Abstract
Riboswitches are genetic control elements that regulate gene expression in a small molecule-dependent way. We developed a two-stage strategy of in vitro selection followed by a genetic screen and identified several artificial small molecule-binding riboswitches that respond to the aminoglycoside neomycin. Structure–function relationships and structural probing revealed that they adopt the general neomycin-binding motif. They display no sequence similarities to in vitro selected neomycin aptamers but contain parts of the decoding site that is the binding site for neomycin on the ribosomal RNA. We propose a model of a composed binding pocket of an internal loop as primary docking site and a terminal flaplike loop structure fixing neomycin in a sandwich-like manner. Such binding pockets characterized by multiple contacts between ligand and RNA are described for both natural and engineered riboswitches. We anticipate that combination of in vitro selection and in vivo screening is a useful strategy to identify RNA molecules with a desired functionality.
Keywords: aptamer, riboswitch, neomycin, conditional gene expression, screening
INTRODUCTION
Riboswitches are RNA-based genetic control elements that regulate a wide set of basic metabolic pathways in prokaryotes (Winkler and Breaker 2003, 2005; Mandal and Breaker 2004; Nudler and Mironov 2004; Tucker and Breaker 2005). These molecular switches consist solely of RNA, sense their ligand in a mostly preformed binding pocket, and undergo restructuring upon metabolite binding, leading to changes in gene expression. Thereby the RNA accomplishes both sensor and regulator functions and integrates the tasks formerly carried out by protein and RNA components together.
In the last few years several engineered riboswitches have been developed that are based on either antisense strategies (Isaacs et al. 2004; Bayer and Smolke 2005), regulatable ribozymes (Yen et al. 2004, 2006), or small molecule-binding aptamers (Werstuck and Green 1998; Suess et al. 2003). The latter control gene expression by insertion of aptamer sequences into the 5′ untranslated region (UTR) of an mRNA. These RNAs function by exploiting the fact that small molecule binding to the RNAs alters their structures in the 5′ UTR and subsequently interferes with translation of the downstream coding regions. Dependent on the insertion site, the aptamer–ligand complex interferes either with binding of the 43S subunit to the cap-structure or the scanning process (Hanson et al. 2003). Thereby, the advantage of such engineered riboswitches is that they can be designed in principle to any ligand of choice. However, the majority of in vitro selected aptamers display no regulatory activity in vivo. Up to now, only five small molecule-binding aptamers have been identified that have the potential to act as riboswitches (Werstuck and Green 1998; Grate and Wilson 2001; Harvey et al. 2002; Suess et al. 2003). In addition, two of these—namely, the theophylline and the tetracycline aptamer—have been used as a binding domain for the engineering of more complex riboswitches with broadened functions and applicability (Buskirk et al. 2004; Desai and Gallivan 2004; Isaacs et al. 2004; Suess et al. 2004; Bayer and Smolke 2005). In vitro selection techniques to isolate small molecule-binding aptamers (Ellington and Szostak 1990; Tuerk and Gold 1990) or regulatable ribozymes (Koizumi et al. 1999; Piganeau et al. 2001) have been well established in the past. However, there is a noticeable lack of methods for the detection of those in vitro selected molecules that in addition show regulatory activity within the cell. To broaden the spectrum of modular synthetic riboswitches by delivering new building blocks, we established an easy method for the isolation of new small molecule-binding aptamers with activity in vivo.
Aminoglycosides are broad-spectrum antibiotics that disturb the fidelity of the tRNA selection step during protein synthesis by binding to the A-site of bacterial 16S rRNA. Aminoglycosides are pseudo-oligosaccharides containing variable numbers of sugar rings and ammonium groups characterized by high flexibility. The positive charges are attracted to the negatively charged RNA backbone. The flexibility of the aminoglycosides facilitates accommodation into a binding pocket within internal loops of RNA helices or into ribozyme cores for making specific contacts. These characteristics predestine this group of antibiotics as universal RNA binders (Walter et al. 1999; Schroeder et al. 2000; Zhao et al. 2005). This prompted us to use neomycin as target molecule to search for artificial riboswitches that can be used for conditional control of gene expression. Neomycin B is composed of four rings. The central 2-deoxystreptamine (ring I) is 4,5-disubstituted and contains ammonium groups at positions 1 and 3 (Fig. 1). It has been shown to inhibit catalytic activity of several ribozymes (von Ahsen et al. 1991; Stage et al. 1995) and has already been exploited to regulate translation termination (Murphy et al. 2006).
FIGURE 1.
In vivo screening for regulatory active neomycin-binding aptamers. (A) Sequence of the 5′ UTR containing the m7G cap structure, the 32- (38-) nt-long sequence of the original 5′ UTR (length depends on which transcriptional start site is used), the restriction sites for AflII and NheI (bold), the constant (underlined) and variable (N74) region of the aptamer pool, and the start codon with the optimized Kozak sequence (italics). (B) Sequence alignment of 30 regulatory active, neomycin-binding aptamers. Bases that correspond to the identified consensus sequence are marked in gray. The numbers in the brackets indicate how often a respective clone was identified. (C) Scheme of the in vivo screening system. (D) Chemical structure of neomycin B. Ring (roman numbers) and atomic (Arabic numbers) numbering system is indicated. The amino group at position 6 of ring I that is substituted in paromomycin is indicated by an arrow.
We successfully built up a selection system and identified aptamers that mediate neomycin-dependent regulation of gene expression. One candidate was further characterized in terms of activity and ligand binding. A structure–function relationship reveals that several nucleotides of the aptamer are involved in ligand binding. We propose a binding pocket that contains both an internal and a terminal loop structure, with neomycin sandwiched between both loop regions. Such composite binding pockets with the RNA recognizing several moieties of its ligand to achieve extraordinary affinity and specificity have already been described for both natural and artificial riboswitches (Gilbert and Batey 2006; Muller et al. 2006). Our study emphasizes the need for a combined approach of in vitro selection and in vivo screening to identify small molecule-binding RNAs that are functional in vivo.
RESULTS
In vivo screening for regulatory active aptamers
We started with an aptamer pool that was enriched for binding neomycin B via six cycles of in vitro selection (Wallis et al. 1995). The 111-nucleotide (nt)-long RNA pool consists of a randomized region of 74 nt flanked by constant parts (Famulok 1994). The pool was inserted directly in front of a constitutively expressed gfp reporter gene in the yeast vector pWHE601 (Fig. 1A; Suess et al. 2003) and amplified in Escherichia coli, generating a starting diversity for in vivo screening of 5 × 104 sequences.
The library was transformed into yeast cells, and screening for neomycin-dependent changes in GFP fluorescence was performed in a two-step process. First, candidates were identified that allowed gene expression in the absence of the ligand, and, second, among these, candidates with neomycin-dependent regulatory properties were searched (Fig. 1C). The first step was used to eliminate candidates in which (1) the ligand-free aptamers interfered with translation; (2) a premature start codon is introduced by the aptamer sequence; or (3) the insertion was not successful (resulting in an AUG-less open reading frame because the start codon is introduced along with the aptamer sequence).
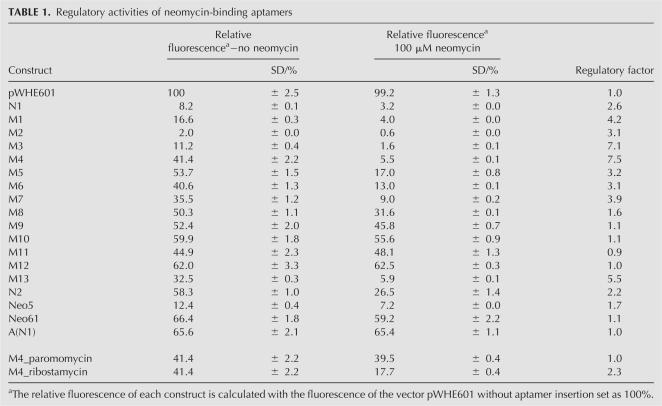
The results of the first screening step demonstrate that only 5% of the clones (2500 of 5 × 104 transformants) show gfp expression in the absence of neomycin. For the second step, 1300 individual fluorescent colonies were transferred into 96-well plates, and GFP fluorescence was analyzed in the absence and presence of 100 μM neomycin. This concentration has no effect on the cell growth of Saccharomyces cerevisiae (data not shown). A regulation factor was determined by calculating the quotient of fluorescence with and without neomycin. Plasmids were isolated from individual candidates with down-regulation of gfp expression and transferred through E. coli, and GFP measurement was repeated. This screening process produced 30 candidates exhibiting a neomycin-dependent decrease in fluorescence. Sequence analysis of these clones yielded 10 individual candidates (Fig. 1B). The alignment shows a 16-nt-long sequence element that is completely conserved in the candidates N1 and N2 (33% of all candidates identified) and partially present in nearly all other candidates (highlighted in Fig. 1B). Interestingly, none of the identified sequences showed sequence similarities to randomly picked neomycin-binding aptamers obtained from the same in vitro selected aptamer pool (Wallis et al. 1995).
We performed truncation analysis of N1 and identified a 47-nt-long minimal construct M1 containing the 16-nt-long consensus sequence that is responsible for the regulatory phenotype. Secondary structure prediction of M1 within the context of the mRNA results in a stem–loop structure with an internal asymmetrical loop (Fig. 2A). We determined gfp expression of N1 and M1 in the presence and absence of neomycin after 48 h of incubation (Fig. 3B; Table 1). Insertion of N1 and M1, respectively, results in lower gfp expression. Addition of 100 μM neomycin then leads to a 2.6-fold decrease in fluorescence for N1. The truncated version M1 shows improved regulation (4.2-fold).
FIGURE 2.
Structural probing of the neomycin-aptamer M1. (A) Secondary structure model of the neomycin–aptamer M1 in the context of the mRNA with the consensus sequence highlighted in gray. Positions with prominent cleavage and neomycin-dependent changes are indicated. (B) Enzymatic cleavage pattern in the absence (−) or presence (+) of 50 μM neomycin with the guanine-specific nuclease T1 and single-strand-specific nuclease S1. C and H represent no and partial digestion with alkali, respectively. Selected fragment bands corresponding to digestion by T1 are identified by nucleotide positions. The closing stem of the aptamer is shown as a black line, and the gray box corresponds to the consensus sequence. Neomycin-dependent changes are indicated.
FIGURE 3.
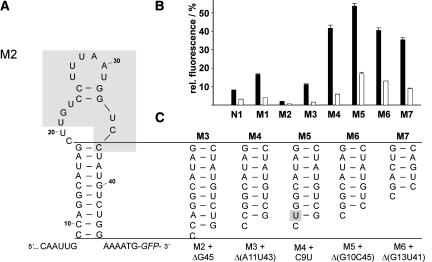
Influence of the stem length of the neomycin–aptamer on regulation. (A) Sequence and secondary structure model of the minimal neomycin-binding aptamer M2 within the context of the 5′ UTR with the consensus sequence highlighted in gray. (B) Measurement of gfp expression of the original identified construct, N1, and several truncated versions, M1 (Fig. 2A), M2 (A), and M3–M7 (C) in the absence (black bars) and presence (white bars) of 100 μM neomycin. The fluorescence emission of GFP expressed by the vector pWHE601 was set to 100%. (C) Different stems analyzed are displayed.
TABLE 1.
Regulatory activities of neomycin-binding aptamers
Analysis of secondary structure and ligand binding by enzymatic probing
Enzymatic probing was performed to elucidate a secondary structure model for the functional RNAs and to determine which nucleotides are involved in ligand recognition. An RNA construct corresponding to the complete 5′ UTR containing M1 and the first 48 nt of the gfp open reading frame was prepared by in vitro transcription. The 136-nt-long RNA was subjected to enzymatic probing using the single-strand-specific nucleases S1 and T1. T1 cleaves between nucleotides at the 3′-phosphate of guanosines. S1 cleaves without any nucleotide preference. Thereby, the cleavage patterns provide information about secondary structure and can also reveal nucleotides participating in tertiary interactions and/or ligand-induced structural changes.
32P-end-labeled RNA was treated with the respective nucleases and the resulting fragments were separated by PAGE. The fragmentation pattern is shown in Figure 2B, and the results of the enzymatic cleavage are summarized schematically onto the secondary structure of the neomycin aptamer in Figure 2A. Several nucleotides of the terminal loop are accessible to the single-strand nuclease S1 (U28, A29, A30), but the proposed internal loop is less clearly defined. U21 is detectable, whereas the remaining nucleotides, C19, U20, G22, and U23, show only marginal S1 cleavage, which implies their involvement in tertiary structure formation. In agreement, U34 and C35 (the 3′ part of this motif) are not detectable at all. Formation of the proposed stem C9–G18/C36–G45 is confirmed by the complete absence of single-strand-specific cleavage of these nucleotides.
The presence of neomycin dramatically changes the cleavage pattern within the conserved 16-nt-long consensus sequence (Fig. 2B, marked with a gray bar). The terminal loop and G22 of the internal loop are completely protected against cleavage, indicating an involvement in ligand-binding and/or ligand-induced structural changes. Neomycin also induces the formation of the two proposed Watson–Crick base-pairs C24/G33 and C25/G32, displayed by the decrease of signal intensity for G32, G33 (both T1), and C24 (S1). This indicates that the main binding site for neomycin is located in the central part of the aptamer and corresponds to the identified 16-nt consensus sequence. Secondary and probably nonspecific binding sites are visible by protection of G60 and G90 against T1 cleavage (data not shown) and may be due to the highly positive charged nature of neomycin. Such weaker binding sites that target primarily the adjacent stem region have already been described for aminoglycoside-binding aptamers (Jiang and Patel 1998; Jiang et al. 1999; Stampfl et al. 2007). S1 and T1 nuclease treatment was also performed on a 200-nt-long RNA fragment containing N1 instead of M1 (data not shown). The results obtained with this RNA showed similar cleavage patterns as the minimal aptamer, indicating that the sequence context does not influence structure formation and ligand-binding properties of the aptamer. Taken together, the aptamer identified adopts an overall stem–loop structure including a terminal and an internal asymmetrical loop previously described as general neomycin-binding fold (Schroeder et al. 2000).
Structure–function analysis of the neomycin-binding core sequence
RNA molecules with influence on biological processes are amenable to the elucidation of basic structure–function relationships by site directed mutagenesis. Based on the sequence alignment (Fig. 1B) and the structural probing data, we hypothesized that both loop elements are necessary for regulation, whereas the stem is important for structure formation but with no sequence constraints. To test this hypothesis and to gain further insight into the role of the conserved region, we varied the stability of the aptamer stem.
Based on the secondary structural model a 37-nt-long minimal motif (nucleotides 9–45 of Fig. 2A) was created with an additional mutation, A43U, resulting in the construct M2 (Fig. 3A). This mutation converts the A11/A43 pair into a Watson–Crick base pair, thereby stabilizing the stem. Gene reporter assays carried out in the absence of neomycin demonstrate that this point mutation results in strong down-regulation of gfp expression (Table 1; Fig. 3B). A stepwise destabilization of the closing stem (M4–M7, displayed in Fig. 3C) results in an increased gfp expression, with maximum gfp expression resulting from M5 (Fig. 3B; Table 1). Maximum regulation was obtained for M4 with 7.5-fold reduction in the presence of neomycin compared to the control lacking neomycin. Therefore, M4 was used as the parent for the following mutagenesis studies.
Mutations within the terminal loop M8–M10 (A30U, deletion of U31 and U27C, U31C) led to a nearly complete loss of function (Fig. 4; Table 1). The same effect was also observed for the M11 mutation (G22C) within the internal loop. These data strongly suggest the importance of both internal and terminal loops for regulation in vivo. Next, one of the two GC base pairs separating the terminal from the internal loop was mutated to test if they are only of structural importance. However, swapping the sequence (M12: C25G/G32C) resulted in a complete loss of regulation. In contrast, swapping of 2 base pairs (bp) in the stem region results in only a moderate reduction in regulation (M13: A15U, U16A, A38U, U39A), implying that stem formation is critical for regulation.
FIGURE 4.
Mutational analysis of the minimal M4 neomycin aptamer. (A) Secondary structure of the minimal neomycin-binding aptamer M4 with the consensus sequence highlighted in gray. Mutations introduced to study structure–function relationships are indicated. (B) Measurement of gfp expression in the absence (black bars) and presence (white bars) of 100 μM neomycin. The fluorescence emission of GFP expressed by the vector pWHE601 was set to 100%.
Taken together, mutational analysis supports the proposed stem–loop structure of the aptamer and underlines the importance of the conserved 16-nt consensus sequence for neomycin binding and regulation since none of the tested mutations within this region is tolerated.
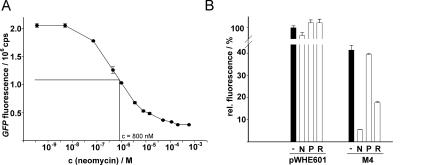
Neomycin aptamer M4 shows a dose-dependent regulation and high ligand specificity
The construct with the best regulatory properties (M4) was used for further characterization. Titration of neomycin into cells expressing GFP under control of M4 results in a dose-dependent decrease of reporter activity with a half-maximal concentration of neomycin needed for efficient regulation of 800 nM (Fig. 5A). We then tested the effect of several neomycin analogs on regulation. The control vector without aptamer shows that all aminoglycosides tested do not affect gene expression. Interestingly, no inhibition was obtained with paromomycin, a closely related analog of neomycin that differs by a single hydroxyl group substitution of the amino group at position 6 at ring I (Fig. 5B). This indicates the specificity of the regulation for neomycin. In contrast, ribostamycin, an analog of neomycin lacking ring IV, still showed regulation. Furthermore, it has been demonstrated that ring IV of neomycin has only minor importance in RNA recognition for a neomycin-binding aptamer R23, which has the same overall fold but no sequence similarities to M4 (Jiang et al. 1997, 1999), or a neomycin-binding RNA motif derived from the decoding site (Fourmy et al. 1998; Francois et al. 2005).
FIGURE 5.
Analysis of dose-dependence and ligand specificity. (A) GFP expression of the neomycin-binding aptamer M4 measured with increasing concentrations of neomycin. The half-maximal concentration needed for efficient neomycin-dependent regulation is indicated. (B) Measurement of fluorescence expression of pWHE601 and M4 in the absence (−) and presence of 100 μM neomycin (N), paromomycin (P) and ribostamycin (R).
Identification of determinants necessary for regulatory activity
The binding of aminoglycosides to RNA has been proposed to be determined more by shape than by sequence (Schroeder et al. 2000). Binding sites for aminoglycosides are primarily located in helical domains that have a widened major groove that serves as docking site for the antibiotics. These sites are formed by an asymmetrical internal loop closed by a set of Watson–Crick and non-Watson–Crick base pairs as in the decoding site (Fourmy et al. 1998; Francois et al. 2005) or by a zippered-up hairpin loop through three GU wobble pairs in the case of several in vitro selected aptamers (Wallis et al. 1995). In both cases, the binding sites adopt the structure of a continuous but disturbed helix as docking platform for rings I and II of aminoglycosides.
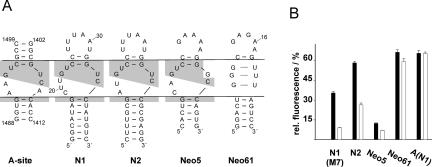
The regulatory active aptamers identified with our in vivo screen are consistent with this model. A comparison of several neomycin-binding RNA motifs (Fig. 6A) reveals that N1 and N2 show sequence similarities to the decoding site of the ribosome but lack the functional adenines A1408, A1492, and A1493 (Fig. 6, cf. A-site and N1/2). Sequence similarities also exist to the neomycin-binding aptamer Neo5 but not to Neo61 (both are from the same in vitro selected aptamer pool but randomly picked) (Wallis et al. 1995). These similarities of the central parts of the motifs prompted us to the test regulatory activity of all five candidates. Therefore, these motifs were attached to a 5-nt-long stem (equivalent to M7), and the A-site construct was capped with the N1 terminal loop.
FIGURE 6.
Comparison of neomycin-binding RNA motifs. (A) Secondary structure of the neomycin-binding site of the ribosomal decoding site (A-site) and the neomycin-binding aptamers N1, N2 (this study), Neo5, and Neo61 (Wallis et al. 1995). The lines dissect the aptamers into an upper part (terminal loop), the internal asymmetrical loop, and the closing stem; conserved nucleotides are highlighted in gray. (B) gfp expression in the absence (black bars) and presence (white bars) of 100 μM neomycin. Fluorescence of a vector without aptamer (pWHE601) was set to 100%. All motifs were constructed with a 5-nt-long stem (from construct M7); the A-site motif was capped with the terminal loop of N1.
In vivo reporter gene assays of N2 and Neo5 demonstrate that these RNAs exhibit neomycin-dependent down-regulation of gfp expression (Fig. 6B) with slightly reduced regulatory efficiency compared to N1. The basal activity of N2 is increased compared to N1. In contrast, it is strongly reduced for Neo5, the reason for which may be the GU wobble instead of the shared UU pair at position 23–34. Furthermore, no neomycin-dependent decrease in reporter activity was detectable for the Neo61 and A-site constructs.
N1 and the A-site construct differ in the lower part of the asymmetrical loop. The aptamer has the nucleotides CUU (nucleotides 19–21) instead of both functional adenines A1492 and A1493 and lacks A1408 on the opposite strand. All three adenines make specific contacts to ring I of neomycin B, whereas rings II and III are mainly coordinated by 3 bp of the upper part of the loop (1496C/1405G, 1495U/1406U, 1494G/1407C) (Francois et al. 2005). This proposes that rings II and III of neomycin are bound by N1 in a manner similar to the A-site, whereas ring I must form different contacts. Thereby, the amine group at position 6 of ring I (marked in Fig. 1D) seems to be crucial since paromomycin, which lacks only this functional group, shows no regulation at all.
DISCUSSION
Model of neomycin-binding pocket
Structural probing and mutational analysis underline the importance of both loop regions for neomycin binding and regulation. The data support a model in which the aptamer recognizes rings II and III of neomycin similarly to the decoding site (Fourmy et al. 1998; Francois et al. 2005). This is supported by the strong neomycin-dependent protection of T1 cleavage at G22, which would correspond to G1494 of the decoding site. In addition, mutagenesis of this position and the neighboring base pair C24/G35 (which corresponds to 1496C/1405G) results in a complete loss of neomycin binding. A crucial interaction of the decoding site with ring I, which stacks against G1491 forming a pseudo base pair to two Watson–Crick sites of the universally conserved A1408, cannot be formed within the aptamer because a nucleotide between positions C35 and C36 (which corresponds to 1408 in the decoding site) is lacking in the aptamers N1, N2, and Neo5, indicating that correct positioning of ring I in the aptamer is crucial for regulation. This is supported by the fact that paromomycin, which lacks only the amine at position 6 of ring I, shows no regulation at all.
Our data also indicate the importance of the terminal loop for neomycin binding (see neomycin-dependent protection in the cleavage pattern) and regulation (all mutants within the loop region are inactive). The NMR structure of a neomycin-binding aptamer R23, which is similar to Neo61 (Jiang et al. 1999), shows important contacts of the terminal loop with the aminoglycoside by a flipped out adenine (A16, which corresponds to A29 in N1). This forms a flaplike structure sandwiching the bound ligand between the zippered-up hairpin loop through three GU wobble pairs and the flipped adenine.
The terminal loop of the regulatory active aptamer is able to form a similar flaplike structure. Therefore, we propose a neomycin-binding pocket that is composed of the internal loop segment coordinating rings II and III, similar to the decoding site, with a flaplike loop structure that closes the binding pocket. Thereby, the terminal and the internal asymmetrical loop represent each half of a clamp in which the neomycin is tightly bound. This complex is then able to interfere with translation initiation.
Combining in vitro selection and in vivo screening creates artificial riboswitches
Previous studies have shown that small molecule-binding aptamers display regulatory activity and can be used as artificial switches to regulate gene expression. However, there are only a few aptamers that have been reported that act as a riboswitch in vivo (Werstuck and Green 1998; Grate and Wilson 2001; Harvey et al. 2002; Hanson et al. 2003). In a previous study, we have shown that only 1 of 15 different tetracycline-binding aptamers is able to mediate ligand-dependent regulation of gene expression (Suess et al. 2003). In vitro selection enriches RNA populations for high affinity binding and specific recognition of their cognate ligands. However, these characteristics seem to be necessary, but not sufficient, to obtain aptamers that are capable of acting as riboswitches.
Therefore, we applied a strategy using the combination of in vitro selection and in vivo screening to identify small molecule-binding regulatory active aptamers. Interestingly, these aptamers show no sequence similarities to randomly analyzed aptamers from the same in vitro selection, demonstrating that in vitro selection does not necessarily result in biologically relevant structures (Wallis et al. 1995). However, the additional step of genetic screening leads to molecules that are capable of controlling gene expression in vivo. In this study, the isolated RNAs possess similarities to natural neomycin-binding sites. Therefore, these results demonstrate that an advanced strategy of in vitro selection and in vivo screening can yield information about biological systems.
We anticipate that combining in vitro selection with genetic screens will be a useful strategy to identify RNAs with desired in vivo functionalities. Thereby, in vitro selection yields RNA molecules able to bind a ligand with high affinity and specificity, but the following selection process in vivo allows us to isolate the subset of molecules with the desired functionality, like the regulation of gene expression. This is underlined by the fact that inactive sequences that form a zippered-up hairpin loop like Neo61 are overrepresented in the in vitro selected neomycin aptamer pool (Wallis et al. 1995). Furthermore, an in vitro selection of an RNA pool of mutagenized A-site motifs for neomycin binding led to Neo61-like, but not to active (N1-like), sequences (Famulok and Hüttenhofer 1996). This impressively demonstrates that selection for high ligand affinity is not sufficient for in vivo activity of aptamers and underlines the need for further selection within the cell.
A recent report by Lynch et al. (2007) presents a similar strategy. The authors used the theophylline aptamer and performed a genetic screen for a respective sequence context turning the aptamer into a functional riboswitch, which is a further example of the tremendous potential of coupling in vitro selection and genetic screens for the isolation of molecules that have a specific in vivo function.
Taken together, the assay described in this report can easily be applied to identify novel regulatory active aptamers binding to any ligand of choice. This will lead to a great variety of novel regulatory active aptamers but also to new building blocks for engineering complex, modularly built riboswitches and opens up the possibility of more customized and applicable riboswitches with binding domains selected for particular needs.
MATERIAL AND METHODS
Plasmids and strains
For all experiments the yeast 2μ plasmid pWHE601 was used to constitutively express the gfp + gene from an Padh promoter (Suess et al. 2003). The vector contains a restriction site for AflII immediately upstream of the start codon with a 5′ UTR length of 38 nt or 44 nt, respectively, depending on the two described transcriptional start sites, and a singular site for NheI directly behind the start codon. For aptamer insertion, the vector was digested with AflII and NheI. Mutations and respective restriction sites were introduced into the aptamer sequences by PCR mutagenesis. The start codon that was cut out of the vector after AflII/NheI digestion was attached 3′ to the aptamer sequence together with an optimized Kozak sequence for yeast. For all experiments, S. cerevisiae strain RS453α was used (Suess et al. 2003). Primer and vector sequences are available upon request.
In vivo screening
The neomycin-binding aptamer pool of round six (Wallis et al. 1995) was amplified by PCR using the primer pair 5′-GATGATCTTAAG GAGCTCAGCCTTCACTG and 5′-GATGATGCTAGC CATTTT GGATCCGACCGTGGTGCC (restriction sites for AflII and NheI, respectively, are shown in bold, the start codon with an optimized Kozak sequence AAAATG is in italics, the underlined sequences anneal to the constant region of the in vitro selected aptamer pool; Wallis et al. 1995). The PCR product was cloned into the yeast plasmid pWHE601 that expresses the gfp + gene from an constitutive Padh promoter (Suess et al. 2003) thereby inserting the aptamer sequences into the 5′ UTR directly in front of the start codon. Yeast cells were transformed with the plasmid pool according to the protocol supplied with the frozen EASY yeast transformation II kit (Zymo Research). Cells were grown at 28°C in minimal medium (0.2% [w/v] yeast nitrogen base, 0.55% ammonium sulfate, 2% [w/v] glucose, 12 μg mL−1 adenine, MEM amino acid; Gibco BRL). Fluorescent colonies were transferred to 96-well plates containing 200 μL minimal medium and incubated for 24 h. Twenty-microliter aliquots of each sample were transferred into fresh medium in new plates with and without the ligand in a final volume of 200 μL. Fluorescence measurements were performed 48 h after inoculation using an excitation wavelength of 484 nm and an emission wavelength of 512 nm with a SpectraFluor Plus fluorescence reader (Tecan, Crailsheim). Additionally, the optical density was determined to correlate the fluorescence to the cell number. The measurements were repeated at least twice. Positive candidates were streaked out to single colonies, and plasmids were prepared according to the protocol supplied with the Zymo prep kit (Zymo Research). After an E. coli passage, yeast cells were retransformed with the plasmids and the fluorescence measurements repeated. Positive candidates were sequenced and subjected to detailed analysis.
GFP measurements
For all measurements S. cerevisiae strain RS453α was transformed according to the protocol supplied with the frozen EASY yeast transformation II kit (Zymo Research). Yeast cells transformed with the respective constructs were grown at 28°C for 48 h in minimal medium in the absence or presence of 100 μM neomycin in a final volume of 5 mL. Cells were harvested by centrifugation and resuspended in 2 mL phosphate-buffered saline (PBS). For each construct three independently grown cultures were analyzed. Fluorescence measurements were carried out at 25°C on a Fluorolog FL3–22, with the excitation wavelength set to 395 nm and an emission wavelength of 510 nm. Additionally, we determined the optical density (OD) at 600 nm to ensure homogeneous cell growth. The vector pVTU102 without the gfp + gene was analyzed in parallel as a blank and its value subtracted from all data. The relative fluorescence of each construct is calculated with the fluorescence of pWH601 either in the presence or absence of 100 μM neomycin set as 100%. All measurements were repeated at least twice.
Enzymatic probing of aptamer RNA
Aptamer RNA was prepared by in vitro transcription using T7 polymerase and the PCR DNA template amplified using the primer pair 5′-TCTAATACGACTCACTATAGGAGCATACA ATCAACTCC and 5′-CAAGAATTGGGACAACTCC with the T7 recognition sequence underlined. The resulting PCR product covers the entire 5′ UTR with the aptamer and the first 48 nt of the gfp coding region. RNA transcripts were dephosphorylated and subsequently 5′-32P-end labeled as previously described (Hanson et al. 2005). RNA (100,000 cpm) was incubated in 50 mM sodium acetate (pH 4.5), 280 mM NaCl, 4.5 mM ZnSO4 (S1), or 50 mM Tris-HCl (pH 7.5) (T1) for 2 min at 56°C and for 10 min at 37°C. Afterward 5 mM MgCl2 were added and the RNA was incubated at room temperature for 5 min without or with 50 μM neomycin. We then added 0.75 U of S1 nuclease (Promega) and 0.75 U of RNase T1 (Roche), respectively, and incubated it for 5 min. Alkaline hydrolysis was carried out in 50 mM sodium carbonate buffer (pH 9.0) for 7 min at 90°C. All reactions were stopped by the addition of an equal volume of sample loading buffer (95% formamide, 0.3% bromophenol blue, 0.3% xylenecyanol, 10 mM EDTA). Denaturing 10% PAGE was used to separate the cleavage products and they were visualized by phosphorimaging.
ACKNOWLEDGMENTS
The studies were carried out in the laboratory of Wolfgang Hillen, whose support is greatly appreciated. We thank Kristian Link and Christian Berens for fruitful discussions and critical reading of the manuscript. Norbert Sauer is acknowledged for the use of the fluorescence stereomicroscope. We are grateful to the Volkswagenstiftung (I/79 950) and the Deutsche Forschungsgemeinschaft (SU 402/1-1) for financial support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.772408.
REFERENCES
- Bayer, T.S., Smolke, C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- Buskirk, A.R., Landrigan, A., Liu, D.R. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Desai, S.K., Gallivan, J.P. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- Ellington, A.D., Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Famulok, M. Molecular recognition of amino acids by RNA-aptamers: An L-citrulline binding RNA motif and its evolution into an L-arginine binder. J. Am. Chem. Soc. 1994;116:1698–1706. [Google Scholar]
- Famulok, M., Hüttenhofer, A. In vitro selection analysis of neomycin binding RNAs with a mutagenized pool of variants of the 16S rRNA decoding region. Biochemistry. 1996;35:4265–4270. doi: 10.1021/bi952479r. [DOI] [PubMed] [Google Scholar]
- Fourmy, D., Recht, M.I., Puglisi, J.D. Binding of neomycin-class aminoglycoside antibiotics to the A-site of 16 S rRNA. J. Mol. Biol. 1998;277:347–362. doi: 10.1006/jmbi.1997.1552. [DOI] [PubMed] [Google Scholar]
- Francois, B., Russell, R.J., Murray, J.B., Aboul-ela, F., Masquida, B., Vicens, Q., Westhof, E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: Role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, S.D., Batey, R.T. Riboswitches: Fold and function. Chem. Biol. 2006;13:805–807. doi: 10.1016/j.chembiol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Grate, D., Wilson, C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer–ligand complex. Bioorg. Med. Chem. 2001;9:2565–2570. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- Hanson, S., Berthelot, K., Fink, B., McCarthy, J.E., Suess, B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- Hanson, S., Bauer, G., Fink, B., Suess, B. Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA. 2005;11:503–511. doi: 10.1261/rna.7251305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, I., Garneau, P., Pelletier, J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs, F.J., Dwyer, D.J., Ding, C., Pervouchine, D.D., Cantor, C.R., Collins, J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Jiang, L., Patel, D.J. Solution structure of the tobramycin–RNA aptamer complex. Nat. Struct. Biol. 1998;5:769–774. doi: 10.1038/1804. [DOI] [PubMed] [Google Scholar]
- Jiang, L., Suri, A.K., Fiala, R., Patel, D.J. Saccharide-RNA recognition in an aminoglycoside antibiotic–RNA aptamer complex. Chem. Biol. 1997;4:35–50. doi: 10.1016/s1074-5521(97)90235-0. [DOI] [PubMed] [Google Scholar]
- Jiang, L., Majumdar, A., Hu, W., Jaishree, T.J., Xu, W., Patel, D.J. Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer. Structure. 1999;7:817–827. doi: 10.1016/s0969-2126(99)80105-1. [DOI] [PubMed] [Google Scholar]
- Koizumi, M., Soukup, G.A., Kerr, J.N., Breaker, R.R. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat. Struct. Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- Lynch, S.A., Desai, S.K., Sajja, H.K., Gallivan, J.P. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Muller, M., Weigand, J.E., Weichenrieder, O., Suess, B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, G.J., Mostoslavsky, G., Kotton, D.N., Mulligan, R.C. Exogenous control of mammalian gene expression via modulation of translational termination. Nat. Med. 2006;12:1093–1099. doi: 10.1038/nm1376. [DOI] [PubMed] [Google Scholar]
- Nudler, E., Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Piganeau, N., Jenne, A., Thuillier, V., Famulok, M. An allosteric ribozyme regulated by doxycyline. Angew. Chem. Int. Ed. Engl. 2001;40:3503. doi: 10.1002/1521-3773(20001201)39:23<4369::AID-ANIE4369>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Schroeder, R., Waldsich, C., Wank, H. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 2000;19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage, T.K., Hertel, K.J., Uhlenbeck, O.C. Inhibition of the hammerhead ribozyme by neomycin. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- Stampfl, S., Lembradl, A., Koehler, G., Schroeder, R. Monovalent ion dependence of neomycin B binding to an RNA aptamer characterized by spectroscopic methods. ChemBioChem. 2007;8:1137–1145. doi: 10.1002/cbic.200700030. [DOI] [PubMed] [Google Scholar]
- Suess, B., Hanson, S., Berens, C., Fink, B., Schroeder, R., Hillen, W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess, B., Fink, B., Berens, C., Stentz, R., Hillen, W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, B.J., Breaker, R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Tuerk, C., Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- von Ahsen, U., Davies, J., Schroeder, R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- Wallis, M.G., von Ahsen, U., Schroeder, R., Famulok, M. A novel RNA motif for neomycin recognition. Chem. Biol. 1995;2:543–552. doi: 10.1016/1074-5521(95)90188-4. [DOI] [PubMed] [Google Scholar]
- Walter, F., Vicens, Q., Westhof, E. Aminoglycoside-RNA interactions. Curr. Opin. Chem. Biol. 1999;3:694–704. doi: 10.1016/s1367-5931(99)00028-9. [DOI] [PubMed] [Google Scholar]
- Werstuck, G., Green, M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Breaker, R.R. Genetic control by metabolite-binding riboswitches. ChemBioChem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Yen, L., Svendsen, J., Lee, J.S., Gray, J.T., Magnier, M., Baba, T., D'Amato, R.J., Mulligan, R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Yen, L., Magnier, M., Weissleder, R., Stockwell, B.R., Mulligan, R.C. Identification of inhibitors of ribozyme self-cleavage in mammalian cells via high-throughput screening of chemical libraries. RNA. 2006;12:797–806. doi: 10.1261/rna.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F., Zhao, Q., Blount, K.F., Han, Q., Tor, Y., Hermann, T. Molecular recognition of RNA by neomycin and a restricted neomycin derivative. Angew. Chem. Int. Ed. Engl. 2005;44:5329–5334. doi: 10.1002/anie.200500903. [DOI] [PubMed] [Google Scholar]