Abstract
MicroRNAs (miRNAs) are ∼22-nucleotide RNAs that are processed from characteristic precursor hairpins and pair to sites in messages of protein-coding genes to direct post-transcriptional repression. Here, we report that the miRNA iab-4 locus in the Drosophila Hox cluster is transcribed convergently from both DNA strands, giving rise to two distinct functional miRNAs. Both sense and antisense miRNA products target neighboring Hox genes via highly conserved sites, leading to homeotic transformations when ectopically expressed. We also report sense/antisense miRNAs in mouse and find antisense transcripts close to many miRNAs in both flies and mammals, suggesting that additional sense/antisense pairs exist.
[Keywords: Drosophila, miR-iab-4, Hox, antisense miRNAs]
Hox genes are highly conserved homeobox-containing transcription factors crucial for development in animals (Lewis 1978; for reviews, see McGinnis and Krumlauf 1992; Pearson et al. 2005). Genetic analyses have identified them as determinants of segmental identity that specify morphological diversity along the anteroposterior body axis. A striking conserved feature of Hox complexes is the spatial colinearity between Hox gene transcription in the embryo and the order of the genes along the chromosome (Duboule 1998). Hox clusters also give rise to a variety of noncoding transcripts, including microRNAs (miRNAs) mir-10 and mir-iab-4/mir-196, which derive from analogous positions in Hox clusters in flies and vertebrates (Yekta et al. 2004). miRNAs are ∼22-nucleotide (nt) RNAs that regulate gene expression post-transcriptionally (Bartel 2004). They are transcribed as longer precursors and processed from characteristic pre-miRNA hairpins. In particular, Hox miRNAs have been shown to regulate Hox protein-coding genes by mRNA cleavage and inhibition of translation, thereby contributing to the extensive regulatory connections within Hox clusters (Mansfield et al. 2004; Yekta et al. 2004; Hornstein et al. 2005; Ronshaugen et al. 2005). Several Hox transcripts overlap on opposite strands, providing evidence of extensive antisense transcription, including antisense transcripts for mir-iab-4 in flies (Bae et al. 2002) and its mammalian equivalent mir-196 (Mainguy et al. 2007). However, the function of these transcripts has been elusive. Here we show that the iab4 locus in Drosophila produces miRNAs from opposite DNA strands that can regulate neighboring Hox genes via highly conserved sites. We provide evidence that such sense/antisense miRNA pairs are likely employed in other contexts and a wide range of species.
Results and Discussion
Our examination of the antisense transcript that overlaps Drosophila mir-iab-4 revealed that the reverse complement of the mir-iab-4 hairpin folds into a hairpin reminiscent of miRNA precursors (Fig. 1A). Moreover, 17 sequencing reads from small RNA libraries of Drosophila testes and ovaries mapped uniquely to one arm of the iab-4 antisense hairpin (Fig. 1B). All reads were aligned at their 5′ end, suggesting that the mir-iab-4 antisense hairpin is processed into a single mature miRNA in vivo, which we refer to as miR-iab-4AS. For comparison, we found six reads consistent with the known miR-iab-4-5p (or miR-iab-4 for short) and one read for its star sequence (miR-iab-4-3p). Interestingly, the relative abundance of mature miRNAs and star sequences for mir-iab-4AS (17:0) and mir-iab-4 (6:1) reflects the thermodynamic asymmetry of the predicted miRNA/miRNA* duplexes (Khvorova et al. 2003; Schwarz et al. 2003). Because they derived from complementary near palindromes, miR-iab-4 and miR-iab-4AS had high sequence similarity, only differing in four positions at the 3′ region (Fig. 1B). However, they differed in their 5′ ends, which largely determine miRNA target spectra (Brennecke et al. 2005; Lewis et al. 2005): miR-iab-4AS was shifted by 2 nt, suggesting targeting properties distinct from those of miR-iab-4 and other known Drosophila miRNAs.
Figure 1.
Drosophila iab-4 contains sense and antisense miRNAs. (A) mir-iab-4 sense and antisense sequences can adopt fold-back stem–loop structures characteristic for miRNA precursors (structure predictions by Mfold [Zuker 2003]; mature miRNAs shaded in blue [miR-iab-4] and red [miR-iab-4AS]). (B) Solexa sequencing reads that uniquely align to the mir-iab-4 hairpin sequence (top) or its reverse complement (bottom; numbers on the right indicate the cloning frequency for each sequence). The mature miRNAs have very similar sequences that are shifted by 2 nt and are different in only four additional positions. (C) Expression of primary transcripts for mir-iab-4 (blue) and mir-iab-4AS (red) in nonoverlapping abdominal segments determined by in situ hybridization (lateral [left panel] and dorsal [right panel] view of embryonic stage 11, anterior is to the left). (D) Lateral views of stage 10/11 embryos in which Ubx and abd-A proteins are visualized (anterior is to the left, and dorsal is upwards).
We confirmed robust transcription of mir-iab-4 sense and antisense precursors by in situ hybridization to Drosophila embryos (Fig. 1C). Both transcripts were detected in abdominal segments in the posterior part of the embryo, but intriguingly in nonoverlapping domains. As described previously (Bae et al. 2002; Ronshaugen et al. 2005), mir-iab-4 sense was expressed highly in abdominal segments A5–A7, showing modulation in levels within the segments: abdominal-A (abd-A)-expressing cells (Fig. 1D; Karch et al. 1990; Macias et al. 1990) appeared to have more mir-iab-4, whereas Ultrabithorax (Ubx)-positive cells appeared to have little or none (Fig. 1D; Ronshaugen et al. 2005). In contrast, mir-iab-4AS transcription was detected in the segments A8 and A9, where Abdominal-B (Abd-B) is known to be expressed (Fig. 1C; Yoder and Carroll 2006). Primary transcripts for mir-iab-4 and mir-iab-4AS were also detected by strand-specific RT–PCR in larvae, pupae, and male and female adult flies (Supplemental Fig. S1), suggesting that both miRNAs are expressed throughout fly development.
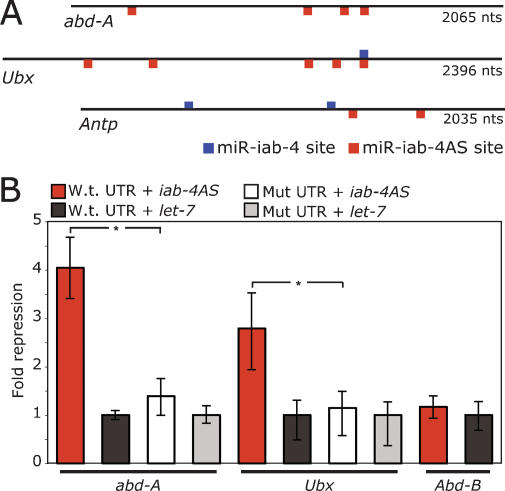
To assess the possible biological roles of the two iab-4 miRNAs, we examined fly genes for potential target sites by searching for conserved matches to the seed region of the miRNAs (Lewis et al. 2005). We found highly conserved target sites for miR-iab-4AS in the 3′ untranslated regions (UTRs) of several Hox genes that are proximal to the iab-4 locus and are expressed in the neighboring more anterior embryonic segments: abd-A, Ubx, and Antennapedia (Antp) have four, five, and two seed sites, respectively, most of which are conserved across 12 Drosophila species that diverged 40 million years ago (Fig. 2A; Supplemental Fig. S2; Drosophila 12 Genomes Consortium 2007; Stark et al. 2007a). More than two highly conserved sites for one miRNA is exceptional for fly 3′ UTRs, placing these messages among the most confidently predicted miRNA targets and suggesting that they might be particularly responsive to the presence of the miRNA. The strong predicted targeting of proximal Hox genes was reminiscent of previously characterized miR-iab-4 targeting of Ubx in flies and miR-196 targeting of HoxB8 in vertebrates (Mansfield et al. 2004; Yekta et al. 2004; Hornstein et al. 2005; Ronshaugen et al. 2005).
Figure 2.
miR-iab-4AS targets neighboring Hox genes. (A) miR-iab-4AS has five 3′ UTR seed sites (red) in Ubx, four in abd-A, and two in Antp of which three, four, and one are conserved across 12 Drosophila species, respectively (Supplemental Fig. S2). miR-iab-4 has one 3′ UTR seed site (blue) in Ubx and two in Antp, while abd-A has no such sites. (B) miR-iab-4AS mediates repression of luciferase reporters through complementary seed sites in 3′ UTRs from abd-A and Ubx, but not Abd-B (Antp was not tested). Luciferase activity in S2 cells cotransfected with plasmid expressing the indicated miRNA with either wild-type luciferase reporters or mutant reporters bearing a single point mutation in the seed. Bars represent geometric means from 16 replicates, normalized to the transfection control and noncognate miRNA control (let-7; see Materials and Methods). Error bars represent the fourth largest and smallest values from 16 replicates ([*] P < 0.0001, Wilcoxon rank-sum test).
To test whether miR-iab4AS is functional and can directly target abd-A and Ubx, we constructed Luciferase reporters carrying the corresponding wild-type 3′ UTRs and control 3′ UTRs in which each seed site was disrupted by point substitutions. mir-iab-4AS potently repressed reporter activity for abd-A and Ubx (Fig. 2B). This repression was specific to the miR-iab-4AS seed sites, as expression of the control reporters with mutated sites was not affected. We also tested whether mir-iab-4AS reduced expression of a Luciferase reporter with the Abd-B 3′ UTR, which has no seed sites. As expected, mir-iab-4AS expression did not affect reporter activity, consistent with a model where miRNAs do not target genes that are coexpressed at high levels (Farh et al. 2005; Stark et al. 2005). In addition to demonstrating specific repression dependent on the predicted target sites, these assays confirmed the processing of the mir-iab-4AS hairpin into a functional mature miRNA.
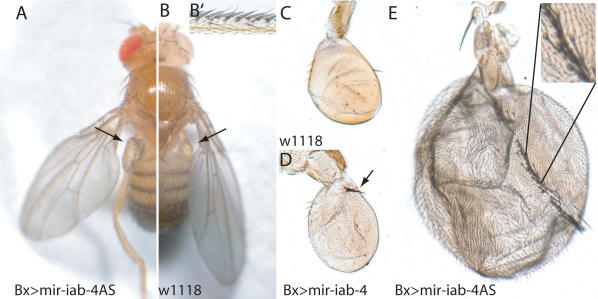
If miR-iab-4AS were able to potently down-regulate Ubx in the fly, its misexpression should result in a Ubx loss-of-function phenotype, a line of reasoning that has often been used to study the functions and regulatory relationships of Hox genes. Ubx is expressed throughout the haltere imaginal disc, where it represses wing-specific genes and specifies haltere identity (Weatherbee et al. 1998). When we expressed mir-iab-4AS in the haltere imaginal disc under bx-Gal4 control, a clear homeotic transformation of halteres to wings was observed (Fig. 3). The halteres developed sense organs characteristic of the wing margin and their size increased severalfold, features typical of transformation to wing (Weatherbee et al. 1998). Consistent with the increased number of miR-iab4AS target sites, the transformation was stronger than that reported for expression of iab-4 (Ronshaugen et al. 2005), for which we confirmed changes in morphology but did not find wing-like growth (Fig. 3D).
Figure 3.
Misexpression of miR-iab-4AS transforms halteres to wings. (A,B) Overview of an adult wild-type Drosophila (B) and an adult expressing mir-iab-4AS using bx-Gal4 (A). The halteres, balancing organs of the third thoracic segment, are indicated by arrows. (C) Wild-type haltere. (D) Expression of mir-iab-4 using bx-Gal4 induces a mild haltere-to-wing transformation. Sensory bristles characteristic of wild-type wing margins (shown in B′) are indicated by an arrow. (E) Expression of mir-iab-4AS using bx-Gal4 induces a strong haltere-to-wing transformation, displaying the triple row of sensory bristles (inset) normally seen in wild-type wings (shown in B′). Note that C–E are at the same magnification.
We conclude that both strands of the iab-4 locus are expressed in nonoverlapping embryonic domains and that each transcript produces a functional miRNA in vivo. In particular, the novel mir-iab-4AS is able to strongly down-regulate neighboring Hox genes. Interestingly, vertebrate mir-196, which lies at an analogous position in the vertebrate Hox clusters, is transcribed in the same direction as mir-iab-4AS and most other Hox genes, and targets homologs of both abd-A and Ubx (Mansfield et al. 2004; Yekta et al. 2004; Hornstein et al. 2005). With its shared transcriptional orientation and homologous targets, mir-iab-4AS appears to be the functional equivalent of mir-196.
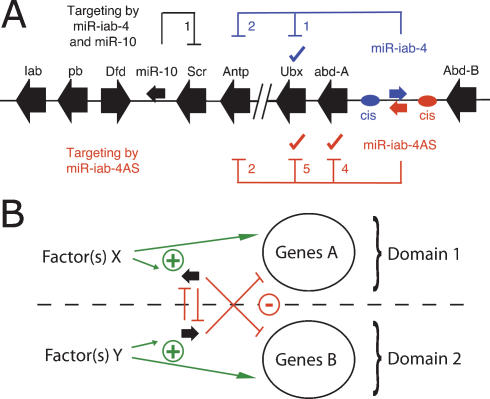
The expression patterns and regulatory connections between Hox genes and the two iab-4 miRNAs show an intriguing pattern in which the miRNAs appear to reinforce Hox gene-mediated transcriptional regulation (Fig. 4A). In particular, miR-iab-4AS would reinforce the posterior expression boundary of abd-A, Ubx, and Antp, supporting their transcriptional repression by Abd-B. mir-iab-4 appears to support abd-A- and Abd-B-mediated repression of Ubx, reinforcing the abd-A/Ubx expression domains and the posterior boundary of Ubx expression. Furthermore, both iab-4 miRNAs have conserved target sites in Antp, which is also repressed by Abd-B, abd-A, and Ubx. The iab-4 miRNAs thus appear to support the established regulatory hierarchy among Hox transcription factors, which exhibits “posterior prevalence,” in that more posterior Hox genes repress more anterior ones and are dominant in specifying segment identity (for reviews, see McGinnis and Krumlauf 1992; Pearson et al. 2005). Interestingly, Abd-B and mir-iab-4AS are expressed in the same segments, and the majority of cis-regulatory elements controlling Abd-B expression are located 3′ of Abd-B (Boulet et al. 1991). This places them near the inferred transcription start of mir-iab-4AS, where they potentially direct the coexpression of these genes. Similarly, abd-A and mir-iab-4 may be coregulated as both are transcribed divergently, potentially under the control of shared upstream elements.
Figure 4.
Regulation of gene expression by antisense miRNAs. (A) miRNA-mediated control in the Drosophila Hox cluster. Schematic representation of the Drosophila Hox cluster (Antennapedia and Bithorax complex) with miRNA target interactions (check marks represent experimentally validated targets). miR-iab-4 (blue) and miR-iab-4AS (red) target anterior neighboring Hox genes and miR-10 (black) targets posterior Sex-combs-reduced (Scr) (Brennecke et al. 2005). abd-A and mir-iab-4 and Abd-B and mir-iab-4AS might be coregulated from shared control elements (cis). Note that mir-iab-4AS is expressed in the same direction as most other Hox genes and its mammalian equivalent, mir-196. (B) General model for defining different expression domains with pairs of antisense miRNAs (black). Different transcription factor(s) activate the transcription of miRNAs and genes in each of the two domains separately (green lines). Both miRNAs might inhibit each other by transcriptional interference or post-transcriptionally (vertical red lines), leading to essentially nonoverlapping expression and activity of both miRNAs. Further, both miRNAs likely target distinct sets of genes (diagonal red lines), potentially re-enforcing the difference between the two expression domains.
Our data demonstrate the transcription and processing of sense and antisense mir-iab-4 into functional miRNAs with highly conserved functional target sites in neighboring Hox genes. In an accompanying study (Bender 2008), genetic and molecular analyses in mir-iab-4 mutant Drosophila revealed that the proposed regulation of Ubx by both sense and antisense miRNAs occurs under physiological conditions and, in particular, the regulation by miR-iab-4AS is required for normal development. These lines of evidence establish miR-iab-4AS as a novel Hox gene, being expressed from within the Hox cluster and regulating Hox genes during development.
The genomic arrangement of two miRNAs that are expressed from the same locus but on different strands might provide a simple and efficient means to create nonoverlapping miRNA expression domains (Fig. 4B). Such sense/antisense miRNAs could restrict each other’s transcription, either by direct transcriptional interference, as shown for overlapping convergently transcribed genes (Shearwin et al. 2005; Hongay et al. 2006), or post-transcriptionally, possibly via RNA–RNA duplexes formed by the complementary transcripts. Sense/antisense miRNAs would usually differ at their 5′ ends and thereby target distinct sets of genes, which might help define and establish sharp boundaries between expression domains. Coupled with feedback loops or coregulation of miRNAs and genes in cis or trans, this arrangement could provide a powerful regulatory switch. The iab-4 miRNAs might be a special case of tight regulatory integration in which miRNAs and proximal genes appear coregulated transcriptionally in cis and repress each other both transcriptionally and post-transcriptionally.
It is perhaps surprising that no antisense miRNA had been found previously, even though, for example, the intriguing expression pattern of the iab-4 transcripts had been reported nearly two decades ago (Cumberledge et al. 1990; Bae et al. 2002), and iab-4 lies in one of the most extensively studied regions of the Drosophila genome. The frequent occurrence of antisense transcripts (Yelin et al. 2003; Katayama et al. 2005) suggests that more antisense miRNAs might exist. Indeed, up to 13% of known Drosophila, 20% of mouse, and 31% of human miRNAs are located in introns of host genes transcribed on the opposite strand or are within 50 nt of antisense ESTs or cDNAs (Supplemental Table S1). These include an antisense transcript overlapping human mir-196 (see also Mainguy et al. 2007). However, because of the contribution of noncanonical base pairs, particularly G:U pairs that become less favorable A:C in the antisense strand, many miRNA antisense transcripts will not fold into hairpin structures suitable for miRNA biogenesis, which explains the propensity of miRNA gene predictions to identify the correct strand (Lim et al. 2003). Nonetheless, in a recent prediction effort, 22 sequences reverse-complementary to known Drosophila miRNAs showed scores seemingly compatible with miRNA processing (Stark et al. 2007b). Deep sequencing of small RNA libraries from Drosophila confirmed the processing of small RNAs from four of these high-scoring antisense candidates (Ruby et al. 2007), and the ovary/testes libraries used here showed antisense reads for an additional Drosophila miRNA (mir-312) (see Supplemental Tables S2, S3). In addition, using high-throughput sequencing of small RNA libraries from mice, we found sequencing reads that uniquely matched the mouse genome in loci antisense to 10 annotated mouse miRNAs. Eight of the inferred antisense miRNAs were supported by multiple independent reads, and two of them had reads from both the mature miRNA and the star sequence (Supplemental Table S2). These results suggest that sense/antisense miRNAs could be more generally employed in diverse contexts and in species as divergent as flies and mammals.
Materials and methods
Plasmids
3′ UTRs were amplified from Drosophila melanogaster genomic DNA and cloned in pCR2.1 for site-directed mutagenesis. The following primer pairs were used to amplify the indicated 3′ UTR: abd-A (tc tagaGCGGTCAGCAAAGTCAACTC; gtcgacATGGATGGGTTCTCGT TGCAG), Ubx (tctagaATCCTTAGATCCTTAGATCCTTAG; ctcgag ATGGTTTGAATTTCCACTGA), and Abd-B (tctagaGCCACCACCT GAACCTTAG; aactcgagCGGAGTAATGCGAAGTAATTG). QuickChange multisite-directed mutagenesis was used to mutate all miR-iab-4AS seed sites from ATACGT to ATAGGT, per the manufacturer’s directions (Stratagene). Wild-type and mutated 3′ UTRs were subcloned into pCJ40 between SacI and NotI sites to make Renilla luciferase reporters. Plasmid pCJ71 contains the abd-A wild-type 3′ UTR, pCJ72 contains the Ubx wild-type 3′ UTR, pCJ74 contains the Abd-B wild-type 3′ UTR, pCJ75 contains the abd-A mutated 3′ UTR, and pCJ76 contains the Ubx mutated 3′ UTR fused to Renilla luciferase. The control let-7 expression vector was obtained by amplifying let-7 from genomic DNA with primers 474 base pairs (bp) upstream of and 310 bp downstream from the let-7 hairpin and cloning it into pMT-puro. To express miR-iab-4 and miR-iab-4AS, a 430-bp genomic fragment containing the miR-iab-4 hairpin was cloned, in either direction, downstream from the tubulin promoter as described in Stark et al. (2005). For the UAS-miR-iab-4 and UAS-miR-iab-4AS constructs, the same 430-bp genomic fragment containing the miR-iab-4 hairpin was cloned downstream from pUAST-DSred2 (Stark et al. 2003) in either direction.
Reporter assays
For the luciferase assays, 2 ng of p2129 (firefly luciferase), 4 ng of Renilla reporter, 48 ng of miRNA expression plasmid, and 48 ng of p2032 (GFP) were cotransfected with 0.3 μL Fugene HD per well of a 96-well plate. Twenty-four hours after transfection, expression of Renilla luciferase was induced by addition of 500 μM CuSO4 to the culture media. Twenty-four hours after induction, reporter activity was measured with the Dual-Glo luciferase kit (Promega), per the manufacturer’s instructions on a Tecan Safire II plate reader.
The ratio of Renilla:firefly luciferase activity was measured for each well. To calculate fold repression, the ratio of Renilla:firefly for reporters cotransfected with let-7 was set to 1. The Wilcoxon rank-sum test was used to assess the significance of changes in fold repression of wild-type reporters compared with mutant reporters. Geometric means from 16 transfections representing four replicates of four independent transfections are shown. Error bars represent the fourth highest and lowest values of each set.
Drosophila strains
UAS-miR-iab-4 and UAS-miR-iab-4AS flies were generated by injection of the corresponding plasmids into w1118 embryos. bxMS1096-GAL4 flies were obtained from the Bloomington Stock Center.
In situ hybridization and protein stainings
Double in situ hybridization for the miRNA primary transcripts was performed as described in Stark et al. (2005). Probes were generated using PCR on genomic DNA with primers TCAGAGCATGCAGAGACAT AAAG, TTGTAGATTGAAATCGGACACG for iab-4 sense and ATTT TACTGGGTGTCTGGGAAAG, TAGAAACTGAGACGGAGAAGCAG for iab-4 antisense. Protein stainings were performed as described in Patel (1994). Antibodies used were mouse anti-Ubx (1:30), mouse anti-abd-A (1:5), and HRP-conjugated goat anti-mouse (Dianova, 1:3000).
RT–PCRs
Total RNA was isolated using Trizol (Invitrogen), treated with RQI DNase (Promega), and used for strand-specific cDNA synthesis with SuperScript III (Invitrogen). Primers for cDNA synthesis were CATATAA CAAAGTGCTACGTG (iab-4 sense) and CTTTATCTGCATTTG GATCCG (iab-4 antisense). Both primers were used for subsequent amplification.
Small library sequencing
Drosophila small RNAs were cloned from adult ovaries and testes as described previously (Brennecke et al. 2007) and sequenced using Solexa sequencing. A total of 657,251 sequencing reads uniquely matched known Drosophila miRNAs (Rfam release 9.2), and the 69 miRNAs with unique matches had 1011 matches on average (Stark et al. 2007b). Two miRNAs had unique matches to the antisense hairpin (Supplemental Tables S2, S3). Mouse small RNAs were cloned from wild-type and c-kit mutant ovaries (Supplemental Table S4; G. Hannon, pers. comm.) and from Comma-Dbgeo cells, a murine mammary epithelial cell line (Ibarra et al. 2007), and were sequenced using Solexa sequencing. A total of 4,217,883 reads uniquely matched known mouse miRNAs (Rfam release 9.2), and the 286 miRNAs with unique reads showed 256 reads on average. Sequencing reads matching to the plus and minus strand of known mouse miRNAs with antisense reads are listed in Supplemental Table S3.
Multiple sequence alignments and target site prediction
The multiple sequence alignments for the indicated Hox 3′ UTRs were obtained from the University of California at Santa Cruz (UCSC) genome browser (Kent et al. 2002) and were slightly manually adjusted. We predicted target sites according to Lewis et al. (2005) by searching for 3′ UTR seed sites (reverse-complementary to miRNA positions 2–8 or matching to “A” + reverse complement of miRNA positions 2–7).
Antisense transcripts near known miRNAs
To assess the fraction of Drosophila, human, and mouse miRNAs that are also putatively transcribed on both strands and might give rise to antisense miRNAs, we determined the number of miRNAs that are near known transcripts on the opposite strand. We obtained the coordinates of all introns of protein-coding genes and all mapped ESTs or cDNAs for the three species from the UCSC genome browser (Kent et al. 2002). We intersected them with the miRNA coordinates from Rfam (release 9.2; Griffiths-Jones et al. 2006), requiring miRNAs and transcripts to be on opposite strands and at a distance of at most 50 nt. For each miRNA, we recorded the number of antisense transcripts and their identifiers. Note that some of the transcripts might have been mapped to more than one place in the genome, such that the intersection represents an upper estimate based on the currently known transcripts.
Acknowledgments
We thank Greg Hannon for providing Solexa sequencing data and support, Juerg Mueller for the anti-Ubx antibody, Thomas Sandmann for Drosophila embryos, and Sandra Mueller for preparing transgenic flies. We thank the Drosophila genome sequencing centers and the UCSC genome browser for access to the 12 Drosophila multiple sequence alignments prior to publication, and Welcome Bender for sharing data prior to publication. A.S. was partly supported by a post-doctoral fellowship from the Schering AG and partly by a post-doctoral fellowship from the Human Frontier Science Program Organization (HFSPO). C.H.J. is an NSF graduate fellow. J.B. thanks the Schering AG for a post-doctoral fellowship. This work was also partially supported by a grant from the NIH.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1613108
References
- Bae E., Calhoun V.C., Levine M., Lewis E.B., Drewell R.A. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bender W. MicroRNAs in the Drosophila bithorax complex. Genes & Dev. 2008 doi: 10.1101/gad.1614208. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A.M., Lloyd A., Sakonju S. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development. 1991;111:393–405. doi: 10.1242/dev.111.2.393. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Zaratzian A., Sakonju S. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 1990;87:3259–3263. doi: 10.1073/pnas.87.9.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Duboule D. Vertebrate hox gene regulation: Clustering and/or colinearity? Curr. Opin. Genet. Dev. 1998;8:514–518. doi: 10.1016/s0959-437x(98)80004-x. [DOI] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34 (Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay C.F., Grisafi P.L., Galitski T., Fink G.R. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Mansfield J.H., Yekta S., Hu J.K., Harfe B.D., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Ibarra I., Erlich Y., Muthuswamy S.K., Sachidanandam R., Hannon G.J. A microRNA fingerprint of mammary epithelial stem cells. Genes & Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F., Bender W., Weiffenbach B. abdA expression in Drosophila embryos. Genes & Dev. 1990;4:1573–1587. doi: 10.1101/gad.4.9.1573. [DOI] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Lewis E.B. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias A., Casanova J., Morata G. Expression and regulation of the abd-A gene of Drosophila. Development. 1990;110:1197–1207. doi: 10.1242/dev.110.4.1197. [DOI] [PubMed] [Google Scholar]
- Mainguy G., Koster J., Woltering J., Jansen H., Durston A. Extensive polycistronism and antisense transcription in the mammalian hox clusters. PLoS ONE. 2007;2:e356. doi: 10.1371/journal.pone.0000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J.H., Harfe B.D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Patel N.H. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Ronshaugen M., Biemar F., Piel J., Levine M., Lai E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes & Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007 doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shearwin K.E., Callen B.P., Egan J.B. Transcriptional interference—A crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R.B., Cohen S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007a;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Kheradpour P., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007b doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee S.D., Halder G., Kim J., Hudson A., Carroll S. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes & Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yelin R., Dahary D., Sorek R., Levanon E.Y., Goldstein O., Shoshan A., Diber A., Biton S., Tamir Y., Khosravi R., et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- Yoder J.H., Carroll S.B. The evolution of abdominal reduction and the recent origin of distinct Abdominal-B transcript classes in Diptera. Evol. Dev. 2006;8:241–251. doi: 10.1111/j.1525-142X.2006.00095.x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]