Abstract
Very little is known regarding regulation of microRNA (miRNA) biogenesis in normal tissues, tumors, and cell lines. Here, we profiled the expression of 225 precursor and mature miRNAs using real-time PCR and compared the expression levels to determine the processing patterns. RNA from 22 different human tissues, 37 human cancer cell lines, and 16 pancreas and liver tissues/tumors was profiled. The relationship between precursor and mature miRNA expression fell into the following four categories: (1) a direct correlation exists between the precursor and mature miRNA expression in all cells/tissues studied; (2) direct correlation of the precursor and mature miRNA exists, yet the expression is restricted to specific cell lines or tissues; (3) there is detectable expression of mature miRNA in certain cells and tissues while the precursor is expressed in all or most cells/tissues; or (4) both precursor and mature miRNA are not expressed. Pearson correlation between the precursor and mature miRNA expression was closer to one for the tissues but was closer to zero for the cell lines, suggesting that processing of precursor miRNAs is reduced in cancer cell lines. By using Northern blotting, we show that many of these miRNAs (e.g., miR-31, miR-105 and miR-128a) are processed to the precursor, but in situ hybridization analysis demonstrates that these miRNA precursors are retained in the nucleus. We provide a database of the levels of precursor and mature miRNA in a variety of cell types. Our data demonstrate that a large number of miRNAs are transcribed but are not processed to the mature miRNA.
Keywords: microRNA biogenesis, microRNA precursor, real-time PCR
INTRODUCTION
The genes encoding microRNAs (miRNAs) are located within introns, intergenic regions, and exons (Kim and Nam 2006). The primary RNA transcript containing the miRNA primary precursor (pri-miRNA) is processed in the nucleus by Drosha/DGCR8 to produce the precursor miRNA (pre-miRNA) (Lee et al. 2003). pre-miRNA is ∼70 nucleotides (nt) in length and contains the characteristic hairpin structure. The pre-miRNA is transported to the cytoplasm by Exportin 5 (Lund et al. 2004), where it is subsequently processed by Dicer to the ∼21-nt mature miRNA.
Very little is known regarding the regulation of miRNA processing. Conceivably, regulation of miRNA biogenesis may occur at the level of Drosha processing, nuclear transport, or Dicer processing. Several studies have shown that miRNAs are present at the level of precursor but are not processed to the mature miRNA. Michael et al. (2003) used Northern blots to show that the precursors to miR-143 and miR-145 are expressed in colorectal tissues and tumors; however, the mature miRNA was detectable only in the normal colorectal tissue. Pre-miR-138-2 was shown by Northern blotting and in situ hybridization to be expressed in HeLa cells, but the mature miR-138 was undetectable (Obernosterer et al. 2006). By using published messenger RNA data to approximate the miRNA precursor expression encoded within introns, Thomson et al. (2006) showed widespread down-regulation of miRNAs in cancer. To our knowledge, no one has addressed the issue of relative differences in precursor and mature miRNA expression by large-scale profiling. This may be due to the fact that the majority of miRNA studies have focused on quantifying the active mature miRNA and not the miRNA precursors.
In 2004, the Schmittgen laboratory developed the first real-time RT PCR method to quantify miRNA (Schmittgen et al. 2004). This method quantified the miRNA precursors with the goal of predicting the expression of mature miRNA. By designing primers to the stem of the miRNA hairpin, the primary precursor and precursor miRNA were simultaneously quantified (Schmittgen et al. 2004). This technology has been applied to profile the expression of over 200 miRNA precursors in a number of human cancer cell lines (Schmittgen et al. 2004; Jiang et al. 2005, 2006) and tissues from patients with pancreatic cancer (Lee et al. 2007). In 2005, a TaqMan-based RT-PCR assay was designed to quantify the mature miRNA (Chen et al. 2005). PCR and Northern blotting demonstrated that the expression of mature and precursor miRNAs generally correlate (Schmittgen et al. 2004; Jiang et al. 2005, 2006; Meng et al. 2006; Lee et al. 2007). However, precursors to miR-1, miR-122a, and miR-133 showed considerable expression by PCR, yet mature miRNA was undetectable by Northern blotting (Jiang et al. 2005). This prompted us to ask the following question: How widespread are alterations in miRNA biogenesis in different tissues, tumors, or cancer cells lines? To address this question, we profiled the expression of over 200 precursor and mature miRNAs using sensitive real-time PCR assays. Our results demonstrate that a wide discrepancy exists between the levels of precursor and mature miRNA, suggesting that unknown mechanisms that control processing play a critical role in regulating the expression of the active, mature miRNA.
RESULTS AND DISCUSSION
The expression of 201 mature and precursor miRNAs was profiled in a panel of 22 human tissues, 37 human cancer cell lines, and pancreas and liver tumors/tissues using real-time PCR assays specific to each class of miRNA. Expression of the active, mature miRNA was either broadly expressed in each of the 22 tissues or was expressed among one, two, or three different tissues. Our mature miRNA data in tissues closely resembled that of Liang et al. (2007), who used the identical TaqMan assay to profile miRNA in 40 normal human tissues. miRNA expression in the tissues is designated as ubiquitous, tissue specific, or tissue enriched (Table 1). Ubiquitous expression is designated as those mature miRNAs that are broadly expressed in all or most tissues. Tissue-specific expression was defined as those miRNAs whose expression is 20-fold or higher in the specific tissues compared with the mean of the other tissues. For many specific miRNAs, the expression in the nonspecific tissues was undetectable, so it was not possible to calculate a fold-change. The tissue-enriched designation was reserved for mature miRNAs whose expression was less than 20-fold in the enriched tissues compared with the mean of the other tissues. Tissues expressing the most specific miRNAs include brain, heart, skeletal muscle, and pancreas (Table 1).
TABLE 1.
microRNAs that display tissue-specific or tissue enriched expression
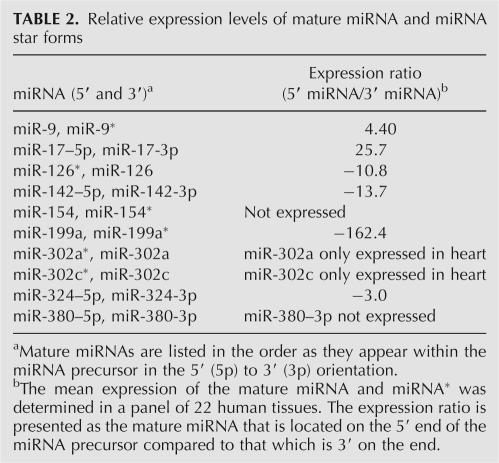
Several miRNAs have the ability to process different mature miRNAs from either end of the same precursor. The mature miRNA is referred to as 5′ or 3′ (e.g., miR-142-5p and miR-142-3p) or by the star designation (e.g., miR-9 and mir-9*) (Griffiths-Jones 2004). TaqMan assays were available to quantify 10 of these miRNAs, thus enabling us to study their processing patterns in a variety of cell lines and tissues. Interestingly, the expression of both the 5′ and 3′ mature miRNA was detectable in most of the cell lines and tissues with no apparent preference to process one or the other miRNA (Table 2). We also compared the relative expression levels of both 5′ and 3′ miRNA. For some miRNAs, the 5′ was expressed at somewhat higher amounts, and for other miRNAs, expression of the 3′ was more predominate (Table 2). In some situations, only one of the two mature miRNAs was detectable, for example, miR-302a and miR-302c but not miR-302a* or miR-302c* (Table 2); however, this was rare. Asymmetric RISC assembly accounts for one strand of the miRNA to accumulate (i.e., guide strand) and the other strand (i.e., passenger strand) to be degraded (Schwarz et al. 2003). Asymmetric RISC assembly likely accounts for one of the two mature miRNAs to accumulate at slightly higher levels. However our data demonstrate that both the 5′ and 3′ miRNAs accumulate in tissues at significant levels, adding to the complexity of the activity of miRNAs.
TABLE 2.
Relative expression levels of mature miRNA and miRNA star forms
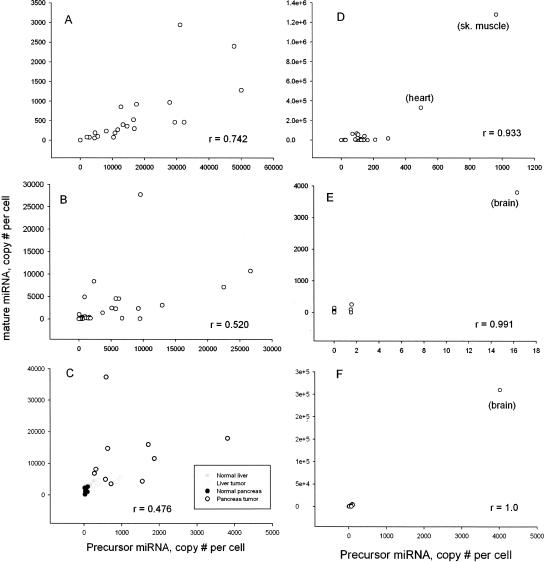
The correlation between the expression levels of the precursor and mature miRNAs was determined from the Pearson coefficient. The Pearson coefficient varied widely among different cells/tissues and miRNA. For many miRNAs, the Pearson's correlation was > 0.6 (P < 0.05), demonstrating good correlation between the expression of precursor and mature miRNA (Supplemental Tables 1–4). For other comparisons, the Pearson value was near zero or negative, demonstrating that the expression of the miRNA precursor did not correlate to the mature miRNA. miR-100 is an example of a miRNA with good correlation between precursor and mature miRNA levels in the cell lines, tissues, and tumors (Fig. 1A–C). One hundred twenty-five miRNAs displayed good correlation (Pearson's ≥ 0.3, P < 0.05) between the expression of the precursor and mature miRNA (Supplemental Tables 1–4).
FIGURE 1.
microRNAs with good correlation in expression levels of precursor and mature. The expression of the precursor and mature miRNAs was determined using real-time PCR assays specific to each class of RNA. Plots of the correlation between the precursor and mature miR-100 in a panel of 22 normal tissues (A), 37 cancer cell lines (B), and the pancreas and liver tumors and normal tissues (C) are shown along with the Pearson correlation coefficient. Also shown is the correlation in the expression of precursor and mature miRNAs in the panel of 22 normal tissues for miR-1 (D), miR-137 (E), and miR-9 (F).
Other situations displayed good correlation between the expression of precursor and mature miRNA in specific tissues, while the expression of both precursor and mature miRNA in the other tissues was very low or undetectable. These tend to be the tissue specific miRNAs listed in Table 1 and were more prevalent among the tissue panel than in the cell lines. Examples include miR-137 and miR-9 in brain and miR-1-1 in skeletal muscle and heart (Fig. 1D–F).
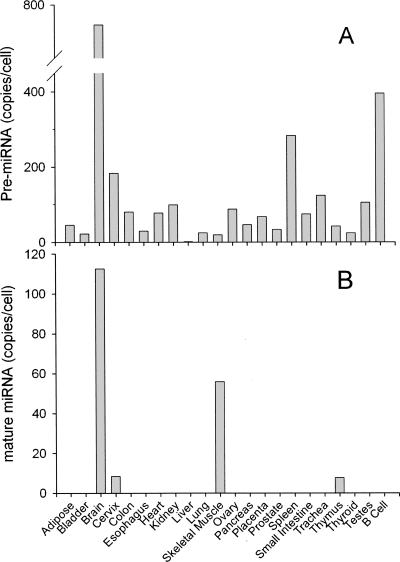
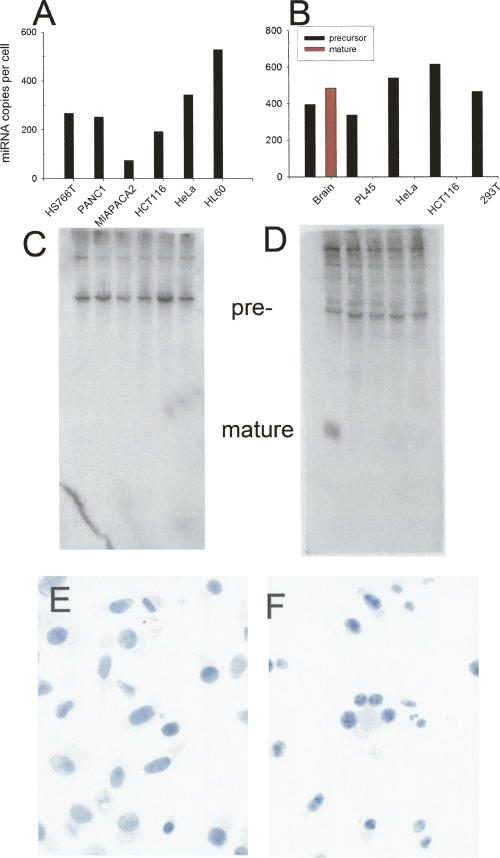
Still other situations exist where high amounts of precursor were present in many samples, yet mature miRNA was undetectable or had very low expression. The precursor to the brain-specific miR-128a was expressed predominately in brain tissue, yet it was expressed at ∼100 copies per cell in many tissues (Fig. 2A). Expression of the mature miRNA was detectable only in brain and skeletal muscle (Fig. 2B). miR-128a precursor was present in all of the cancer cell lines at levels of several hundred copies per cell, yet mature miR-128a was not expressed in any of these cell lines (Fig. 3B). miR-105 had detectable precursor expression, at several hundred copies per cell, in HS766T, Panc1, MiaPaca2, HCT-116, HeLa, and HL-60 cells, but these cells had undetectable mature miRNA (Fig. 3A).
FIGURE 2.
Expression of miR-128a in 22 normal tissues and cells. The expression of miR-128a precursor (A) and mature miR-128a (B) was determined by quantitative real-time PCR in 22 normal human tissues and cells.
FIGURE 3.
Lack of microRNA processing in cancer cell lines. The expression of precursor and mature miR-105 (A) and miR-128a (B) was assayed in several human cancer cell lines by real-time PCR. Mir-128a was also detected in mouse brain tissue. Detection of the primary precursor (black columns) was apparent in all cell lines; expression of mature miRNA (red column) was detectable only in the brain tissue. With the exception of mouse brain tissue, Northern blots to miR-105 (C) and miR-128a (D) reveal the presence of the precursor only. Cell lines are the same as shown for the quantitative PCR. In situ RT-PCR results for pre-miR-105 in HeLa cells (E, note intense nuclear localization) and pre-miR-128a in HS766T cells (F).
The precursor PCR assay simultaneously measures the pri- and pre-miRNA (Schmittgen et al. 2004). We asked the following question: Is the mature miRNA absent in some of these cell lines because the pri-miRNA is not processed by Drosha to the pre-miRNA? To answer this question, we used Northern blotting to detect the pre-miRNA. Our experiments required the enhanced sensitivity of LNA probes to visualize many of the miRNA precursors. The precursor to miR-128a was present in four cancer cell lines and mouse brain; however, the mature miR-128a was present only in mouse brain (Fig. 3D). Precursor to miR-105 was clearly visible by Northern blotting in 6 cancer cell lines, but mature miR-105 was not (Fig. 3C).
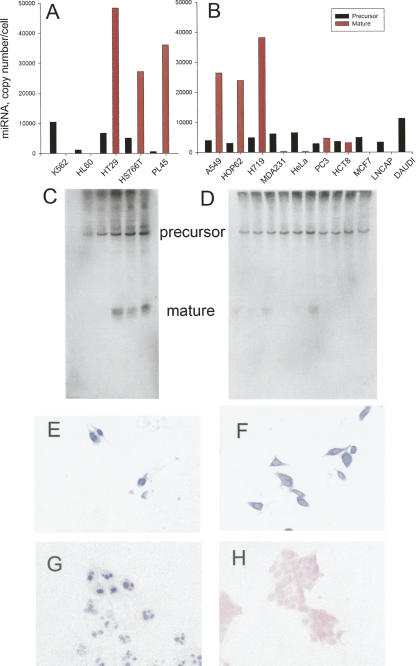
miR-31 was of particular interest because it displayed the greatest amount of differential processing of any of the miRNAs studied. This differential processing was validated by both real-time PCR and Northern blotting (Fig. 4). Mature miR-31 was detectable by real-time PCR and was clearly visible by Northern blotting in the cancer cell lines HS766T and PL45 (pancreas) and HT-29 (colorectal) but not in the hematological cell lines (K562 or HL-60) (Fig. 4C,D). Mature miR-31 was detectable at low to high levels in A549, HOP62, H719, MDA231, HeLa, PC3, and HCT8 cells but was undetectable in MCF-7, LNCaP, and Daudi cells (Fig. 4). Interestingly the miR-31 precursor was detectable by Northern blotting in each of these cell lines (Fig. 4C,D). Our data clearly demonstrate situations where the miRNA primary precursors are transcribed and processed to the precursor miRNA yet are not processed to mature miRNA.
FIGURE 4.
miR-31 expression in cancer cell lines. The expression of precursor and mature miR-31 was assayed in 15 cancer cell lines. Detection of the primary precursor (black columns) was apparent in all cell lines by real-time RT-PCR (A, B); however, the mature miR-31(red columns) was present at varying levels or not at all. (C, D) Northern blot confirmation of precursor and mature miR-31 in cell lines. Note the presence of the ∼70-nt precursor in each of the cell lines. Each column of the Northern blot corresponds to the identical cell lines listed in panels A and B. In situ RT-PCR to miR-31 precursor in HS766T cells (E) and MCF-7 cells (G). In situ hybridization using an LNA probe to mature miR-31 in HS766T cells (F) and MCF-7 cells (H). Magnification, ×400.
Precursor miRNA is generated by Drosha in the nucleus and then transported to the cytoplasm by Exportin 5. Since Dicer processing occurs in the cytoplasm, we asked the following question: Are certain miRNA precursors not processed to the mature miRNA because they remain in the nucleus rather than being transported to the cytoplasm? To answer this question, we applied in situ RT-PCR and in situ hybridization to detect the precursor and mature miRNAs, respectively. For miRNAs that are processed to mature (e.g., miR-31 in HS766T cells), the precursor staining was apparent in both the nucleus and cytoplasm, while the mature miRNA was present only in the cytoplasm (Fig. 4E,F). For miRNAs that are not processed to the mature form, the precursor staining was primarily nuclear and/or nucleolar (Figs. 3E,F, 4G). This phenomena is particularly striking for miR-31 precursor expression in MCF-7 cells; extensive positive staining for pre-Mir-31 was apparent in the nucleolus only (Fig. 4G). MCF-7 cells are negative for mature Mir-31 (Fig. 4H). For miR-31, miR-105, and miR-128a in these cell lines, processing to the mature form does not proceed because the miRNA precursor is retained in the nucleus. The in situ PCR signal is for the precursor miRNA (as well as the primary precursor) because the precursor miRNAs were detected by Northern blotting (Figs. 3, 4).
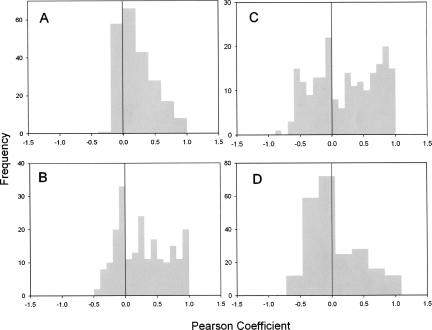
The Pearson correlation was compared among the cell lines, tissues, and tumors by plotting the frequency versus the Pearson coefficient (Fig. 5). The cell lines produced a unimodal distribution centering near zero (Fig. 5A). The histogram for the 22 normal tissues produced a bimodal distribution with one mode centered near zero and the other greater than zero (Fig. 5B). The pancreas and liver tissues also produced conflicting histograms, with the liver specimens displaying a unimodal distribution centering at zero while the pancreas tissues were bimodal (Fig. 5C,D). A high positive Pearson correlation demonstrates that the expression of precursor and mature miRNAs are directly proportional and suggests efficient processing of precursor to mature miRNA. A low Pearson coefficient suggests no relationship between the two, one explanation being that precursors are not being processed to mature miRNA. A negative Pearson suggests an inverse relationship between mature and precursor miRNA. It should be emphasized that of over 800 comparisons, only 27 negative Pearson coefficients were significant (P < 0.05). Our data suggest that processing of many precursor miRNAs is somehow deficient or defective in cancer cell lines and liver tissues. However in 22 normal tissues and pancreas tissues, there is a population of miRNAs that are processed efficiently. This trend parallels the miRNA expression pattern in tumors. In pancreas cancer, there is a predominate increase in the expression of the miRNA (Lee et al. 2007), while in liver cancer, the opposite is true (Gramantieri et al. 2007; Jiang et al. 2007). This suggests that in pancreatic cancer, the increased expression of miRNA is regulated at the level of transcription with no apparent alterations in processing, but in liver cancer, the reduced expression of miRNA in the tumors results from a reduction in miRNA processing.
FIGURE 5.
Pearson correlation among the various samples studied. The frequency of the Pearson's correlation coefficient describing the relationship between the precursor and mature miRNA expression is plotted for the cell lines (A), 22 normal tissues (B), normal pancreas and pancreas tumors (C), and normal liver and liver tumors (D).
It was recently demonstrated that mutations (Calin et al. 2005) or SNPs (Duan et al. 2007) within the miRNA precursors can cause interference with miRNA processing. While we believed this to be an unlikely explanation to describe the lack of miRNA processing, since the phenomena was so widespread (Figs. 3, 4), we sequenced +100 bp on either side of the miR-31 hairpin in HeLa and MCF-7 cell lines to look for evidence of mutations or SNPs. The sequences were unchanged compared with wild type, demonstrating that reduced levels of mature Mir-31 in these cells was not due to reduced processing caused by a mutation or SNP.
The Pearson's correlation data along with the relative expression levels of precursor and mature miRNA demonstrated that biogenesis of miRNAs fit into different categories. In one group of miRNAs, the precursor and mature miRNAs are expressed in most tissues and cell lines, and the precursor expression correlates with the mature miRNA expression. These miRNAs typically had a Pearson correlation coefficient > 0.3 and P < 0.05. Examples of these miRNAs include many of the ubiquitously expressed miRNAs such as miR-100, miR-155, and miR-125a. Expression of these miRNAs would presumably be regulated by transcription alone with no regulation of processing events (i.e., all miRNA expressed as primary transcripts are processed to mature miRNA).
Another group of miRNAs, such as many of the tissue-specific miRNAs, have a high amount of expression of both the precursor and mature in specific tissues only. So expression of these miRNAs is regulated by transcription in specific tissues with no apparent regulation of miRNA biogenesis. Examples of these miRNAs include miR-9, miR-1, and miR-216. Still another group of miRNAs expressed the precursor at significant levels in all or most of the cells or tissues; however, the mature miRNA is not present. Examples include miR-105, miR-128a, and miR-31 in cancer cell lines (Figs. 3, 4) and represent miRNAs that are expressed at the primary transcript level yet for some unknown reason are unable to be processed to the mature miRNA.
Our data clearly show that a large number of miRNAs are transcribed as precursors but are not processed to mature miRNA. Conceivably any of the steps involved in miRNA biogenesis (nuclear transport or processing by Drosha or Dicer) could be involved to reduce the mature miRNA levels. Reduced levels of Dicer were shown in lung tumors (Karube et al. 2005), and many of the cancer cell lines studied here had reduced mature miRNA. It is unlikely that a whole-scale reduction in Drosha or Dicer processing accounts for the reduced levels of mature miRNA in cells that express the miRNA precursors since other miRNAs are efficiently processed to mature in these same cell lines or tissues. One explanation to account for reduced nuclear transport or processing by Drosha or Dicer in some miRNAs but not others, is that other specific factors bind to the miRNA precursors and either promote or interfere with processing or transport. In fact, this inhibitory model was proposed by Obernosterer et al. (2006) to account for a lack of mature miR-138 in HeLa cells. It was recently shown that hnRNP A1 is required for Drosha processing of miR-18 from the primary precursor (Guil and Caceres 2007). Conceivably, other hnRNPs may be involved in regulating steps of miRNA biogenesis. While we did not use Northern blotting and in situ hybridization to study all of the miRNAs with reduced processing, the ones that were studied in several cancer cell lines showed that the miRNA was processed to the precursor but was not transported from the nucleus to the cytoplasm. This suggests an intriguing, unknown mechanism to guide the subcellular distribution of miRNA precursors and prevent them from being processed to the active mature miRNA. It is our hope that with the availability of this data set, many future studies will be initiated to unravel the various mechanisms that control miRNA biogenesis.
MATERIALS AND METHODS
Cells, tissues and RNA
Human cancer cell lines were obtained from ATCC or other investigators. Human total RNA from 20 tissues was from Ambion. CD19+ human B cells were purchased from AllCells. RNA from pancreas and liver tumor and adjacent benign pancreas were obtained from our prior studies (Jiang et al. 2007; Lee et al. 2007). RNA was isolated from all samples using Trizol per the manufacturer's instructions. RNA purchased from Ambion was isolated using ToTALLY RNA (Ambion). Both Trizol and ToTALLY RNA are similar to the widely used guanidine thiocyanate, acid phenol:chloroform procedure of Chomczynski and Sacchi (1987) and will contain mature miRNA. RNA integrity was evaluated by analyzing 50–100 ng of total RNA using the Agilent 2100 Bioanalyzer.
Real-time RT PCR, miRNA precursors, and mature miRNA
Expression data on 205 miRNA precursors in human cancer cell lines, pancreas, and liver tissues were as previously reported (Jiang et al. 2005, 2007; Lee et al. 2007). The expression of 201 miRNA precursors in the 21 normal human tissues was determined using real-time RT PCR as previously described (Schmittgen et al. 2004; Jiang et al. 2005). Mature miRNA expression in the normal tissues, cell lines, pancreas, and liver tissues was determined using a real-time PCR assay to mature miRNA as described (Chen et al. 2005) with several modifications. cDNA to the mature miRNA was synthesized using gene specific looped primers. Primers were grouped into five portions of 48-plex each. Mature miRNAs were selected only to match the miRNAs on which we had precursor data. Five different cDNAs were synthesized for each sample using one of the five RT 48 plex primer mix as described (Chen et al. 2005).
Real-time RT PCR, absolute copy number for precursor and mature miRNA
Absolute copy number of miR-155 precursor was determined by constructing a standard curve of pre-miR-155. Pre-miRNA RNA was synthesized using in vitro transcription using primers that annealed to ∼50 base pairs (bp) upstream of and downstream from the miR-155 hairpin. The T7 promoter sequence was added to 5′ end of the forward primer. Amplification of the pri-miRNA gene was performed by amplifying genomic DNA (Roche) using standard procedures. The PCR product served as a template for in vitro transcription. The transcription product was gel purified, and the concentration was measured using a Nanodrop spectrophotometer. The reverse transcription reaction was performed as described above, and standard curves of the log of pre-Mir-155 copy number versus the CT were made. We also generated a standard curve for mature miR-155 as described (Chen et al. 2005). A minimum of 10 copies/cell was used as the selection criteria for cutoff. The raw data set for the precursor and mature miRNA expression levels is presented in the supplemental data section.
Northern blotting
Northern blotting was performed as previously described (Lau et al. 2001). LNA probes specific to the mature miRNA were purchased from Exiqon.
In situ detection of miRNA
RT in situ PCR to detect the miRNA precursors was used as previously described (Lee et al. 2007). The identical primers used to detect the miRNA precursors by real-time PCR were used for the in situ RT-PCR. The only exception was miR-31; solution PCR used miR-31 primary precursor primers, while the in situ PCR used precursor miR-31 primers. The chromogen is nitroblue tetrazolium and bromochloroindolyl phosphate with nuclear fast red as the counterstain. Therefore positive signal will appear blue, and a negative signal will appear pink. Mature miRNA was detected by in situ hybridization using LNA probes specific to the mature miRNA (Exiqon).
Data analysis
Pearson product moment correlation coefficients were determined for each pair of precursor and mature miRNA using Microsoft Excel. The significance (probability) of the correlation coefficients for each pair was determined from the t-statistic. The probability of the t-statistic indicates whether the observed correlation coefficient occurred by chance if the true correlation is zero. Correlation coefficient was considered to be significantly different from zero at the 0.05 level (P ≤ 0.05). P values for all of the correlations may be located in the supplemental data section.
SUPPLEMENTAL DATA
Supplemental Materials can be found at www.pharmacy.ohio-state.edu/programs/ceut/faculty/schmittgen/supp_materials.cfm.
ACKNOWLEDGMENTS
We thank Caifu Chen at Applied Biosystems for his assistance with the TaqMan microRNA assays. This work was supported by grant R21 CA114304 to T.D.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.804508.
REFERENCES
- Calin, G.A., Ferracin, M., Cimmino, A., Di Leva, G., Shimizu, M., Wojcik, S.E., Iorio, M.V., Visone, R., Sever, N.I., Fabbri, M., et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Chen, C., Ridzon, D.A., Broomer, A.J., Zhou, Z., Lee, D.H., Nguyen, J.T., Barbisin, M., Xu, N.L., Mahuvakar, V.R., Andersen, M.R., et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Duan, R., Pak, C., Jin, P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- Gramantieri, L., Ferracin, M., Fornari, F., Veronese, A., Sabbioni, S., Liu, C.G., Calin, G.A., Giovannini, C., Ferrazzi, E., Grazi, G.L., et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil, S., Caceres, J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Lee, E.J., Gusev, Y., Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Lee, E.J., Schmittgen, T.D. Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer. 2006;45:103–106. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Gusev, Y., Aderca, I., Mettler, T.A., Nagorney, D.M., Brackett, D.J., Roberts, L.R., Schmittgen, T.D. microRNA expression in hepatocellular carcinoma is associated with hepatitis infection, cirrhosis and patient's survival. Clin Cancer Res. 2007 doi: 10.1158/1078-0432.CCR-07-0523. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karube, Y., Tanaka, H., Osada, H., Tomida, S., Tatematsu, Y., Yanagisawa, K., Yatabe, Y., Takamizawa, J., Miyoshi, S., Mitsudomi, T., et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V.N., Nam, J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee, E.J., Gusev, Y., Jiang, J., Nuovo, G.J., Lerner, M.R., Frankel, W.L., Morgan, D.L., Postier, R.G., Brackett, D.J., Schmittgen, T.D. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Ridzon, D., Wong, L., Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E., Guttinger, S., Calado, A., Dahlberg, J.E., Kutay, U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Meng, F., Henson, R., Lang, M., Wehbe, H., Maheshwari, S., Mendell, J.T., Jiang, J., Schmittgen, T.D., Patel, T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Michael, M.Z., van Holst, S.M.O.C., Pellekaan, N.G., Young, G.P., James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- Obernosterer, G., Leuschner, P.J., Alenius, M., Martinez, J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D., Jiang, J., Liu, Q., Yang, L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:E43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Thomson, J.M., Newman, M., Parker, J.S., Morin-Kensicki, E.M., Wright, T., Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]