Abstract
β-Catenin plays important roles in cell adhesion and gene transcription, and has been shown recently to be essential for the establishment of a bipolar mitotic spindle. Here we show that β-catenin is a component of interphase centrosomes and that stabilization of β-catenin, mimicking mutations found in cancers, induces centrosome splitting. Centrosomes are held together by a dynamic linker regulated by Nek2 kinase and its substrates C-Nap1 (centrosomal Nek2-associated protein 1) and Rootletin. We show that β-catenin binds to and is phosphorylated by Nek2, and is in a complex with Rootletin. In interphase, β-catenin colocalizes with Rootletin between C-Nap1 puncta at the proximal end of centrioles, and this localization is dependent on C-Nap1 and Rootletin. In mitosis, when Nek2 activity increases, β-catenin localizes to centrosomes at spindle poles independent of Rootletin. Increased Nek2 activity disrupts the interaction of Rootletin with centrosomes and results in binding of β-catenin to Rootletin-independent sites on centrosomes, an event that is required for centrosome separation. These results identify β-catenin as a component of the intercentrosomal linker and define a new function for β-catenin as a key regulator of mitotic centrosome separation.
[Keywords: β-Catenin, centrosome, Nek2, mitosis, C-Nap1, Rootletin]
β-Catenin is a multifunctional protein that plays key roles in the organization and maintenance of cells in tissues. β-Catenin is an effector of the Wnt pathway that regulates cell proliferation and gene expression in development, and is a component of the cell adhesion complex that regulates cell sorting and tissue organization (Dierick and Bejsovec 1999; Eastman and Grosschedl 1999; Nelson and Nusse 2004). β-Catenin levels are tightly controlled by a destruction complex of adenomatous polyposis coli (APC), axin, casein kinase I (CKI), and GSK3β, which bind, phosphorylate, and target β-catenin to the proteasome (Peifer and Polakis 2000; Moon et al. 2002; Nelson and Nusse 2004). Mutations that stabilize β-catenin are common in cancer, and are thought to be the earliest event in colon cancer (Bienz and Clevers 2000; Peifer and Polakis 2000; Behrens 2005). Recently, it has been shown that β-catenin localizes to centrosomes in mitosis and has a role in establishing a bipolar spindle (Kaplan et al. 2004); however, the regulation and function for β-catenin at centrosomes in normal cells and how this function is perturbed in cancers are not understood.
Centrosomes undergo a highly regulated duplication cycle in interphase cells so that at the onset of mitosis a cell has two centrosomes that can separate to establish a bipolar spindle (Hinchcliffe and Sluder 2001; Tsou and Stearns 2006). In general, mechanisms of centrosome cohesion and separation are not well understood. It is thought that in interphase the two centrosomes, each with a pair of centrioles, are held together by a dynamic physical linker composed of C-Nap1 (centrosomal Nek2-associated protein 1) and Rootletin (Bahe et al. 2005; Yang et al. 2006). In mitosis, activation of the NIMA-related centrosomal kinase Nek2A results in the phosphorylation of C-Nap1 and Rootletin, which is thought to cause their dissociation from centrosomes and enable centrosome separation (Fry et al. 1998a; Mayor et al. 2002; Bahe et al. 2005; Hayward and Fry 2006; Yang et al. 2006). “Centrosome separation” describes the cell cycle-regulated separation of centrosomes at the onset of mitosis, while “centrosome splitting” describes the increased distance between parental centrioles in interphase cells due to decreased cohesion (Meraldi and Nigg 2001). Loss of C-Nap1 and Rootletin is sufficient to induce centrosome splitting in interphase cells (Bahe et al. 2005); however, C-Nap1 dissociates from unseparated centrosomes in monopolar spindles caused by expression of kinase-dead (KD) Nek2 in mitotic cells (Faragher and Fry 2003). Thus, Nek2 activity is not essential for loss of C-Nap1 from mitotic centrosomes, but is required for centrosome separation, likely through the phosphorylation of additional unknown substrates.
Depletion of β-catenin induces monopolar spindles with unseparated centrosomes (Kaplan et al. 2004), a phenotype strikingly similar to that caused by kinase-dead Nek2 (Faragher and Fry 2003). Here we show that stabilization of β-catenin, mimicking mutations found in cancer, induces centrosome splitting, similar to ectopic Nek2 activity (Fry et al. 1998b). Using an unbiased approach, we show that β-catenin is a substrate and binding partner for Nek2 kinase in vitro and in vivo, and colocalizes with the other Nek2 substrates, Rootletin and C-Nap1, in between centrosomes. C-Nap1 and Rootletin are required for localization of β-catenin in between centrosomes in interphase, whereas β-catenin has Rootletin-independent binding sites on centrosomes at mitotic spindle poles. In response to ectopic expression of active Nek2 in interphase cells, Rootletin is strongly reduced at interphase centrosomes and β-catenin localizes to Rootletin-independent sites on centrosomes, an event that is required for centrosome separation in mitosis. These results provide the first evidence detailing the function of β-catenin in centrosome separation and give mechanistic insight into how Nek2 regulates β-catenin during centrosome separation.
Results
β-Catenin localizes to interphase and mitotic centrosomes via its armadillo repeat domain
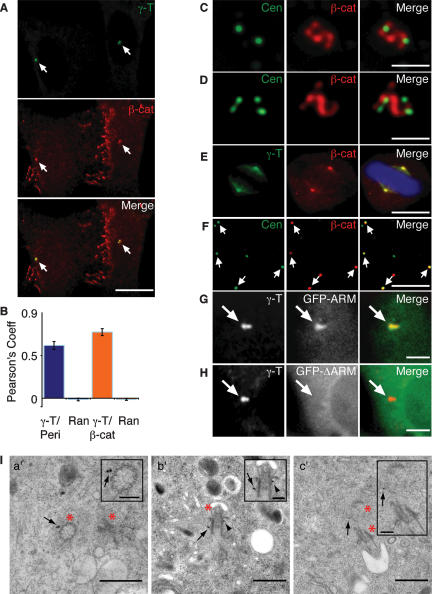
β-Catenin localized to sites of cell–cell adhesion and to cytoplasmic puncta, some of which colocalized with γ-tubulin-labeled centrosomes in U-2 OS cells (Fig. 1A, arrows). We tested whether localization of β-catenin to centrosomes was statistically significant using Pearson’s correlation coefficient (Manders et al. 1993) for β-catenin and γ-tubulin, and for γ-tubulin and another known centrosome component, pericentrin (Doxsey et al. 1994). Pearson’s coefficient for γ-tubulin and β-catenin was 0.77 ± 0.04, and 0.62 ± 0.05 for γ-tubulin and pericentrin (Fig. 1B), indicating that the localization of β-catenin to centrosomes was as specific as that of pericentrin. Randomizing the γ-tubulin channel 100 times in a 200 × 200-pixel square near the centrosome in the β-catenin or pericentrin channel resulted in identical, low Pearson’s coefficients for both proteins (−0.015 ± 0.011) (Fig. 1B), indicating that β-catenin localization to centrosomes is not a random effect of high cytoplasmic β-catenin.
Figure 1.
β-Catenin associates with interphase and mitotic centrosomes via its armadillo domain. (A) U-2 OS cells costained for γ-tubulin (γ-T; green) and β-catenin (β-cat; red). Arrows indicate colocalization of β-catenin with γ-tubulin. Bar, 10 μm. (B) Pearson correlation coefficient quantified for colocalization of γ-tubulin with pericentrin or β-catenin and for randomization of γ-tubulin channel in a 200 × 200-pixel square in the centrosome region and subsequent colocalization with pericentrin or β-catenin (n = 10). (C,D) Immunostaining of centrin (green) and β-catenin (red). G1/S-phase cell with two centrioles (C) or S/G2-phase cell with four centrioles (D). Bar, 2 μm. See also Supplemental Movie S1. (E) Immunostaining of mitotic U-2 OS cell with γ-tubulin (green), β-catenin (red), and DAPI (blue). Bar, 10 μm. (F) Centrosome-enriched fractions from nocodazole-treated U-2 OS cells costained with centrin (green) and β-catenin (red). Arrows mark colocalization of β-catenin with centrin-marked centrosomes. Note centrioles without β-catenin are also present, which indicates either that β-catenin was lost from centrioles during the purification procedure or that β-catenin localizes to a subset of centrioles. Bar, 10 μm (G,H) U-2 OS cells immunostained for γ-tubulin (red in merged image) and transiently expressing GFP-tagged Armadillo domain of β-catenin (G) or GFP-tagged ΔARM (H). Arrows indicate γ-tubulin-labeled centrosomes. Bar, 5 μm. (I) Immunogold labeling of β-catenin localization relative to centrioles in RPE-1 cells. Serial sectioning was performed in the centrosome region of 20 cells. β-Catenin label was found at the proximal (16%) and distal (60%) ends of centrioles, and between centrioles (4%), while labeling in surrounding areas was minimal. Twenty percent of the label on centrioles could not be assigned to either end. A single section is shown for each example. Insets show higher magnification of centrioles in panels a′–c′. Asterisks mark centrioles. (Panel a′) Cross-section of a centriole showing β-catenin near the “pinwheel” at the proximal end of the centriole (arrow). (Panel b′) Longitudinal section of a centriole showing β-catenin at the distal end (arrowhead), as identified by the subdistal and distal appendages. Arrow marks β-catenin at the proximal end of the centriole, relative to the appendages. (Panel c′) Example of β-catenin between centrioles. Bars: 0.5 μm; inset, 0.15 μm.
To follow the localization of β-catenin during centrosome duplication, we immunostained U-2 OS cells for the centriolar marker centrin. G1 cells have two centrioles with two centrin spots, while S/G2 cells with fully duplicated centrioles have four centrin spots (Fig. 1C,D; Supplemental Movie S1). β-Catenin was observed at centrosomes in 100% of U-2 OS cells, localizing in between centrin-marked centrioles in 78% of cells (n = 9) (Fig. 1C,D; Supplemental Movie S1). β-Catenin also localized to mitotic centrosomes at spindle poles (Fig. 1E; Kaplan et al. 2004). Identical results were obtained with two different β-catenin antibodies in U-2 OS cells, and in MDCK and HeLa cells (Supplemental Fig. S1A–C).
β-Catenin cosedimented with enriched centrosome fractions from asynchronous HeLa cells (Kaplan et al. 2004). We found that β-catenin colocalized with centrin in these preparations from nocodazole-treated asynchronous U-2 OS (Fig. 1F, arrows) and MDCK (Supplemental Fig. S1D) cells that were active in microtubule assembly (Louie et al. 2004).
We identified the domain that mediates β-catenin localization to centrosomes. A mutant deleted of the N and C termini but containing the complete armadillo repeat domain (GFP-ARM) localized to centrosomes (Fig. 1G), but one containing a deletion of the armadillo repeat domain (GFP-ΔARM) (Elul et al. 2003) did not (Fig. 1H).
Finally, we used immuno-electron microscopy to localize β-catenin at centrioles (Fig. 1I). Gold labeling of β-catenin was observed in the centriole region, while labeling over distant areas of the cytoplasm remained minimal at various antibody dilutions (Fig. 1I, panels a′–c′). Using pinwheel, distal appendages, and the primary cilium as markers, we found that β-catenin localized to the proximal (Fig. 1I, arrows in panels a′,b′) and distal regions of the centriole (Fig. 1I, arrowhead in panel b′) and between centrioles (Fig. 1I, arrow in panel c′). Similar results were found with two different antibodies, and were consistent with our immunofluorescence data (see Fig. 1C,D).
Taken together, these results demonstrate that β-catenin colocalizes with γ-tubulin- and centrin-labeled centrosomes throughout interphase and mitosis and cosediments with centrosome-enriched fractions, and that centrosome binding is mediated by the armadillo domain. The centrosome location of β-catenin is specific, statistically significant, and not random; was found in four different cell lines with two different antibodies at the level of light and electron microscopy; and is independent of microtubules (Supplemental Fig. S2A). We conclude that β-catenin is an integral component of centrosomes throughout the cell cycle.
β-Catenin is a dynamic component of interphase centrosomes
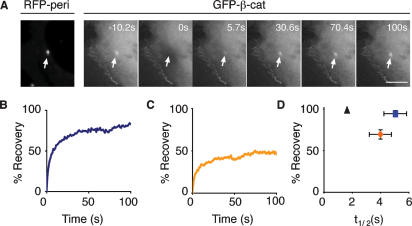
Centrosomal proteins are characteristically dynamic (Khodjakov and Rieder 1999; Hames et al. 2005). To examine the mobility of β-catenin at centrosomes, we measured fluorescence recovery after photobleaching (FRAP) of GFP-tagged wild-type β-catenin (GFP-β-cat), using RFP-pericentrin to mark the centrosome. GFP-β-cat fluorescence recovered to 94.5 ± 3.8% with a t1/2 of 5.03 ± 0.80 sec (Fig. 2A,B, summarized in D), similar to known centrosomal proteins such as Nek2 (Hames et al. 2005). Mutations of GSK3β phosphorylation sites in β-catenin, which are found in cancer, increase β-catenin stability and protein level in cells (Polakis 1999; Peifer and Polakis 2000). Therefore, we measured the FRAP of a stabilized form of β-catenin (referred to as β-cat*) that also localizes to centrosomes (Supplemental Fig. S2B). The mobile fraction of GFP-β-cat* had a t1/2 of 3.98 ± 0.78 sec, similar to that of GFP-β-cat. However, GFP-β-cat* recovered to only 69.6 ± 5.6% (Fig. 2C, summarized in D). The increased immobile fraction of GFP-β-cat* may be due to stable interactions of β-cat* with centrosomal binding partners. These data indicate that β-catenin exchanges between centrosomal and noncentrosomal pools with kinetics similar to those of other centrosome components, while stabilized β-cat* has decreased turnover at centrosomes.
Figure 2.
Dynamics of β-catenin and stabilized β-catenin (β-cat*) at centrosomes. (A) FRAP of GFP-β-catenin (gray montage) at centrosomes marked by RFP-pericentrin. Arrow marks photobleach region. Bar, 5 μm. (B) Fluorescence recovery profile of data in A. (C) Representative fluorescence recovery profile of GFP-β-catenin mutated in four GSK3β phosphorylation sites (β-cat*) at centrosomes. (D) Summary of average fluorescence recovery and t1/2 of GFP-β-catenin (blue; n = 4) and GFP-β-cat* (orange; n = 6) at centrosomes. Diffusion alone is shown as a black triangle (n = 2). Average of experiments is shown ± standard deviation. Student’s t-test was performed for recovery of β-cat and β-cat* with P = 0.011.
β-Catenin is a direct binding partner and substrate of centrosomal Nek2 kinase
To understand the function of β-catenin at centrosomes, we took an unbiased approach to identify centrosomal binding partners of β-catenin. We purified centrosomes (Mitchison and Kirschner 1986) from a stable HeLa cell line expressing myc-tagged β-catenin (Kaplan et al. 2004), immunoprecipitated centrosome-associated β-catenin-myc, and identified coimmunoprecipitating proteins by SDS-PAGE and MALDI TOF/TOF mass spectrometry (Supplemental Fig. S3). This analysis identified the kinase Nek2 as a putative centrosomal binding partner of β-catenin. Nek2 is a centrosomal kinase involved in centrosome cohesion (Fry et al. 1998b; Hayward and Fry 2006). Significantly, disruption of Nek2 function results in monopolar spindles with unseparated centrosomes and chromosome rosettes, a phenotype strikingly similar to what we observe in cells depleted of β-catenin (Supplemental Figs. S4, S7B; Kaplan et al. 2004). We also found that endogenous Nek2 coimmunoprecipitated with endogenous β-catenin from isolated centrosomes in HeLa cells (Fig. 3A). This interaction was further verified by coimmunoprecipitation of GFP-tagged Nek2 with endogenous β-catenin from transiently transfected HEK 293T cells (Fig. 3B). Together, these data show that β-catenin and Nek2 interact at the centrosome.
Figure 3.
β-Catenin binds to and is phosphorylated by Nek2 in vitro and in vivo. (A; left) Centrosomes were isolated from HeLa cells (Kaplan et al. 2004) and sucrose fractions from the final purification step were subjected to immunoblot analysis for β-catenin and the centrosomal marker γ-tubulin. (right) β-Catenin was immunoprecipitated from fractions 16 and 17 and the immunoprecipitates were analyzed by immunoblot for β-catenin and Nek2. Data are from one experiment that is representative of two independent experiments. (B) Immunoblot of lysates from HEK 293T cells transfected with control or GFP-Nek2 expression vectors is shown at left. (Right) Lysates were incubated with control or β-catenin antibodies and immunoprecipitates were subjected to immunoblot analysis for β-catenin or Nek2. Data are from one experiment that is representative of four independent experiments. (C) HA-tagged wild-type (WT), activated (NB), or kinase-dead (KD) Nek2A, or wild-type CKIε (CK1), were immunoprecipitated from transfected HEK 293T cells and used in the in vitro kinase assays with γ-32PATP and recombinant β-catenin. (Top panel) Autoradiograph (32P). (Middle panel) Coomassie brilliant blue-stained SDS-PAGE gel (CBB). (Bottom panel) Anti-HA immunoblot (IB:HA). Data are from one experiment that is representative of five independent experiments. (D) Recombinant GST or GST-β-catenin bound to glutathione sepharose beads was incubated with recombinant His-Nek2A. Immunoblot was performed for β-catenin and Nek2 on the precipitated material (P) and the supernatants (S). Data are from one experiment that is representative of two independent experiments. (E) Recombinant full-length β-catenin (GST-β-cat) and the Armadillo domain of β-catenin (ARM) were incubated with recombinant His-Nek2A for 0, 10, or 60 min in the presence of γ-32PATP and separated by SDS-PAGE. A 60-min reaction containing His-Nek2A without substrate shows Nek2A autophosphorylation (Nek2 lane). A representative autoradiograph of the kinase assays is shown in the top panel (32P) and the corresponding Coomassie brilliant blue-stained gel is shown in the bottom panel (CBB). Data are from one experiment that is representative of four independent experiments. (F) HEK 293T cells were transfected with control vector (Con), wild-type GFP-Nek2 (WT), or kinase-dead GFP-Nek2 (KD), and the electrophoretic mobility of endogenous β-catenin was assessed by SDS-PAGE, followed by an immunoblot for β-catenin (IB:β-cat). Nek2 expression is shown in the bottom panel (IB:Nek2). Wild-type, but not kinase-dead Nek2, results in decreased electrophoretic mobility of β-catenin (denoted by asterisk at top left of shifted band), which is reversed by λ-phosphatase treatment (+). Data are from one experiment that is representative of two independent experiments.
Next, we determined whether β-catenin is a substrate for Nek2 kinase. Purified β-catenin was phosphorylated by HA-tagged Nek2 or HA-Nek2NB (Nek2 mutated in the protein phosphatase 1-binding site, resulting in increased activity [Eto et al. 2002]) that had been immunoprecipitated from transfected HEK 293T cells, but not by a kinase-dead mutant of Nek2 (HA-Nek2KD) (Fig. 3C). Nek2 phosphorylated β-catenin with an efficiency similar to that of CKIε, a well-characterized β-catenin kinase (Fig. 3C; Gao et al. 2002).
To determine whether binding between β-catenin and Nek2 is direct, we used purified β-catenin and Nek2 in an in vitro binding assay. Indeed, immobilized GST-β-catenin bound directly to His-Nek2 (Fig. 3D). Furthermore, His-Nek2 directly phosphorylated full-length GST-β-catenin in vitro (Fig. 3E), as well as the β-catenin armadillo repeat domain (ARM) (Fig. 3E), which is necessary and sufficient for β-catenin localization to centrosomes (see Fig. 1G,H).
In some cases, phosphorylation decreases the electrophoretic mobility of proteins in SDS-PAGE. Indeed, we detected a slower migrating form of endogenous β-catenin from HEK 293T cells expressing wild-type, but not kinase-dead, Nek2 (Fig. 3F). Treatment of the immunoprecipitates with λ-phosphatase resulted in loss of this slower migrating β-catenin band (Fig. 3F), confirming that the gel shift was due to protein phosphorylation. Thus, β-catenin is a binding partner and substrate in vitro and in vivo for the centrosomal kinase Nek2, an important regulator of centrosome cohesion.
β-Catenin stabilization and depletion affect centrosome cohesion
Kaplan et al. (2004) had shown previously that depletion of β-catenin resulted in monopolar spindles with duplicated but unseparated centrosomes and distinct chromosome rosettes (see also Supplemental Figs. S4, S7B), phenotypes identical to those reported in cells expressing kinase-dead Nek2 (Faragher and Fry 2003). Additionally, depletion of β-catenin resulted in a delay in prometaphase in live cells, consistent with a role for β-catenin in the establishment of a bipolar spindle (Supplemental Fig. S4; Supplemental Movies S2, S3). We asked if the converse happened—that is, whether the stabilized form of β-catenin (β-cat*), which has decreased turnover at centrosomes (see Fig. 2), results in decreased centrosome cohesion, similar to overexpressing Nek2. In asynchronous interphase cells expressing β-cat* under the tetR doxycycline (dox)-repressible promoter and with two centrosomal structures marked by γ-tubulin and pericentrin, we found a significantly greater distance between centrosomal structures (β-cat*, 5.2 ± 0.2 μm) (Fig. 4A–C) than that in parental (Parental, 3.2 ± 0.4 μm) or dox-repressed (+Dox, 3.3 ± 0.2 μm) cells.
Figure 4.
Increased distance between centrosome and centrioles in stabilized β-catenin-expressing cells. (A,B) DNA content and immunostains, γ-tubulin (green) and DAPI (blue), of unsynchronized parental and stabilized β-catenin (β-cat*)-expressing cells. G1/S, 37%; S, 41%; S/G2, 17% in both cell lines. Percentage of cells in each phase was determined by the Dean-Jett-Fox algorithm (Bauer et al. 1998). Bar, 5 μm. (C) Average distance between γ-tubulin- and pericentrin-marked centrosomes in parental (n = 413), dox-repressed (+Dox; n = 300), and β-cat* (n = 424) cells. (D) Histogram showing intercentrosomal distance in parental, dox-repressed, and β-cat*-expressing cells. (E,F) DNA content and immunostains, glutamylated tubulin (GT335; green) and DAPI (blue), of parental and stabilized β-catenin (β-cat*)-expressing cells synchronized in G1/S by double-thymidine block. Parental: G1/S, 86.1%; G2/M, 8.7%. β-cat*: G1/S, 83.6%; G2/M, 7.6%. Bar, 5 μm. (G) Average distance between centrioles in parental (n = 173), dox-repressed (+Dox; n = 204), and β-cat*-expressing (n = 247) cells. (H) Histogram showing intercentriolar distance in parental, dox-repressed, and β-cat*-expressing cells. Average of three independent experiments ± standard error is shown. Student’s t-test was performed for wild-type (WT)/+D (combined) and β-cat* with P < 0.01.
It is known that centrosome distance is regulated by proteins that link mother–daughter centriole pairs in G1/S and parental centrioles between centrosomes after centriole duplication in S/G2 (see below; Fry et al. 1998a; Mayor et al. 2000; Bahe et al. 2005; Yang et al. 2006). As overexpression of Nek2A results in increased centriole distance in G1/S (Faragher and Fry 2003), we tested whether β-cat* affected the distance between centrioles specifically in G1/S using cells synchronized in G1/S by double-thymidine block and immunostained for glutamylated tubulin to mark centrioles (Fig. 4E–G). The average distance between centrioles in the presence of β-cat* was 7.2 ± 0.34 μm compared with 3.75 ± 0.75 μm in dox-repressed cells and 3.15 ± 0.30 μm in parental cells (Fig. 4G). Even more striking was the finding that the percentage of cells containing centrosomes or centrioles separated by ≥8 μm was much greater in the presence of β-cat* than wild-type β-catenin (Fig. 4D,H); 32 ± 2% of centrioles and 25 ± 3% of centrosomes in β-cat*-expressing cells were split by >8 μm compared with 7% in either dox-repressed or parental cells. We conclude that β-cat* increased the distance between centrosomes in asynchronous cells and between centrioles in G1/S phase of the cell cycle. Thus, the effects of depletion of β-catenin or expression of β-cat* on centrosome cohesion are comparable with decreasing (Faragher and Fry 2003) or increasing Nek2 (Fry et al. 1998b) activity, respectively. These data indicate that like Nek2, β-catenin is a negative regulator of centrosome cohesion.
β-Catenin colocalizes with C-Nap1 and Rootletin, and is in a complex with Rootletin
β-Catenin localized in between centrosomes in interphase cells (see Fig. 1), suggesting that β-catenin may colocalize and interact with the other known centrosomal linker proteins and substrates of Nek2, C-Nap1 and Rootletin (Fry et al. 1998a; Mayor et al. 2000; Bahe et al. 2005; Yang et al. 2006). C-Nap1 localizes to the proximal end of centrioles (Mayor et al. 2000) and is thought to provide binding sites for Rootletin, which forms fibers in between centrosomes (Bahe et al. 2005; Yang et al. 2006). C-Nap1 partially colocalized with β-catenin at centrosomes, although β-catenin distribution extended in between C-Nap1 puncta in 10 of 13 cells observed, and in short flares beyond C-Nap1 puncta (Fig. 5A; Supplemental Movies S4a,b; see also β-catenin between centrioles in Fig. 1C,D; Supplemental Movie S1). Additionally, overexpression of C-Nap1 at centrosomes resulted in increased colocalization and puncta of endogenous β-catenin in the centrosome region marked by γ-tubulin (Fig. 5B). Furthermore, we were able to observe a reproducible and above-background, albeit weak, interaction between β-catenin and C-Nap1 by coimmunoprecipitation (S. Bahmanyar and D.D. Kaplan, unpubl.). Thus, C-Nap1 appears to be in a complex with β-catenin at centrosomes, but the interaction between these proteins may be indirect, and is likely mediated by Rootletin (see below).
Figure 5.
β-Catenin partially colocalizes with C-Nap1 and Rootletin, and is in a complex with Rootletin. (A) Projection of deconvolved sections of U-2 OS cells immunostained with C-Nap1 (green) and β-catenin (red). See also Supplemental Movies S4, a and b. (B) Projection of deconvolved sections of U-2 OS cells overexpressing GFP-C-Nap1 (green) in centrosome region and immunostained with γ-tubulin (blue) and β-catenin (red). (C) Three-dimensional rendering of deconvolved sections of U-2 OS cells immunostained with C-Nap1 (blue), β-catenin (red), and Rootletin (green). Two angles of a counterclockwise rotation are shown. See also Supplemental Movies S5, a and b. (D) MDCK lysates were incubated with control or Rootletin antibodies and immunoprecipitates were subjected to immunoblot analysis for β-catenin or Rootletin (shown at right).
To determine the distribution of β-catenin relative to C-Nap1 and Rootletin in centrosomes that were not split, we triple-immunostained cells for C-Nap1, β-catenin, and Rootletin. Using three-dimensional reconstructions of images, we found that both β-catenin and Rootletin colocalized partially with C-Nap1 in 12 of 19 cells observed (Fig. 5C). β-Catenin coaligned with Rootletin that extended between C-Nap-1-positive puncta (Fig. 5C; Supplemental Movies S5a,b). Immunoprecipitation with an antibody to Rootletin resulted in coimmunoprecipitation of β-catenin (Fig. 5D), and β-catenin may interact with Rootletin directly as seen by Far Western blotting (Supplemental Fig. S5). Thus, β-catenin localizes to and interacts with the protein linker in between centrosomes that also contains Nek2, C-Nap1, and Rootletin. However, similar to Nek2 but unlike C-Nap1 and Rootletin, β-catenin is a negative regulator of centrosome cohesion, suggesting that the β-catenin–Rootletin complex may play a regulatory, rather than a structural, role in centrosome cohesion and separation (see Discussion).
Rootletin and C-Nap1 regulate localization of β-catenin in between centrosomes in interphase cells
To determine whether the distribution of β-catenin in between centrosomes is affected by depletion of C-Nap1 or Rootletin from centrosomes, we used siRNA complexes against either C-Nap1 or Rootletin. C-Nap1 and Rootletin siRNA cause centrosome splitting (Bahe et al. 2005). Thus, to directly test for protein levels at centrosomes, we compared the average fluorescence intensity of C-Nap1, Rootletin, and β-catenin at individual centrosomes in unsplit pairs with their fluorescence intensities at individual centrosomes within split pairs (see Materials and Methods). It had been reported previously that different levels of knockdown of C-Nap1 caused either a reduction of Rootletin from the intercentrosomal linker region or formation of long Rootletin fibers from split centrosomes (Bahe et al. 2005), indicating that C-Nap1 is required for proper localization and organization of Rootletin at the intercentrosomal linker region. Using siRNA duplexes against C-Nap1 (Bahe et al. 2005) in U-2 OS cells, we found that 85% of cells (n = 28) had split centrosomes with reduced C-Nap1 (Supplemental Fig. S6A), similar to published results (Bahe et al. 2005). In the 34% of C-Nap1 knockdown cells in which Rootletin was no longer localized to the intercentrosomal linker region, β-catenin also did not localize in between centrosomes (Fig. 6B, top panel, cf. control siRNA-treated cells in A). In the remaining 66% of C-Nap1 knockdown cells, Rootletin formed long continuous fibers, and β-catenin had a broad punctate distribution in the centrosome region (Fig. 6B, bottom panel). Similar results were found in C-Nap1 knockdown HeLa cells in which β-catenin puncta had striking colocalization along Rootletin fibers (Supplemental Fig. S6B,C). Thus, in cells depleted of C-Nap1, the distribution of β-catenin at centrosomes reflects the changes in Rootletin localization at centrosomes, providing further evidence that these proteins are in a complex at interphase centrosomes (see also Fig. 5; Supplemental Fig. S5).
Figure 6.
C-Nap1 and Rootletin regulate binding of β-catenin in between centrosomes. (A–C) U-2 OS cells immunostained for C-Nap1 (gray), Rootletin (green), and β-catenin (red). Merged images show C-Nap1 in blue. Control and siRNA images were taken at equal exposure times for each channel. (A) Control siRNA-treated cell with localization of β-catenin (red) to intercentrosomal linker region (arrow). (B, top panel) C-Nap1-siRNA-treated cell in which Rootletin (green) and β-catenin (red) are not localized to intercentrosomal linker region between centrosomes (arrows). (Bottom panel) C-Nap1 siRNA-treated cell with residual C-Nap1 at centrosomes (arrows). Rootletin (green) forms long continuous fibers and β-catenin (red) has broad punctate distribution. See Supplemental Figure S6 for additional examples of C-Nap1 siRNA-treated HeLa cells with striking colocalization of β-catenin puncta along Rootletin fibers, and quantitative fluorescence data showing loss of C-Nap1 from split centrosomes in C-Nap1 siRNA-treated cells. (C) Rootletin siRNA-treated cell in which β-catenin (red) is not localized to intercentrosomal linker region between centrosomes and is decreased at centrosomes (arrows). Bars, 5 μm. Graph shows mean fluorescence intensity per square micron for Rootletin (green) and β-catenin (red) in control (Con; n = 26 centrosomal puncta marked by C-Nap1) and Rootletin siRNA-treated (Root; n = 37 centrosomal puncta marked by C-Nap1) cells normalized for C-Nap1 (see Materials and Methods), which is unaffected by Root siRNA (Supplemental Fig. S6A; Bahe et al. 2005). Rootletin fluorescence intensity is significantly decreased at centrosomes in cells treated with Root siRNA as compared with control ([**] P < 0.0001), and the fluorescence intensity of β-catenin at centrosomes is significantly reduced in Root siRNA as compared with control siRNA-treated cells ([*] P = 0.0005). Mean ± standard error is shown.
To test directly whether Rootletin depletion affects localization of β-catenin in between centrosomes, we treated cells with Rootletin siRNA (Bahe et al. 2005). Eighty percent (n = 20) of Rootletin-depleted cells had increased distances between centrosomes, and the mean fluorescence intensity of C-Nap1 in split centrosomes was unchanged when compared with the averaged fluorescence intensity of C-Nap1 in unsplit centrosomes of control cells (Supplemental Fig. S6A), as expected (Bahe et al. 2005). We noticed that β-catenin had decreased localization at centrosomes and did not localize in between centrosomes in any of Rootletin-depleted cells with split centrosomes (Fig. 6C). We measured the mean fluorescence intensity per square micron of Rootletin and β-catenin at centrosomes and found that β-catenin was significantly decreased at centrosomes depleted for Rootletin as compared with control siRNA-treated cells (Fig. 6C, graph). Thus, Rootletin regulates the binding of β-catenin to centrosomes in interphase cells, likely through direct interactions with β-catenin (see Fig. 5D; Supplemental Fig. S5). C-Nap1 likely provides binding sites for both Rootletin and β-catenin that organize these proteins at the intercentrosomal linker region in interphase. Binding of β-catenin to Rootletin at the intercentrosomal linker in interphase cells may reserve a pool of β-catenin that is readily available for centrosome separation at the onset of mitosis (see Discussion). Note that although there was an overall decrease of β-catenin at centrosomes in the absence of Rootletin, some β-catenin remained on centrosomes in Rootletin siRNA cells, which could be associated with C-Nap1 or Rootletin-independent β-catenin-binding sites on centrosomes (see below).
Regulation of β-catenin localization to centrosomes by Nek2
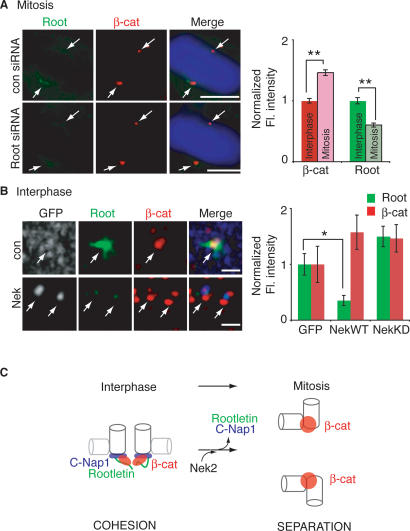
In contrast to C-Nap1 and Rootletin, endogenous β-catenin accumulates to centrosomes at mitotic spindle poles (Figs. 1E, 7A; Kaplan et al. 2004), indicating that β-catenin has binding sites on centrosomes in mitosis that are independent of C-Nap1 and Rootletin (Kaplan et al. 2004). In all cases, β-catenin was significantly increased at mitotic centrosomes in control cells lacking Rootletin at mitotic centrosomes (Fig. 7A, top panel and graph), and in mitotic cells treated with Rootletin siRNA (Fig. 7A, bottom panel). It is known that Nek2 activity increases at the onset of mitosis, which is required for centrosome separation (Fry et al. 1998a; Mayor et al. 2002; Bahe et al. 2005; Hayward and Fry 2006; Yang et al. 2006). We showed that β-catenin is a substrate for Nek2 (see Fig. 3) but, unlike C-Nap1 and Rootletin, β-catenin localizes to mitotic centrosomes and is required for centrosome separation. In order to test whether increased Nek2 activity allows β-catenin to bind to Rootletin-independent sites on centrosomes, we overexpressed wild-type GFP-Nek2 (GFP-Nek2WT) in HeLa cells. Because GFP-Nek2WT overexpression causes centrosome splitting, the fluorescence intensities of individual centrosomes in split centrosome pairs were compared with average fluorescence intensities of individual centrosomes in unsplit pairs from GFP and GFP-Nek2-kinase-dead (GFP-Nek2KD)-expressing cells (see Materials and Methods). As expected, GFP-Nek2WT overexpression caused centrosome splitting (Fry et al. 1998b) and a significant loss of Rootletin from split interphase centrosomes (n = 10) (Fig. 7B, bottom panel and graph; Bahe et al. 2005), when compared with enriched Rootletin at centrosomes in control GFP (n = 15) (Fig. 7B, top panel and graph) and GFP-Nek2KD-expressing cells (n = 9) (Fig. 7B, graph). Importantly, in contrast to Rootletin, there was an increase in localization of β-catenin on split centrosomes in the presence of active Nek2 (n = 8) (Fig. 7B, graph) as compared with GFP-expressing cells (n = 12), and β-catenin localized on rather than in between centrosomes (Fig. 7B, bottom panel). Rootletin-independent localization of β-catenin to centrosomes is dependent on Nek2 activity because in GFP-Nek2KD-expressing cells (Fig. 7B, graph) and in Rootletin siRNA-treated cells (see Fig. 6C), changes in β-catenin fluorescence intensity at centrosomes correlated with that of Rootletin. Thus, consistent with localization of β-catenin to Rootletin-independent sites on centrosomes at spindle poles in mitosis, when Nek2 activity is increased, β-catenin localizes on split interphase centrosomes even if Rootletin is strongly reduced. Furthermore, consistent with the interaction of Rootletin and β-catenin in between centrosomes, when Rootletin is dissociated from centrosomes in the presence of Nek2, β-catenin localizes on rather than in between centrosomes. Taken together, these results suggest a model in which Nek2 activity inhibits the interaction of Rootletin with centrosomes, thereby releasing a pool of β-catenin that can bind to Rootletin-independent sites on centrosomes, an event that is required for centrosome separation in mitosis (Fig. 7C).
Figure 7.
Regulation of β-catenin localization to centrosomes by Nek2. (A) Mitotic U-2 OS cells treated with control siRNA (top panel) and Rootletin siRNA (bottom panel). Arrows point to β-catenin (red) localization to centrosomes at spindle poles in the absence of Rootletin (green). DAPI (blue) marks chromosomes at metaphase plate. Bars, 10 μm. Graph shows normalized mean fluorescence intensity of β-catenin and Rootletin at interphase and mitotic centrosomes marked by γ-tubulin. A significant increase in β-catenin intensity at mitotic centrosomes (n = 21; [**] P < 0.0001) correlates with a significant decrease in Rootletin intensity at mitotic centrosomes (n = 21; [**] P < 0.0001). (B) Deconvolved section of GFP-expressing (top panel) and GFP-Nek2WT-expressing (bottom panel) HeLa cells (gray in single; blue in merge) immunostained for Rootletin (green) and β-catenin (red). Arrow in the top panel points to localization of β-catenin in between centrosomes. Arrows in the bottom panel point to β-catenin localization on, and not in between, centrosomes in the presence of GFP-Nek2WT. Bar, 2 μm. Graph shows normalized fluorescence intensity measurements of β-catenin and Rootletin in control, Nek2WT-expressing, and Nek2KD-expressing cells. Rootletin is significantly decreased from split centrosomes in Nek2WT-expressing cells as compared with control cells ([*] P = 0.016), whereas β-catenin is slightly increased. (C) Model for localization and regulation of β-catenin during centrosome cohesion and separation. In interphase, C-Nap1 provides sites that anchor and organize Rootletin and β-catenin to the intercentrosomal linker region. In mitosis, Nek2 activity results in the loss of Rootletin and C-Nap1 from centrosomes and binding of β-catenin to Rootletin-independent sites on centrosomes, an event that is required for centrosome separation.
Discussion
β-Catenin is a well-studied protein with known functions in the nucleus in a complex with the transcriptional coactivator T-cell factor/lymphocyte enhancer factor-1 (TCF/Lef-1), and at the plasma membrane in a complex with the cell–cell adhesion protein E-cadherin (Nelson and Nusse 2004). We show that β-catenin localizes and functions at a new subcellular location, the centrosome. Our results provide seven independent experiments in support of this claim: (1) β-Catenin was detected at centrosomes with two different antibodies in four different cell lines at both the resolution of light and electron microscopy, and this distribution was shown to be as statistically significant as that of several well-characterized centrosome proteins. (2) Endogenous β-catenin copurified with centrosomes, and was coimmunoprecipitated from centrosomes with bone fide centrosomal proteins. (3) Exogenous GFP-β-catenin localized to centrosomes, and centrosome-binding site(s) in β-catenin are located in the armadillo domain. (4) β-Catenin was identified as a binding partner and substrate for the centrosome-localized Nek2 kinase in vivo and in vitro. (5) The distribution of β-catenin at centrosomes overlapped that of C-Nap1 and Rootletin, two known components of the intercentrosome linker, and β-catenin is in a complex with Rootletin. (6) β-Catenin localized with Rootletin in between centrosomes in interphase, and to Rootletin-independent sites on centrosomes in mitosis that were regulated by Nek2. (7) Consistent with a role for β-catenin in centrosome cohesion, stabilized β-catenin decreased centrosome cohesion and increased intercentrosome distance in interphase cells, while loss of β-catenin increased centrosome cohesion in mitosis. These results, therefore, not only identify β-catenin as a centrosomal protein, but also define a role for β-catenin in regulating centrosome cohesion by connecting it to a specific subset of known centrosomal proteins that form the intercentrosome linker.
Our data show that β-catenin localizes in the intercentrosomal linker region of interphase centrosomes, and accumulates on mitotic centrosomes at spindle poles (Fig. 7C; see also Kaplan et al. 2004). In interphase cells, we show by immunoprecipitation that β-catenin is in a complex with Rootletin and, by overexpression and depletion experiments, we show that this complex forms in between centrosomes. Depletion of C-Nap1 caused loss of Rootletin and β-catenin from the intercentrosomal linker region and partial depletion of C-Nap1 caused disorganization of Rootletin and β-catenin at centrosomes (Fig. 6B; Supplemental Fig. S6; Bahe et al. 2005). It has been suggested that partially reducing C-Nap1 decreases the number of binding sites available for Rootletin at centrosomes, causing increased binding of Rootletin to itself and possibly other interactors (Bahe et al. 2005), such as β-catenin. Thus, C-Nap1 likely provides sites to organize Rootletin and β-catenin in between centrosomes. The parallel effects on Rootletin and β-catenin when C-Nap1 is reduced likely reflect that Rootletin and β-catenin are in a complex and may bind directly to each other. In the absence of Rootletin at interphase centrosomes, β-catenin is reduced at centrosomes and does not localize in between centrosomes, whereas C-Nap1 levels at centrosomes are not significantly affected. Together, these results support a model in which C-Nap1 recruits Rootletin to centrosomes, which in turn provides sites for β-catenin at the intercentrosomal linker region in between centrosomes in interphase (Fig. 7C).
By depletion and stabilization studies, we demonstrate that β-catenin acts as a negative regulator of centrosome cohesion, similar to the function of Nek2 (Fry et al. 1998b; Faragher and Fry 2003). This is consistent with the observation that β-catenin remains on centrosomes upon centrosome separation and increased Nek2 activity, and depletion of β-catenin results in unseparated centrosomes in mitosis. Conversely, C-Nap1 and Rootletin are positive regulators of centrosome cohesion; they dissociate from centrosomes upon centrosome separation and their depletion results in increased distance between centrosomes in interphase (Fry et al. 1998a; Bahe et al. 2005; Yang et al. 2006). It has been suggested that C-Nap1 and Rootletin are structural components of the intercentrosome linker required for centrosome cohesion, and we suggest that β-catenin has a regulatory role promoting centrosome separation in mitosis (see below). It is known that the Armadillo repeat domain of β-catenin, which we show is necessary and sufficient for localization of β-catenin to centrosomes and is a substrate for Nek2, mediates the mutually exclusive binding of β-catenin to its diverse partners (Hulsken et al. 1994; Tao et al. 1996; von Kries et al. 2000; Huber and Weis 2001). We propose that, similar to how the binding of β-catenin to E-cadherin prevents β-catenin from associating with its other binding partners (Hulsken et al. 1994), the binding of β-catenin to Rootletin in between centrosomes in interphase cells may prevent β-catenin from binding other sites on centrosomes. In this manner, Rootletin complexed with β-catenin in interphase cells may reserve a pool of β-catenin that can be readily available for binding to other sites on centrosomes in mitosis that are required for centrosome separation and regulated by Nek2 (Fig. 7C). In support of this model, our data show that centrosomal loss of Rootletin in interphase cells results in decreased binding of β-catenin at centrosomes; however, β-catenin accumulates on mitotic centrosomes independent of Rootletin. Furthermore, we demonstrate that increased Nek2 activity, which uniquely occurs at the onset of mitosis, disrupts the association of Rootletin with centrosomes, and causes β-catenin to localize on centrosomes independent of Rootletin.
Since Rootletin and β-catenin interact and colocalize in between centrosomes in interphase cells (Figs. 5, 6), we hypothesize that Nek2 activity in mitosis may disrupt the interaction between Rootletin and β-catenin and/or increase the interaction of β-catenin with Rootletin-independent sites on centrosomes (Fig. 7C). In vitro incubation of the coimmunoprecipitated Rootletin–β-catenin complex with Nek2 resulted in loss of phosphorylated β-catenin from the complex (Supplemental Fig. S7C); however, further studies are required to dissect the regulation of the Rootletin–β-catenin complex by Nek2. Thus, at the onset of mitosis when Nek2 activity is increased, a pool of β-catenin is released upon Rootletin dissociation from centrosomes, which in turn could bind to Rootletin-independent sites on centrosomes that promote centrosome separation (Fig. 7C).
Although β-catenin does not have enzymatic activity that could itself promote separation, β-catenin is well known for its role as an adaptor protein that organizes functional protein complexes such as its interaction with E-cadherin and α-catenin in cell–cell adhesion, and its binding to TCF/Lef-1 and components of the transcriptional machinery in the nucleus (Huber et al. 1997, 2001; Graham et al. 2000; Huber and Weis 2001). At the onset of mitosis when Nek2 is active, binding of β-catenin to Rootletin-independent sites on centrosomes may promote the scaffolding of a complex required for centrosome separation. Similarly, stabilization of β-cat* at interphase centrosomes may prematurely bind and induce scaffolding of this complex, due to limited Rootletin-binding sites for β-catenin in between centrosomes, and in turn cause premature centrosome splitting. An analogous function for β-catenin occurs in the nucleus: In the absence of β-catenin, TCF/Lef-1 transcription factors are bound to promoter sequences but remain transcriptionally inactive, but upon binding to β-catenin an active TCF/Lef complex forms and initiates transcription (Hurlstone and Clevers 2002).
The requirement of a positive activity involving Nek2/β-catenin that induces centrosome separation in mitosis, either independent of or in addition to the loss of linker proteins, is supported by the finding that in cells expressing kinase-dead Nek2, C-Nap1 dissociates from centrosomes in mitosis, but the centrosomes remain unseparated (Faragher and Fry 2003). Moreover, C-Nap1 and Rootletin dissociate from monopolar spindles containing unseparated centrosomes caused by depletion of β-catenin (Supplemental Fig. S7A,B). Therefore, dissociation of C-Nap1 and Rootletin from centrosomes at spindle poles in the absence of active Nek2 or β-catenin is not sufficient to cause centrosome separation in mitosis. Instead, increased Nek2 activity may be required to alter the interaction between β-catenin and Rootletin (Supplemental Fig. S7C), releasing a pool of β-catenin that is able to bind Rootletin-independent sites on centrosomes, which in turn promotes centrosome separation (Fig. 7C). Consistent with a role for β-catenin as a positive regulator of centrosome separation, stabilized β-cat* prematurely induces centrosome splitting, which is likely due to increased binding of β-cat* to Rootletin-independent sites on centrosomes. One potential binding partner for β-catenin on centrosomes is the microtubule-binding protein APC, which localizes to centrosomes (Louie et al. 2004), binds stably to β-cat* (Barth et al. 1997, 1999), and is involved in centrosome positioning in Drosophila (Yamashita et al. 2003). We propose that the phosphorylation state of β-catenin regulates the pool of β-catenin available for activating centrosome separation: Centrosomes do not separate when either β-catenin (Supplemental Figs. S4, S7A,B; Kaplan et al. 2004) or Nek2 kinase activity (Faragher and Fry 2003) is absent, whereas centrosome cohesion is decreased when either β-catenin is stabilized (Fig. 4) or Nek2 kinase activity is higher (Fry et al. 1998b).
Mutations that inhibit the regulation of β-catenin degradation occur in many cancers and are the earliest event in colon cancer (Polakis 1999; Bienz and Clevers 2000; Peifer and Polakis 2000; Moon et al. 2002; Behrens 2005), but it is not completely understood how this contributes to tumor progression. Increased centrosome number and abnormal spindles occur in precancerous lesions and aggressive, low-grade tumors and correlate with chromosomal instability (D’Assoro et al. 2002; Pihan et al. 2003), which is common in colon cancers with stabilized β-catenin (Hadjihannas and Behrens 2006; Hadjihannas et al. 2006). Interestingly, APC, GSK3β, and components of the SCF ubiquitin ligase that regulate β-catenin stability are centrosome components (Freed et al. 1999; Wakefield et al. 2003; Louie et al. 2004; Bobinnec et al. 2006), and mutations in some of these components induce supernumerary centrosomes and multipolar spindles (Wojcik et al. 2000; Fodde et al. 2001). Furthermore, cells overexpressing β-catenin have increased numbers of γ-tubulin puncta in the centrosome region (Ligon et al. 2001; S. Bahmanyar, unpubl.), and impaired microtubule anchoring at centrosomes (Huang et al. 2007). Although a direct link between abnormal centrosome cohesion and centrosome number and function is not yet understood, altering Nek2 activity also induces extra centrosomal structures (Faragher and Fry 2003) and Nek2 protein levels are increased in many types of human cancer-derived cell lines (Hayward et al. 2004). In addition to directly contributing to centrosome amplification, stabilized β-catenin or altered Nek2 activity may inhibit clustering of supernumerary centrosomes in mitosis, an intrinsic cellular protection mechanism against centrosome amplification, and may thereby promote spindle multipolarity. It will be necessary to determine whether changes in stability of β-catenin could induce abnormal centrosome organization and function, which could synergize with other known effects of mutant β-catenin on gene transcription and cell–cell adhesion to induce cell transformation.
Materials and methods
Cell lines
MDCK type II/G cell lines, HeLa, and HEK 293T cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS). U-2 OS cells were grown in McCoy’s 5A media (Sigma) supplemented with 10% FBS. The parental MDCK cell line T23 and dox-repressible stabilized β-catenin MDCK cells have been described (Barth et al. 1999). To repress β-cat* expression, cells were thawed into culture medium containing 20 ng/mL dox (Sigma). For transient transfection of cDNAs, Lipofectamine 2000 reagent was used as described by the manufacturer (Invitrogen). Flow cytometry on a Becton Dickinson FACScan has been described (Bauer et al. 1998). Gating and Dean-Jett-Fox algorithms were used in FlowJo software (Tree Star, Inc.). For double-thymidine block experiments, MDCK cell lines were plated onto coverslips, and the next day thymidine (Sigma) was added to a final concentration of 2 mM. Cells were incubated with thymidine for 16–18 h, thymidine was washed out three times, and cells were incubated without thymidine for 5 h; cells were then treated with 2 mM thymidine and incubated for another 16–18 h.
DNA constructs
Expression vectors for β-cat* have been described previously (Barth et al. 1999). GFP-tagged ΔARM (Elul et al. 2003) was a gift from T. Elul (Touro University), GFP-tagged C-Nap1 (Mayor et al. 2002) was a gift from E. Nigg (Max Planck Institute), and HA-WT, NB, and KD-Nek2 (Eto et al. 2002) constructs were a gift from D. Brautigan (University of Virginia). For transient transfections of GFP-tagged wild-type Nek2, BamH1/Xho1 fragments were removed from pRKHA3-WTNek2 and cloned into BglII/SalI-cut pEGFP-C1. For transient transfection experiments of GFP-tagged β-cat, β-cat*, and ARM, SacII/BamHI fragments were removed from pUDH10-3 (Barth et al. 1997) and cloned into pEGFP-C1 (Clontech). RFP-pericentrin was a gift from Sean Munro (MRC, Cambridge, England).
β-Catenin, C-Nap1, and Rootletin siRNA
For β-catenin siRNA, HeLa cells were transfected with β-catenin or GL2 luciferase control siRNA duplexes (Thompson et al. 2002; Kaplan et al. 2004) and, ∼50 h post-transfection, coverslips were mounted in Rose chambers and imaged as described previously (DeLuca et al. 2002). For C-Nap1 (oligo 239, 5′-CTGGAAGAGCGTCTAACTGATdtdt-3′) (Bahe et al. 2005) and Rootletin (oligo 222, 5′-AAGCCAGTCTAGACAAGGA dtdt-3′) (Bahe et al. 2005) (Dharmacon) siRNA, U-2 OS cells were transfected for 48 h prior to fixation as described previously (Bahe et al. 2005).
Fluorescence intensity measurements
ImageJ (http://rsb.info.nih.gov/ij) was used to circle the centrosome area and an area near the centrosome (background), and to measure the mean fluorescence intensity. Unsplit centrosomes were measured together, whereas split centrosomes were measured separately. Exposure times were equal between different samples. Background intensities were subtracted from each measurement in each channel. In Figure 7B and Supplemental Figure S6, average intensities of individual centrosomes within unsplit centrosome pairs were used for comparison with intensities from individual centrosomes within split pairs. To do this, intensities of unsplit pairs, which were measured together, were divided by 2 to get the average intensity at each centrosome. In Figure 6C, Rootletin and β-catenin intensities were divided by the fluorescence intensity of C-Nap1 in the same cell, to normalize for variations due to centrosome splitting, focal plane, centrosome angle, and size. In Figure 7A, measurements were taken of centrosomal β-catenin and Rootletin and corrected for γ-tubulin in cells triple-stained for β-catenin, Rootletin, and γ-tubulin.
Fixation and antibodies
Cells were extracted with 0.5% Triton X-100 in 80 mM PIPES (pH 6.8), 1 mM MgCl2, and 1 mM EGTA for 30 sec and fixed in −20°C methanol for 5 min. Extraction prior to fixation more clearly shows the centrosomal localization of β-catenin using the polyclonal β-catenin antibody (Nathke et al. 1994) or the monoclonal β-catenin antibody (BD/Transduction Labs), although centrosomal localization with the polyclonal β-catenin antibody (Nathke et al. 1994) can be detected without pre-extraction provided that optical section and deconvolution microscopy are used. The following antibodies were used: rabbit polyclonal β-catenin (Nathke et al. 1994), mouse monoclonal β-catenin (BD/Transduction Labs), C-Nap1 and Nek2 mouse monoclonal (BD/Transduction Laboratories), γ-tubulin clone GTU88 (Sigma), and rabbit polyclonal pericentrin (Covance/BabCO). Chicken and rabbit Rootletin (Yang et al. 2006) were provided by Tiansen Li (Harvard Medical School); rabbit polyclonal centrin was provided by T. Mitchison (Harvard Medical School) and mouse monoclonal centrin (Sanders and Salisbury 1994) was provided by J. Salisbury (Mayo Clinic Foundation); mouse monoclonal glutamylated tubulin (Wolff et al. 1992) was a gift from P. Denoulet (University Pierre and Marie Curie). Secondary antibodies against mouse, rat, rabbit, or chicken IgG with minimal species cross-reactivity coupled to FITC, rhodamine, or Cy5 were used (Jackson ImmunoResearch). Cells were analyzed by epifluorescence, deconvolution microscopy, and three-dimensional rendering methods as described elsewhere (Louie et al. 2004).
Electron microscopy
Retina primary epithelial (RPE) cells were grown to near-confluency on tissue culture plates, trypsinized for 1–2 min at room temperature, centrifuged at 400g for 2 min, rinsed in PBS, resuspended in 150 mM mannitol in PBS, and centrifuged into a soft pellet. Aliquots of the cell pellet were cryofixed in a BAL-TEC HPM-010 high-pressure freezer, stored under liquid nitrogen, then freeze-substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone for 4 d at 80°C. After gradual warming to −20°C, the preparations were infiltrated with Lowicryl HM20 at −20°C, and loaded into BEEM capsules for polymerization under ultraviolet light at 45°C. Ribbons of ∼20 serial sections, each 60 nm thick, were picked up on Formvar-coated nickel slot grids. For immunolabeling, the grids were exposed to a blocking solution consisting of 1% nonfat dry milk in PBS containing 0.1% Tween 20 (PBST), labeled for 2 h with a 1:3 or a 1:20 dilution of the purified polyclonal C-terminal β-catenin antibody (Nathke et al. 1994) in the PBST/milk blocking solution, rinsed with PBST and incubated in goat–anti-rabbit 15-nm gold conjugate (Ted Pella) diluted 1:20 in PBST/milk. The grids were then rinsed sequentially in PBST and distilled water, dried, and finally stained with uranyl acetate and lead citrate. The specimens were examined in a Philips CM10 (FEI Co.) or an FEI Technai F20 operating at 80 kV. Images were captured using a Gatan CCD camera at a 0.76-nm or 2.3-nm pixel.
Pearson’s correlation coefficient
rp = ∑(Ri − Raver)*(Gi − Gaver)/∑(Ri − Raver)2*(Gi − Gaver)2 where Ri = voxel intensity in red channel and Gi = voxel intensity in the green channel (Manders et al. 1993). ImageJ (http://rsb.info.nih.gov/ij) was used to calculate Pearson’s correlation coefficient (rp) with or without randomization.
Live-cell imaging
β-Catenin turnover at centrosomes was measured with the FRAP module of the Marianas system (Yamada et al. 2005) (Intelligent Imaging, Inc). Equivalent laser intensity, repetition, and region size were used for FRAP on wild-type and stabilized β-cat* samples. Fluorescence intensity at the centrosome was analyzed with ImageJ using a 20 × 20-pixel square. Because the contribution of cytoplasmic diffusion was small, much faster, and had lower intensity compared with recovery at centrosomes, the recovery curve was dominated by reaccumulation of proteins at centrosomes.
Centrosome isolation, immunoprecipitation of centrosomal β-catenin, and identification of binding partners
Centrosome-enriched fractions were prepared as described previously from HeLa cells, or a HeLa cell line stably expressing Myc-tagged β-catenin (Kaplan et al. 2004). Centrosome-containing sucrose gradient fractions were diluted in buffer containing 20 mM Tris (pH 7.7), 120 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease inhibitor cocktail, then concentrated using a Centriprep-30 (Millipore) filter unit. β-Catenin-Myc or endogenous β-catenin was immunoprecipitated from the concentrated centrosome fractions using anti-c-Myc agarose (Sigma) or anti-β-catenin antibody (BD/Transduction Labs) coupled to protein A/G sepharose 4B (GE Healthcare), respectively. Immunoprecipitates were washed extensively, followed by heating of precipitated material in Laemmli sample buffer. Precipitated proteins were separated using SDS-PAGE and analyzed by MALDI TOF/TOF mass spectrometry (to identify centrosomal β-catenin-interacting proteins) or immunoblotting (to confirm interactions identified by mass spectrometry).
Coimmunoprecipitation of β-catenin, Nek2, and Rootletin
HEK 293T cells (transfected with GFP-Nek2 in the case of β-catenin/Nek2 coimmunoprecipitations) were scraped in cold lysis/IP buffer (20 mM Tris at pH 7.5, 120 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5% NP-40, plus Complete EDTA-free protease inhibitor cocktail [Roche]) and phosphatase inhibitor cocktails 1 and 2 (Sigma) and were lysed using a dounce homogenizer. Nuclei and unbroken cells were pelleted, the supernatant was precleared by incubation with protein A/G sepharose 4B (GE Healthcare), and the precleared supernatant was incubated with control antibody (anti-GST; Santa Cruz Biotechnology) or anti-β-catenin antibody (Nathke et al. 1994) followed by addition of protein A/G sepharose 4B. Beads were washed four times using lysis/IP buffer with 500 mM NaCl, followed by heating of precipitated material in Laemmli sample buffer. Precipitated proteins were separated using SDS-PAGE and analyzed by immunoblotting. For Rootletin immunoprecipitation, previously described protocols were followed (Yang et al. 2006).
Kinase assays
For kinase assays using HA-Nek2 or HA-CKIε produced in HEK 293T cells, the cells were transfected with wild-type, NB, of KD forms of HA-Nek2 or wild-type HA-CKIε and scraped in lysis buffer (50 mM HEPES at pH 7.5, 100 mM NaCl, 10 mM MgCl2, 5 mM MnCl2, 5 mM KCl, 2 mM EDTA, 5 mM EGTA, 1 mM DTT, 0.1% NP-40, protease inhibitor cocktail) 2 d later, and kinases were immunoprecipitated using monoclonal anti-HA antibody (clone 12CA5, Roche) coupled to protein A/G sepharose beads (GE Healthcare). Immunoprecipitated kinases were washed twice in lysis buffer with 500 mM NaCl, once in lysis buffer, and once in kinase buffer without ATP or substrate (50 mM Tris at pH 7.7, 10 mM MgCl2, 1 mM DTT, protease inhibitor cocktail). For kinase assays, washed kinases were incubated with 300 nM recombinant β-catenin (Kaplan et al. 2001), 150 nCi/nmol γ32PATP with a total ATP concentration of 100 nM, and 50 nM microcystin in kinase buffer. Kinase reactions were incubated for 15 min at 30°C, then heated in Laemmli sample buffer, followed by SDS-PAGE and autoradiography. Kinase assays utilizing recombinant His-Nek2 were performed as described above, but using 200 ng of recombinant His-Nek2 (BPS Bioscience) per reaction as the source of kinase and 300 nM recombinant GST-β-catenin (Yamada et al. 2005) or GST-β-catenin ARM domain (Huber et al. 1997) as substrate. Reactions were incubated for 0, 10, or 60 min at 30°C. For addition of Nek2 kinase to immunoprecipitated complexes, 0.2 μg of purified Nek2 was added to immunoprecipitates and incubated with up to 50 of μL kinase buffer rotating for 30 min at 30°C.
In vitro binding assays
Equimolar amounts of recombinant GST (0.6 μg per reaction) or GST-β-catenin (2.5 μg per reaction) (Yamada et al. 2005) were incubated with recombinant His-Nek2 (1.0 μg per reaction; BPS Bioscience) in binding buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1% NP-40) with a total reaction volume of 200 μL on a rocking platform for 2 h at 4°C. Fifty microliters of glutathione sepharose 4B bead slurry (GE Healthcare) were then added to each reaction, followed by rocking for an additional 1 h at 4°C. Beads were washed four times using wash buffer (50 mM Tris at pH 7.4, 500 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1% NP-40) followed by heating in Laemmli sample buffer. Bound and unbound proteins were separated using SDS-PAGE and analyzed by immunoblotting.
Electrophoretic mobility assay
HEK 293T cells were transfected and scraped 2 d later in lysis buffer (20 mM Tris at pH 7.7, 120 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5% NP-40, Complete protease inhibitor cocktail [Roche]) with (for nonphosphatase-treated samples) or without (for λ-phosphatase-treated samples) phosphatase inhibitor cocktails 1 and 2 (Sigma). For λ-phosphatase treatment, lysates were incubated in phosphatase buffer with MnCl2 plus 1.5 μL of λ-Phosphatase (400 U) for 30 min at 30°C. Samples were heated in Laemmli sample buffer, followed by SDS-PAGE using 3%–8% TAC gels (Bio-Rad) and immunoblotting for endogenous β-catenin.
Acknowledgments
We thank J. Stamos and W. Weis (Stanford University) for purified β-catenin proteins. We are grateful to the Duke University Proteomics Center for performing mass spectrometry, C. Weinbaum for technical assistance, W. Anderson for FACS analysis, and S. Yamada, J. Benjamin, and J.M. Gendron for helpful discussion. This work was supported by grants from the NIH to W.J.N. (GM078270), M.W. (GM074746), and P.J.C. (CA100869); a Howard Hughes Medical Institute Predoctoral Fellowship and American Cancer Society Post-doctoral Fellowship to D.D.K. (PF-06-026-01-DDC); and a grant (T32GM07276-27) from the Cell and Molecular Biology Training Program and a Predoctoral Fellowship from the Department of Defense Breast Cancer Research Program to S.B. (BC050431).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1596308
References
- Bahe S., Stierhof Y.D., Wilkinson C.J., Leiss F., Nigg E.A. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A.I., Pollack A.L., Altschuler Y., Mostov K.E., Nelson W.J. NH2-terminal deletion of β-catenin results in stable colocalization of mutant β-catenin with adenomatous polyposis coli protein and altered MDCK cell adhesion. J. Cell Biol. 1997;136:693–706. doi: 10.1083/jcb.136.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A.I., Stewart D.B., Nelson W.J. T cell factor-activated transcription is not sufficient to induce anchorage-independent growth of epithelial cells expressing mutant β-catenin. Proc. Natl. Acad. Sci. 1999;96:4947–4952. doi: 10.1073/pnas.96.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A., Lickert H., Kemler R., Stappert J. Modification of the E-cadherin–catenin complex in mitotic Madin-Darby canine kidney epithelial cells. J. Biol. Chem. 1998;273:28314–28321. doi: 10.1074/jbc.273.43.28314. [DOI] [PubMed] [Google Scholar]
- Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem. Soc. Trans. 2005;33:672–675. doi: 10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- Bienz M., Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Bobinnec Y., Morin X., Debec A. Shaggy/GSK-3β kinase localizes to the centrosome and to specialized cytoskeletal structures in Drosophila. Cell Motil. Cytoskeleton. 2006;63:313–320. doi: 10.1002/cm.20128. [DOI] [PubMed] [Google Scholar]
- D’Assoro A.B., Lingle W.L., Salisbury J.L. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Moree B., Hickey J.M., Kilmartin J.V., Salmon E.D. hNuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierick H., Bejsovec A. Cellular mechanisms of wingless/Wnt signal transduction. Curr. Top. Dev. Biol. 1999;43:153–190. doi: 10.1016/s0070-2153(08)60381-6. [DOI] [PubMed] [Google Scholar]
- Doxsey S.J., Stein P., Evans L., Calarco P.D., Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Eastman Q., Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- Elul T.M., Kimes N.E., Kohwi M., Reichardt L.F. N- and C-terminal domains of β-catenin, respectively, are required to initiate and shape axon arbors of retinal ganglion cells in vivo. J. Neurosci. 2003;23:6567–6575. doi: 10.1523/JNEUROSCI.23-16-06567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto M., Elliott E., Prickett T.D., Brautigan D.L. Inhibitor-2 regulates protein phosphatase-1 complexed with NimA-related kinase to induce centrosome separation. J. Biol. Chem. 2002;277:44013–44020. doi: 10.1074/jbc.M208035200. [DOI] [PubMed] [Google Scholar]
- Faragher A.J., Fry A.M. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde R., Kuipers J., Rosenberg C., Smits R., Kielman M., Gaspar C., van Es J.H., Breukel C., Wiegant J., Giles R.H., et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- Freed E., Lacey K.R., Huie P., Lyapina S.A., Deshaies R.J., Stearns T., Jackson P.K. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes & Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., Nigg E.A. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998a;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Meraldi P., Nigg E.A. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998b;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z.H., Seeling J.M., Hill V., Yochum A., Virshup D.M. Casein kinase I phosphorylates and destabilizes the β-catenin degradation complex. Proc. Natl. Acad. Sci. 2002;99:1182–1187. doi: 10.1073/pnas.032468199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T.A., Weaver C., Mao F., Kimelman D., Xu W. Crystal structure of a β-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- Hadjihannas M.V., Behrens J. CIN by WNT: Growth pathways, mitotic control and chromosomal instability in cancer. Cell Cycle. 2006;5:2077–2081. doi: 10.4161/cc.5.18.3282. [DOI] [PubMed] [Google Scholar]
- Hadjihannas M.V., Bruckner M., Jerchow B., Birchmeier W., Dietmaier W., Behrens J. Aberrant Wnt/β-catenin signaling can induce chromosomal instability in colon cancer. Proc. Natl. Acad. Sci. 2006;103:10747–10752. doi: 10.1073/pnas.0604206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R.S., Crookes R.E., Straatman K.R., Merdes A., Hayes M.J., Faragher A.J., Fry A.M. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell. 2005;16:1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D.G., Fry A.M. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Hayward D.G., Clarke R.B., Faragher A.J., Pillai M.R., Hagan I.M., Fry A.M. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004;64:7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Sluder G. ‘It takes two to tango’: Understanding how centrosome duplication is regulated throughout the cell cycle. Genes & Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Huang P., Senga T., Hamaguchi M. A novel role of phospho-β-catenin in microtubule regrowth at centrosome. Oncogene. 2007;26:4357–4371. doi: 10.1038/sj.onc.1210217. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Weis W.I. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Nelson W.J., Weis W.I. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Stewart D.B., Laurents D.V., Nelson W.J., Weis W.I. The cadherin cytoplasmic domain is unstructured in the absence of β-catenin. A possible mechanism for regulating cadherin turnover. J. Biol. Chem. 2001;276:12301–12309. doi: 10.1074/jbc.M010377200. [DOI] [PubMed] [Google Scholar]
- Hulsken J., Birchmeier W., Behrens J. E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J. Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlstone A., Clevers H. T-cell factors: Turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.D., Meigs T.E., Casey P.J. Distinct regions of the cadherin cytoplasmic domain are essential for functional interaction with Gα 12 and β-catenin. J. Biol. Chem. 2001;276:44037–44043. doi: 10.1074/jbc.M106121200. [DOI] [PubMed] [Google Scholar]
- Kaplan D.D., Meigs T.E., Kelly P., Casey P.J. Identification of a role for β-catenin in the establishment of a bipolar mitotic spindle. J. Biol. Chem. 2004;279:10829–10832. doi: 10.1074/jbc.C400035200. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon L.A., Karki S., Tokito M., Holzbaur E.L. Dynein binds to β-catenin and may tether microtubules at adherens junctions. Nat. Cell Biol. 2001;3:913–917. doi: 10.1038/ncb1001-913. [DOI] [PubMed] [Google Scholar]
- Louie R.K., Bahmanyar S., Siemers K.A., Votin V., Chang P., Stearns T., Nelson W.J., Barth A.I. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J. Cell Sci. 2004;117:1117–1128. doi: 10.1242/jcs.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M.M., Verbeek F.J., Aten J.A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993;169:8. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Mayor T., Stierhof Y.D., Tanaka K., Fry A.M., Nigg E.A. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T., Hacker U., Stierhof Y.D., Nigg E.A. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J. Cell Sci. 2002;115:3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Nigg E.A. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J. Cell Sci. 2001;114:3749–3757. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- Mitchison T.J., Kirschner M.W. Isolation of mammalian centrosomes. Methods Enzymol. 1986;134:261–268. doi: 10.1016/0076-6879(86)34094-1. [DOI] [PubMed] [Google Scholar]
- Moon R.T., Bowerman B., Boutros M., Perrimon N. The promise and perils of Wnt signaling through β-catenin. Science. 2002;296:1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- Nathke I.S., Hinck L., Swedlow J.R., Papkoff J., Nelson W.J. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J. Cell Biol. 1994;125:1341–1352. doi: 10.1083/jcb.125.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W.J., Nusse R. Convergence of Wnt, β-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Polakis P. Wnt signaling in oncogenesis and embryogenesis—A look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- Polakis P. The oncogenic activation of β-catenin. Curr. Opin. Genet. Dev. 1999;9:15–21. doi: 10.1016/s0959-437x(99)80003-3. [DOI] [PubMed] [Google Scholar]
- Sanders M.A., Salisbury J.L. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y.S., Edwards R.A., Tubb B., Wang S., Bryan J., McCrea P.D. β-Catenin associates with the actin-bundling protein fascin in a noncadherin complex. J. Cell Biol. 1996;134:1271–1281. doi: 10.1083/jcb.134.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B., Townsley F., Rosin-Arbesfeld R., Musisi H., Bienz M. A new nuclear component of the Wnt signalling pathway. Nat. Cell Biol. 2002;4:367–373. doi: 10.1038/ncb786. [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Stearns T. Controlling centrosome number: Licenses and blocks. Curr. Opin. Cell Biol. 2006;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- von Kries J.P., Winbeck G., Asbrand C., Schwarz-Romond T., Sochnikova N., Dell’Oro A., Behrens J., Birchmeier W. Hot spots in β-catenin for interactions with LEF-1, conductin and APC. Nat. Struct. Biol. 2000;7:800–807. doi: 10.1038/79039. [DOI] [PubMed] [Google Scholar]
- Wakefield J.G., Stephens D.J., Tavare J.M. A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment. J. Cell Sci. 2003;116:637–646. doi: 10.1242/jcs.00273. [DOI] [PubMed] [Google Scholar]
- Wojcik E.J., Glover D.M., Hays T.S. The SCF ubiquitin ligase protein slimb regulates centrosome duplication in Drosophila. Curr. Biol. 2000;10:1131–1134. doi: 10.1016/s0960-9822(00)00703-x. [DOI] [PubMed] [Google Scholar]
- Wolff A., de Nechaud B., Chillet D., Mazarguil H., Desbruyeres E., Audebert S., Edde B., Gros F., Denoulet P. Distribution of glutamylated α and β-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 1992;59:425–432. [PubMed] [Google Scholar]
- Yamada S., Pokutta S., Drees F., Weis W.I., Nelson W.J. Deconstructing the cadherin–catenin–actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Jones D.L., Fuller M.T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Yang J., Adamian M., Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol. Biol. Cell. 2006;17:1033–1040. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]