (primes 450 bp upstream of the CDC5 START codon, with the EcoRI site in bold) together with 5′-GCGGGATCCCTTTATTCGACTTGA TCTTACTCTGTTCTTTCTTCTCC-3′ (primes just upstream of the ATG; BamHI site in bold); CDC5 deleted from the first 70 amino acids (region containing CDC5 two destruction-box sequences) was amplified with 5′-GCGGGATCCATGCCACCT TCATTAATCAAAACAAGAGG-3′, which primes at the 71st amino acid codon sequence (a BamHI site and the ATG [bold] have been introduced in the sequence) together with 5′-GGA ATTCGGTAAACCGCGGATGTACCAAG-3′, which primes 420 bp downstream from the STOP codon (the EcoRI site is in bold). The two PCR products were then cloned into Yiplac211 (Gietz and Sugino 1988). The plasmid obtained was used for transforming a diploid CDC5/cdc5Δ yeast strain. The number of integrants was assessed by Southern blot analysis. Growth conditions for individual experiments are described in the figure legends.
Immunoblot and kinase analysis
Cells were lysed in 50 mM Tris (pH 7.5), 1 mM EDTA, 1 mM PNP, 50 mM DTT, 1 mM PMSF, and 2 μg/mL pepstatin with glass beads for 1 min and boiled in 1× sample buffer. Immunoblot analysis of the total amount of Clb2, Sic1, and Kar2 was performed as described in Cohen-Fix et al. (1996). Immunoblot analysis of the total amount of endogenous Cdc5 was performed using the Cdc5 (sc-6733) antibodies from Santa Cruz Biotechnology diluted 1:500. Clb2 kinase activity was assayed as described in Amon et al. (1994).
Fluorescence microscopy
Indirect in situ immunofluorescence methods and antibody concentrations were as described in Visintin et al. (1999). Cells were analyzed on a Zeiss Axioplan 2 microscope, and images were captured with a Hamamatsu camera controller. Openlab 3.0.2. software was used to process immunofluorescence images. In Figures 3 and 5, Cdc14 was detected using a goat anti-Cdc14 antibody (Santa Cruz Biotechnology), which was used at 1:300 dilution; anti-goat antibodies were used at 1:500 (Santa Cruz Biotechnology). The fraction of cells with Cdc14 partially released from the nucleolus was <20% and was combined with the fraction of cells with Cdc14 fully released from the nucleolus.
Acknowledgments
We thank Chao Zhang and Kevan Shokat for the cdc15-as1 inhibitor and the cdc5-as1 allele; Ly-sha Ee for help with strain constructions; and Frank Uhlmann and Kyung Lee for strains. We are grateful to members of the Amon laboratory for their critical reading of the manuscript. J.P. is funded by an NSF predoctoral fellowship. Work in the Amon laboratory was supported by a National Institute of Health grant GM 56800. A.A. is also an investigator of the Howard Hughes Medical Institute. R.V. is supported by an Armenise-Harvard foundation career development program grant and a grant from the Associazione Italiana Ricerca sul Cancro (AIRC).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1601308
References
- Amon A., Irniger S., Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Azzam R., Chen S.L., Shou W., Mah A.S., Alexandru G., Nasmyth K., Annan R.S., Carr S.A., Deshaies R.J. Phosphorylation by cyclin B–Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Amon A. Men and sin: What’s the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bishop A.C., Buzko O., Shokat K.M. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- Charles J.F., Jaspersen S.L., Tinker-Kulberg R.L., Hwang L., Szidon A., Morgan D.O. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J.M., Kirschner M.W., Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Walker P.A., Johnston L.H., Sedgwick S.G. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 2003;278:14591–14594. doi: 10.1074/jbc.C300059200. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Campbell K., Simpson C., Grindlay J., Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Burton J.L., Solomon M.J. The anaphase-promoting complex: It’s not just for mitosis any more. Genes & Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Hu F., Elledge S.J. Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle. 2002;1:351–355. [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S.J. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., Tinker-Kulberg R.L., Morgan D.O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Bartel B., Seufert W., Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Peters J.M., Uhlmann F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Novartis Found. Symp. 2001;237:113–133. doi: 10.1002/0470846666.ch10. [DOI] [PubMed] [Google Scholar]
- Pereira G., Tanaka T.U., Nasmyth K., Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p–Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Manson C., Grindlay J., Schiebel E. Regulation of the Bfa1p–Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 2002;157:367–379. doi: 10.1083/jcb.200112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M. The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- Queralt E., Lehane C., Novak B., Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Ross K.E., Cohen-Fix O. A role for the FEAR pathway in nuclear positioning during anaphase. Dev. Cell. 2004;6:729–735. doi: 10.1016/s1534-5807(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Schwab M., Lutum A.S., Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shah R., Jensen S., Frenz L.M., Johnson A.L., Johnston L.H. The Spo12 protein of Saccharomyces cerevisiae: A regulator of mitotic exit whose cell cycle-dependent degradation is mediated by the anaphase-promoting complex. Genetics. 2001;159:965–980. doi: 10.1093/genetics/159.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R., Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Toth A., Galova M., Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol J.H., Shevchenko A., Baskerville C., Moazed D., Chen Z.W., Jang J., Shevchenko A., Charbonneau H., Deshaies R.J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Huang J., Rahal R., Zmolik J., Moazed D., Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr. Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Amon A. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell. 2001;12:2961–2974. doi: 10.1091/mbc.12.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Visintin R., Stegmeier F., Amon A. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol. Biol. Cell. 2003;14:4486–4498. doi: 10.1091/mbc.E03-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shirogane T., Liu D., Harper J.W., Elledge S.J. Exit from exit: Resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell. 2003;112:697–709. doi: 10.1016/s0092-8674(03)00121-1. [DOI] [PubMed] [Google Scholar]
- Wasch R., Cross F.R. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Toh-e A. Budding yeast Cdc5 phosphorylates Net1 and assists Cdc14 release from the nucleolus. Biochem. Biophys. Res. Commun. 2002;294:687–691. doi: 10.1016/S0006-291X(02)00544-2. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zhang C., Kenski D.M., Paulson J.L., Bonshtien A., Sessa G., Cross J.V., Templeton D.J., Shokat K.M. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]