Abstract
Effective turnover of many incorrectly processed RNAs in yeast, including hypomodified tRNAi Met, requires the TRAMP complex, which appends a short poly(A) tail to RNA designated for decay. The poly(A) tail stimulates degradation by the exosome. The TRAMP complex contains the poly(A) polymerase Trf4p, the RNA-binding protein Air2p, and the DExH RNA helicase Mtr4p. The role of Mtr4p in RNA degradation processes involving the TRAMP complex has been unclear. Here we show through a genetic analysis that MTR4 is required for degradation but not for polyadenylation of hypomodified tRNAi Met. A suppressor of the trm6-504 mutation in the tRNA m1A58 methyltransferase (Trm6p/Trm61p), which causes a reduced level of tRNAi Met, was mapped to MTR4. This mtr4-20 mutation changed a single amino acid in the conserved helicase motif VI of Mtr4p. The mutation stabilizes hypomodified tRNAi Met in vivo but has no effect on TRAMP complex stability or polyadenylation activity in vivo or in vitro. We further show that purified recombinant Mtr4p displays RNA-dependent ATPase activity and unwinds RNA duplexes with a 3′-to-5′ polarity in an ATP-dependent fashion. Unwinding and RNA-stimulated ATPase activities are strongly reduced in the recombinant mutant Mtr4-20p, suggesting that these activities of Mtr4p are critical for degradation of polyadenylated hypomodified tRNAi Met.
Keywords: DExH helicase, tRNA degradation, polyadenylation, ATPase
INTRODUCTION
Eukaryotic pre-tRNA is extensively processed before becoming functional in translation (Wolin and Matera 1999). The processing of pre-tRNA includes 5′-end cleavage, intron removal, 3′-end trimming, 3′-end CCA addition, and ribose or nucleotide modification (Hopper and Phizicky 2003). One such modified nucleotide, 1-methyladenosine (m1A), at position 58 of the TΨC loop, is synthesized by a methyltransferase encoded by two essential genes, TRM6 and TRM61 (Anderson et al. 1998). Hypomodified tRNAi Met lacking m1A58 is preferentially degraded in a process involving Trf4p, a poly(A) polymerase that creates short poly(A) tails (Vanacova et al. 2005). The role of poly(A) tails in RNA processing or degradation is not well understood.
Recent evidence indicates a role of the nuclear exosome, a multiprotein complex composed of 3′–5′ riboexonucleases, in the degradation of RNAs with short poly(A) tails (Mitchell et al. 1997; van Hoof et al. 2000; Mitchell 2001). The nuclear exosome is related to the cytoplasmic exosome, but employs different accessory factors including Rrp47, Mtr4p (Mitchell and Tollervey 2003), and Rrp6p (Mitchell et al. 1997; van Hoof et al. 2000). MTR4 was identified and named in a genetic screen for genes required for mRNA transport in Saccharomyces cerevisiae (Kadowaki et al. 1994). MTR4 is allelic with DOB1, which was identified as an essential component of the 3′–5′ processing of pre-rRNA (de la Cruz et al. 1998). Mtr4p functions together with Trf4p [poly(A) polymerase] and Air2p (RING finger protein that presumably binds RNA) in the oligomeric TRAMP complex, which has been implicated in the polyadenylation and facilitation of RNA processing or degradation by the exosome (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006). The role of Mtr4p in the degradation of RNAs with short poly(A) tails is unclear, and it is an open question whether Mtr4p functions in the polyadenylation step, the degradation step, or both.
Mtr4p belongs to the DExH/D protein family. Proteins from this family are thought to remodel RNAs and RNA–protein complexes in an ATP-dependent fashion and are often referred to as RNA helicases (Tanner and Linder 2001). Accordingly, more than 60 individual DExH/D proteins have been shown to unwind RNA duplexes in vitro (Jankowsky and Fairman 2007). DExH/D proteins share at least eight characteristic sequence motifs, which are highly conserved from bacteria to humans (Rocak and Linder 2004). Based on sequence characteristics, DExH/D proteins are further divided into three subfamilies—the DEAD, DEAH, and DExH helicases—named after the conserved helicase motif II (in single-letter amino acid code) (Rocak and Linder 2004). Recent evidence suggests that proteins from these subfamilies use distinct mechanisms for RNA and RNP remodeling (Yang and Jankowsky 2006; Jankowsky and Fairman 2007). Mtr4p is a DExH protein, with high sequence similarities to other cellular DExH proteins (e.g., Ski2p, Brr2p, Suv3p) and to viral DExH proteins such as NPH-II and HCV NS3 (Tanner and Linder 2001). While RNA unwinding by these viral enzymes is comparably well understood, little is known about unwinding characteristics of cellular DExH proteins (Jankowsky and Fairman 2007).
Here we show that Mtr4p is required for the degradation but not for polyadenylation of hypomodified tRNAi Met lacking m1A58. Biochemical studies of Mtr4p reveal RNA-dependent ATPase and RNA-unwinding activity with a 3′-to-5′ polarity. RNA-dependent ATPase and RNA-unwinding activities are greatly reduced in a mutant Mtr4p that is functional for polyadenylation but not for RNA degradation. Collectively, our findings suggest that RNA- dependent ATP hydrolysis and presumably also RNA unwinding by Mtr4p are critical for degradation of polyadenylated hypomethylated tRNAi Met, raising the possibility that RNAs with 3′-poly(A) tails are substrates for Mtr4p.
RESULTS
Identification of sup3 as MTR4
We previously reported that a mutation in the gene encoding one subunit of the tRNA m1A methyltransferase, trm6-504, confers a ts− growth phenotype at 37°C and resistance to the amino acid analog 3-aminotriazole (3-ATr). The ts− phenotype of trm6-504 can be suppressed by multicopy plasmids bearing IMT1-4, genes encoding initiator tRNAMet (tRNAi Met) (Anderson et al. 2000), and by increasing the steady-state amount of tRNAi Met. We selected for spontaneous reversion of the trm6-504 ts− phenotype to reveal genetic suppressors, and this analysis yielded at least three complementation groups (Kadaba et al. 2004). Identification of the wild-type (WT) genes representing complementation groups 1 (TRF4) and 2 (RRP44) led to the understanding that tRNAi Met lacking m1A58 (hypomodified) is polyadenylated and degraded in the nucleus of yeast cells (Kadaba et al. 2004).
As the nuclear exosome had been implicated in the degradation of hypomethylated tRNAi Met (Kadaba et al. 2004), we tested whether the third complementation group represented a gene encoding one of the core components of the exosome, or an accessory factor that stimulates tRNA degradation by the exosome. First, we examined whether the sup3 mutant phenotypes could be complemented by the exosome components RRP6, RRP4, or the TRF4 paralog TRF5, in single-copy plasmids. No complementation was observed (data not shown).
Next, we tested if the sup3 mutant could be complemented by MTR4, an accessory factor of the nuclear exosome (van Hoof et al. 2000). We introduced a low-copy plasmid bearing a WT HA-tagged MTR4 into the Sup3 strain. The transformants were tested for complementation of the sup3 mutant by replica plating onto selective minimal media and selective minimal media containing 3-AT. Transformants carrying WT MTR4 could not grow at 37°C and grew on 3-AT plates, while the same strain transformed with an empty plasmid grew at 37°C and did not grow on plates containing 3-AT (Fig. 1A). The complementation of Sup3 by MTR4 suggested that sup3 is MTR4. To confirm the identity of sup3 as MTR4, we integrated a selectable marker (URA3) near the MTR4 locus in the Sup3 mutant strain using DNA outside the MTR4 open reading frame (ORF) as the targeting sequence. Stable transformants were screened for the presence of plasmid DNA at the targeted site near the MTR4 locus using PCR (data not shown). In order to demonstrate a physical linkage between the selectable marker and sup3, we created a diploid strain homozygous at trm6-504 and heterozygous at sup3 by mating the sup3∷URA3-containing strain to a trm6-504 isogenic strain of opposite mating type. Tetrad analysis was done with the trm6-504/trm6-504 sup3/SUP3+ diploid, and the selectable marker and suppressor phenotypes cosegregated in all 20 tetrads tested (data not shown). These results linking sup3 to the MTR4 locus are consistent with the results of the complementation experiments. Collectively, the above data identify sup3 as MTR4.
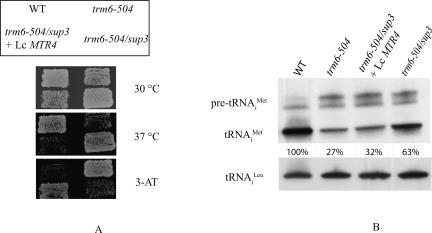
FIGURE 1.
A low-copy plasmid bearing MTR4 complements the mutant phenotypes of a trm6-504/sup3/gcn2-101 strain. (A) Wild-type (Y200), trm6-504 (Y190), and trm6-504/sup3 (Sup3) strains were transformed with YCpLac33 or a low-copy number MTR4 plasmid. Strains were grown as patches on synthetic complete (SC) plates lacking uracil at 30°C and replica-plated to SC plates lacking uracil and histidine supplemented with 30 mM 3-aminotriazole (3-AT) or SC plates lacking uracil and incubated 30°C and 37°C, respectively. (B) Northern blot analysis of total RNA (10 μg) isolated from the same strains described in A grown at 30°C in SC medium lacking uracil. Hybridization with probes JA11 (tRNAi Met) and JA151 (tRNACCA Leu) were performed as described in Materials and Methods, followed by autoradiography or PhosphorImager analysis and quantification using Image Quant software. tRNAi Met was normalized to the amount of tRNACCA Leu in the same sample and is expressed as percentage of the wild type.
To further characterize the effect of sup3 on tRNAi Met instability, we examined the tRNAi Met steady-state level in the mtr4/trm6-504 strain by conducting Northern blot analysis of total RNA from WT, trm6-504, trm6-504/sup3 carrying empty control plasmids, and trm6-504/sup3 carrying a low-copy number (lc) MTR4 plasmid (Fig. 1B). The steady-state level of mature tRNAi Met in the trm6-504 strain was 27% of WT, but the presence of sup3 along with trm6-504 led to an increase in the level of tRNAi Met to 63%. This was in contrast to the trm6-504/sup3 strain bearing lcMTR4, where the level of tRNAi Met was 32% of WT and not significantly different from that in trm6-504. This result provides further evidence that Mtr4p participates in the degradation of hypomodified tRNAi Met.
The mtr4-20 mutation maps to the helicase motif VI
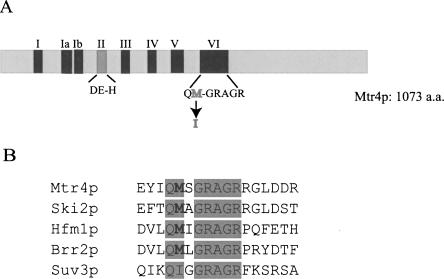
To determine the location of the trm6-504 suppressor mutation in MTR4, the entire MTR4 ORF was amplified by PCR from trm6-504/sup3 genomic DNA, cloned, and sequenced. This analysis revealed one point mutation, a C → G change at nucleotide 1620 of the MTR4 ORF, which changed methionine 540 to isoleucine (Fig. 2A). We designated this mutation mtr4-20. The mtr4-20 mutation was further confirmed by DNA sequence analysis of five independent 1-kb PCR products that included nucleotide 1620 of the mtr4-20 mutation. Each of these PCR products contained the identical C → G change. The mutated methionine 540 is located in the highly conserved helicase motif VI. A methionine in this position is conserved in the other DExH proteins from yeast (Fig. 2B), and in many other DExH proteins (data not shown).
FIGURE 2.
Schematic representation of MTR4. (A) (Light gray box) The MTR4 ORF; (black and dark gray boxes) the conserved motifs found in most RNA helicases. The conserved amino acids in motif II and motif VI are shown under the boxes. The amino acid mutated in mtr4-20 is indicated in motif VI (methionine to isoleucine). (B) Alignment of Motif VI between members of the DExH subfamily in yeast. The shaded background designates identical amino acids in motif VI. The methionine (bold), which is mutated in mtr4-20p to isoleucine and highly conserved among members of the Ski2p subfamily, is indicated in bold.
Hypomodified tRNAi Met is polyadenylated but not efficiently degraded in the trm6-504/mtr4-20 mutant strain
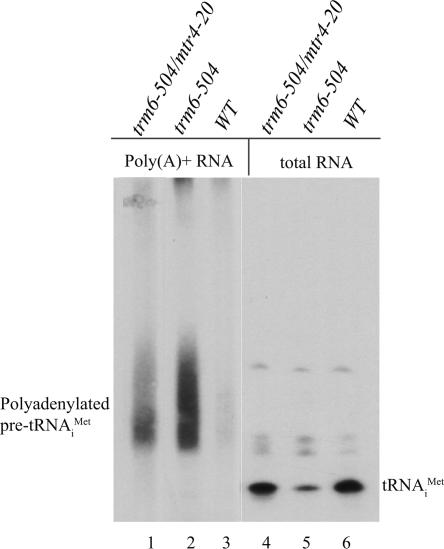
As mentioned above, it is thought that hypomodified pre-tRNAi Met is degraded by the nuclear exosome, but only after initial polyadenylation by the TRAMP complex that contains Trf4p, Air2p, and Mtr4p (Kadaba et al. 2004). Mtr4p has been implicated in the turnover of polyadenylated RNAs through the observation that mutation or depletion of Mtr4p leads to accumulation of poly(A)+ snoRNAs and rRNAs that are not normally polyadenylated (Liang et al. 1996; van Hoof et al. 2000). We wished to test whether mtr4-20 blocks polyadenylation of hypomethylated tRNAi Met or if it inhibits degradation of the poly(A)+ tRNAi Met. First, we tested whether hypomethylated tRNAi Met is polyadenylated in a trm6-504/mtr4-20 mutant. Poly(A)+ RNA was selected from total RNA isolated from trm6-504, trm6-504/mtr4-20, and WT strains, and Northern blot analysis was used to detect the presence of poly(A)+ tRNAi Met. The strong presence of tRNAi Met in the poly(A)+ pool from trm6-504 and trm6-504/mtr4-20 mutants, and its complete absence from WT yeast shows that hypomethylated tRNAi Met is polyadenylated in the mtr4-20 mutant (Fig. 3). This observation suggests that Mtr4p does not function in polyadenylation, but at a subsequent step, at or after the initiation of RNA degradation by the exosome.
FIGURE 3.
Pre-tRNAi Met lacking m1A58 is polyadenylated in trm6-504/ mtr4-20. (Lanes 4–6) Total RNA (5.0 μg) or (lanes 1–3) poly(A)+ RNA (2.0 μg) isolated from the indicated strains cultured at 30°C was separated by denaturing polyacrylamide gel electrophoresis and transferred to a membrane. Hybridization was performed with radiolabeled probe JA11 (tRNAi Met), and tRNAi Met was visualized by autoradiography.
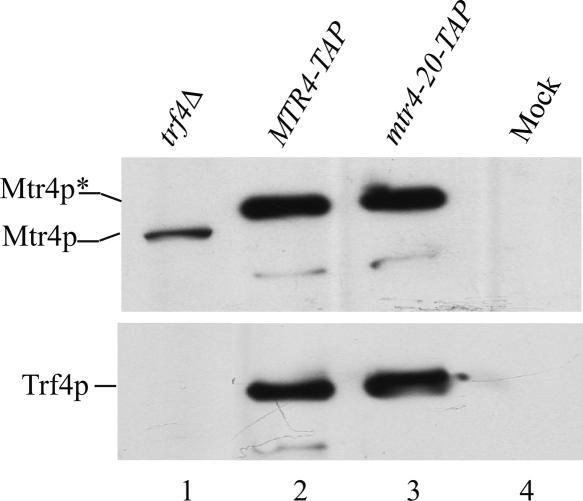
Budding yeast contains two nuclear poly(A) polymerases, Trf4p and Trf5p, that are likely to function on different substrates (Houseley and Tollervey 2006). It was thus possible that hypomodified tRNAi Met was polyadenylated in the mtr4-20 mutant not by TRAMP4–Trf4p but by the functionally distinct TRAMP5 complex containing Trf5p and Air1p. To test whether TRAMP4 is able to polyadenylate an RNA in the presence of Mtr4-20p, we purified TRAMP4 complexes from Mtr4p–TAP and Mtr4-20p–TAP tandem-affinity-tagged strains, and measured RNA polyadenylation in vitro with these complexes (see below). The components of the purified complexes were visualized by SDS-PAGE and Coomassie-blue staining (data not shown), and the presence of Mtr4p and Trf4p in complexes was confirmed by immunoblot analysis using polyclonal antibodies against Mtr4p or Trf4p (Fig. 4). There was no significant difference in the amount of Trf4p coprecipitating with TAP-tagged Mtr4p or Mtr4-20p proteins, indicating that TRAMP4 does not undergo a gross alteration in structure due to the presence of the mutant Mtr4-20p.
FIGURE 4.
Mtr4-20p and Trf4p are in a complex. Tap-tagged Mtr4p (Y358) and Mtr4-20p (Y359) were purified from whole cell extracts (Materials and Methods). Equivalent amounts of affinity-purified Mtr4p, as determined by immunoblot using anti-Mtr4p polyclonal serum, and whole cell extracts (40 μg) were separated by SDS-PAGE and transferred to a membrane for immunoblot. Rabbit anti-Trf4p or anti-Mtr4p polyclonal serums were used to probe for the respective proteins followed by hrp-coupled goat anti-rabbit IgG antibodies and ECL for detection. Mtr4p with altered mobility due to the presence of the Tap-tag is indicated with an asterisk. (Lane 1) trf4Δ (F22), (lanes 2,3) purified Mtr4p complexes from Tap-Mtr4 (Y358) and Tap-mtr4-20, and (lane 4) the equivalent amount of protein found in lanes 2 and 3 except purified using extract containing untagged Mtr4p (Y200).
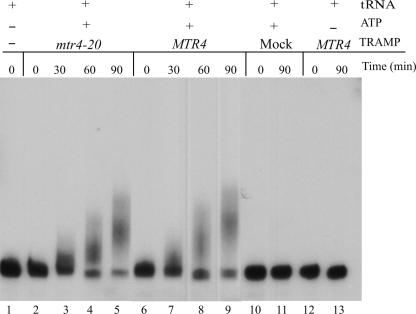
To measure polyadenylation by these purified complexes, we utilized an in vitro transcribed, 5′-end-radiolabeled yeast tRNAi Met. Equal amounts of TRAMP4 complexes, as judged from the Mtr4p level by immunoblot, were purified from the two different strains and incubated at 30°C with the radiolabeled tRNAi Met substrate and ATP. Purified Mtr4-20p TRAMP complexes polyadenylated tRNAi Met as efficiently as Mtr4p TRAMP (Fig. 5). This result shows that TRAMP4 complexes with Mtr4-20p retain polyadenylation activity in vitro.
FIGURE 5.
Mutant and wild-type Mtr4p complexes possess robust polyadenylation activity in vitro. Polyadenylation of radiolabeled synthetic initiator tRNA was conducted with purified complexes containing wild-type Mtr4p (MTR4), Mtr4–20p (mtr4-20), or proteins purified from an extract containing no TAP-tag (Mock). Reactions were incubated from 0 to 90 min with (1 mM) or without ATP at 30°C and once stopped by addition of formamide loading dye separated by denaturing polyacrylamide electrophoresis, dried, and detected by autoradiography. (Lane 1) A control reaction with no protein added.
Taken together, our data thus indicate that TRAMP complexes containing Mtr4-20p are functional in polyadenylation in vivo and in vitro. Consequently, the mutation in Mtr4-20p should be deficient in either initiating or propagating the exosome-mediated degradation of polyadenylated hypomodified tRNAi Met, suggesting a function of Mtr4p in either one or both of these steps.
Mtr4p unwinds RNA duplexes with 3′-to-5′ polarity in an ATP-dependent fashion
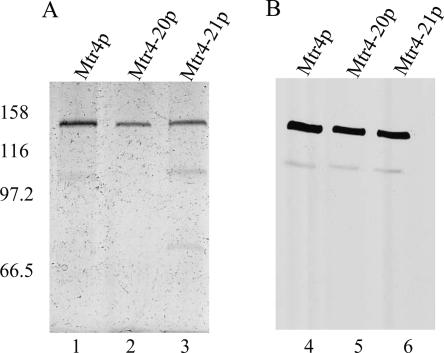
The location of the Mtr4-20p mutation in one of the conserved helicase domains raised the possibility that the observed RNA degradation defect was due to an impaired ability of the mutant protein to hydrolyze ATP and/or to unwind RNA duplexes. To address this question, we measured RNA unwinding, ATP hydrolysis, and RNA binding by WT and mutant proteins. We expressed and purified wild-type Mtr4p, mutant Mtr4-20p, and, as a nonfunctional control, Mtr4-21p (Fig. 6). Mtr4-21p contained mutations in helicase motif II (D, E → A) that abolish RNA-unwinding activity of various other DExH/DEAD-box helicases (Pause and Sonenberg 1992; Iost et al. 1999).
FIGURE 6.
Purification of recombinant wild-type and mutant Mtr4p. 6xHis-tagged Mtr4p was expressed in E. coli and purified using TALON metal affinity resin and anion exchange chromatography (Materials and Methods). Equal amounts of purified protein, as determined by Bradford assay, were resolved on 4%–12% gradient SDS-PAGE gel. (A) Silver staining and (B) Western blot with anti-His antibody were used to visualize and quantify the amount of purified protein in each preparation. (Lanes 1,4) WT Mtr4p; (lanes 2,5) Mtr4-20p; (lanes 3,6) Mtr4-21p contains the changes DEvH to AAvH.
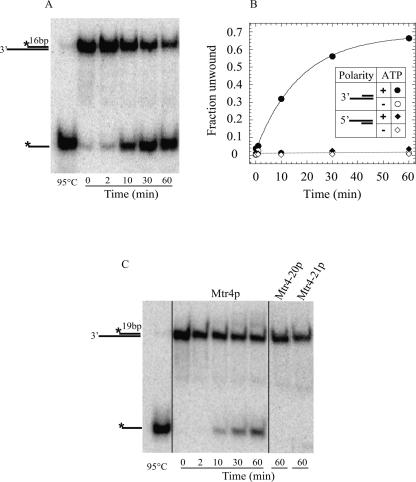
We first examined whether Mtr4p unwound RNA duplexes with a preferred polarity. We measured strand separation with substrates containing a single-stranded region either 3′ or 5′ to the duplex (Fig. 7A). Mtr4p unwound substrates with a single strand 3′ to the duplex region, but not those with an overhang at the 5′ position (Fig. 7A,B). No unwinding was seen in the absence of ATP (Fig. 7B) or with the nonhydrolyzable ATP analog AMPPNP (data not shown). These data indicate that Mtr4p unwinds RNA duplexes with a preferred 3′-to-5′ polarity, utilizing ATP to drive the reaction.
FIGURE 7.
Mtr4p but not Mtr4-20p and Mtr4-21p unwinds RNA duplexes with a 3′-to-5′ polarity in an ATP-dependent fashion. (A) Representative PAGE of an unwinding reaction of a 16-bp duplex [S16-3′] with a 3′-single-stranded overhang by wild-type Mtr4p. Cartoons on the left represent the mobility of duplex and the single-stranded radiolabeled RNA; the asterisk indicates the radiolabel. Reactions were assembled with Mtr4p and without ATP, and an aliquot removed at time zero. After ATP addition, aliquots were removed at the times shown. Complete dissociation of the duplex is shown after incubating the RNA briefly at 95°C. (B) Time courses of unwinding reactions by wild-type Mtr4p for 16-bp duplexes with 3′- and 5′-single-stranded overhangs with and without ATP (inset). Sequences of duplexes and single stranded overhangs were identical (Materials and Methods). Significant unwinding was seen only for the substrate with the 3′-overhang (•). The solid line through these points indicates the best fit to a first-order reaction (Yang and Jankowsky 2005), yielding the reaction amplitude A = 0.65 ± 0.02, and the observed unwinding rate constant k′unw = 0.057 ± 0.003 min−1. (C) Representative PAGE of unwinding reactions with a 19-bp substrate with a 3′-overhang [S19-3′] with wild-type Mtr4p and Mtr4-20p and Mtr4-21p. For the mutant proteins, only aliquots removed after 60 min are shown. Wild-type Mtr4p unwinds the 19-bp substrate with an observed rate constant (k′unw = 0.054 ± 0.007 min−1) highly similar to that seen with the 16-bp duplex, but the reaction amplitude is lower (A = 0.39 ± 0.03) than for the 16-bp duplex.
Next, we tested whether the mutant proteins Mtr4-21p (negative control, motif II mutation) and Mtr4-20p were able to unwind an RNA duplex with a 3′-end extension. No significant strand separation was observed with either protein (Fig. 7B). Thus, unwinding activity was significantly impaired in Mtr4-20p. This observation suggests that an ATP-dependent RNA or RNP remodeling activity of Mtr4p may be involved in the degradation of hypomodified tRNAi Met.
Mtr4-20p binds RNA but does not exhibit ATPase activity
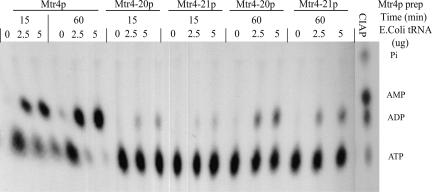
To examine whether the inability of Mtr4-20p to unwind duplex RNA was a result of defective ATPase activity or a failure to bind RNA, we measured ATPase and RNA binding by WT and mutant Mtr4p. WT Mtr4p exhibited a robust RNA-dependent ATPase activity, completely hydrolyzing the input ATP within 60 min (Fig. 8). The Mtr4-20p and Mtr4-21p mutants had little ATPase activity under the same conditions (Fig. 8). Presumably, the vastly reduced ATPase activity of Mtr4-20p also diminishes the ability of the protein to unwind RNA duplexes.
FIGURE 8.
Mtr4-20p is defective in tRNA-dependent ATPase activity. Purified recombinant Mtr4p was incubated with [α-32P]ATP in the presence of 2.5 μg, 5 μg, or absence of E. coli total tRNA. Aliquots of reactions were collected and stopped by the addition of 0.1% SDS, 5 mM EDTA after 15 min and 60 min at 30°C. Visualization of ATP, ADP, AMP, and Pi was accomplished by incubating 1.0 μCi of [α-32P]ATP and 0.025 U of CIAP together for 15 min at 37°C. Samples were resolved by thin layer chromatography (Materials and Methods), and reaction products were visualized by autoradiography. The positions of ATP, ADP, AMP, and Pi are indicated.
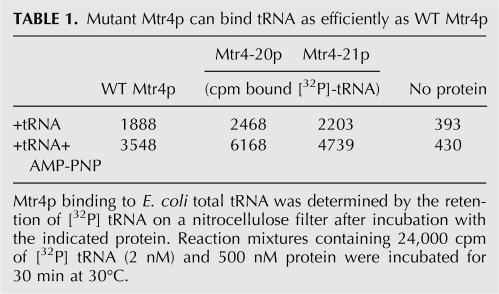
To test whether tRNA binding is affected in the Mtr4p mutants, binding of WT or mutant Mtr4p proteins to radiolabeled Escherichia coli tRNA was measured using a filter-binding assay (Materials and Methods). No significant differences in tRNA binding between the WT and mutant Mtr4p were detected (Table 1). The affinity of all Mtr4p proteins tested increased threefold when AMPPNP was included in the reactions, suggesting that all Mtr4p proteins were able to bind ATP. Collectively, RNA-binding and ATPase assays showed that the mutation in Mtr4-20p does not affect RNA and presumably ATP binding, making it likely that the isoleucine substitution in motif VI blocks ATP hydrolysis.
TABLE 1.
Mutant Mtr4p can bind tRNA as efficiently as WT Mtr4p
DISCUSSION
In this study, we have revealed the involvement of the DExH RNA helicase Mtr4p in the degradation of hypomethylated tRNAi Met. Our screen for suppressors of the trm6-504 ts− phenotype identified MTR4. Mtr4p is part of the TRAMP complex, which appends short poly(A) tails to RNAs designated for decay (LaCava et al. 2005; Vanacova et al. 2005), but Mtr4p has also been linked to the function of the nuclear exosome in processes other than the degradation of hypomethylated tRNAi Met (de la Cruz et al. 1998; van Hoof et al. 2000). It has thus been unclear whether Mtr4p functions during polyadenylation or at a later step of the RNA degradation process. Here, we have identified a mutant Mtr4p, Mtr4-20p, that impairs degradation of hypomethylated tRNAi Met, but not polyadenylation by the TRAMP complex in vivo or in vitro (Figs. 3, 5). We conclude from these observations that Mtr4p does not function during polyadenylation but facilitates a step at or after initiation of exosome degradation. Such a role would be consistent with the link of Mtr4p to the nuclear exosome (de la Cruz et al. 1998; van Hoof et al. 2000).
Our results raise the possibility that Mtr4p either targets the exosome to the polyadenylated tRNAi Met substrate, and/or disrupts RNA structures in the polyadenylated tRNAi Met to facilitate effective degradation by the exosome. More research is necessary to distinguish between these possibilities. However, our data clearly show that the physiological function of Mtr4p in the degradation of polyadenylated tRNAi Met correlates with the ability of the enzyme to hydrolyze ATP and to unwind RNA duplexes in vitro, as the mutant Mtr4-20p does not display either activity. The ability of Mtr4p to bind RNA, which is also seen in the Mtr4-20p mutant, is also not sufficient for the physiological function of the enzyme in the degradation of hypomethylated tRNAi Met.
The ATP-dependent RNA-unwinding activity of Mtr4p would be consistent with a function of the enzyme in disrupting the structure of the polyadenylated tRNAi Met. Importantly, we have observed a 3′-to-5′ polarity of Mtr4p in the unwinding reaction (Fig. 7). This is notable, as the TRAMP complex polyadenylates pre-tRNAi Met at the 3′-end (Kadaba et al. 2006), thus producing a pre-tRNAi Met with a 30–50-nucleotide (nt) 3′-extension. It is not unreasonable to speculate that polyadenylated hypomodified tRNAi Met is a potential substrate for ATP-driven unwinding by Mtr4p. Future studies will not only need to specifically test this hypothesis, but also probe a potential ability of Mtr4p to recruit the exosome to the polyadenylated tRNAi Met. In fact, it may be advantageous to coordinate putative tRNA unwinding with the exosome activity, to prevent futile rounds of ATP-driven tRNA unfolding by Mtr4p. It would be an elegant solution if one protein controlled both steps.
The observed 3′-to-5′ unwinding polarity of Mtr4p suggests that this cellular DExH protein may function like the viral DExH helicases NPH-II and HCV NS3, both of which unwind RNA duplexes also with a strict 3′-to-5′ polarity (Jankowsky et al. 2000; Seth et al. 2006). Although it would be premature to draw any mechanistic conclusions from the data presented in this study, especially with respect to a possible translocation of Mtr4p, the identified polarity is noteworthy as the DEAD-box proteins generally do not display a preferred unwinding polarity (Yang and Jankowsky 2006; Jankowsky and Fairman 2007). Thus, Mtr4p may unwind RNA duplexes by a mechanism that differs in at least some aspects from the unwinding mode of DEAD-box proteins. The polarity seen with Mtr4p may represent a mechanistic adaptation of the enzyme for directional RNA unfolding to facilitate degradation by the exosome, which also acts in the 3′-to-5′ direction.
MATERIALS AND METHODS
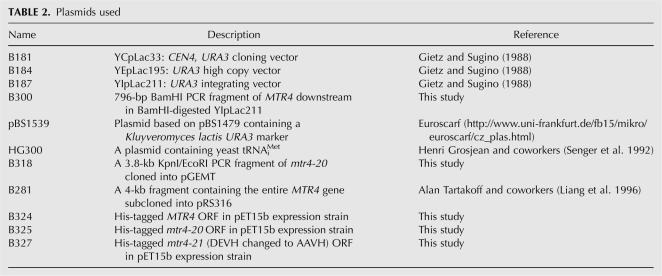
Plasmid construction and mtr4-20 mutant mapping
The plasmids used in this study are given in Table 2. pBS1539 used for PCR reactions in TAP-tagging was kindly provided by the Bertrand Séraphin (Center de Génétique Moléculaire, France). B281 was a generous gift from Alan Tartakoff (Case Western Reserve University, Cleveland, Ohio). HG300 was provided by Henri Grosjean (Laboratoire d'Enzymologie et Biochimie Structurales, France). A 796-base-pair (bp) BamHI fragment downstream from the MTR4 open reading frame (ORF) was obtained by PCR amplification. B300 was created by cloning this fragment into BamHI-digested YIpLac211 (B187). The entire MTR4 ORF together with 500 bp upstream and 300 bp downstream (3.8 kb) was amplified by PCR with Pfu-turbo polymerase (Stratagene) using primers JA283 and JA284 and Sup3 genomic DNA as a template. The PCR product was cloned into pGEM-T (Promega) to create B318. Cloned DNAs were propagated through transformation of E. coli DH10B cells (Invitrogen) by electroporation. B318 was subjected to DNA sequence analysis (MWG Biotech). Five separate PCR reactions encompassing the putative mutation position were conducted with Pfu-turbo using primers JA 280 and JA 297 (Table 4), with Sup3 genomic DNA as a template. The five 1.0-kb PCR products were subjected to DNA sequencing analysis. Plasmids B324 and B325 were created by subcloning the entire ORFs of MTR4 and mtr4-20, obtained by PCR amplification using WT and Sup3 genomic DNA as templates, to the expression vector pET15b (Novagen) using an XhoI site. Mutations in the DEvH-box were created by a PCR-based Quick Change XL Site-directed mutagenesis kit (Stratagene) using plasmid B324 as a template and following the manufacturer's specifications. The resulting plasmid, B327, was subjected to DNA sequence analysis to confirm the presence of the desired mutations.
TABLE 2.
Plasmids used
TABLE 4.
Oligonucleotides used
Yeast strains and media
Standard genetic techniques and culture media were employed (Sherman et al. 1974). The S. cerevisiae strains used in this study are listed in Table 3. Yeast cells were transformed using a lithium chloride method (Ito et al. 1983). Y339 was generated by transformation of trm6-504/sup3 (Sup3) with EcoNI-digested B300 to integrate the URA3 gene within the UBX locus downstream of MTR4. Ura+ transformants were selected on synthetic complete medium lacking uracil (Sc-Ura) medium, and correct integrants were identified by PCR screening with primers JA 239, JA 240, and JA 255 (Table 4). The C-terminal TAP-tagged MTR4 strains Y358 and Y359 were created from Y200 and Sup3 according to the protocol provided by the Séraphin Group (Rigaut et al. 1999). The correct integration of the TAP-tag was verified by PCR analysis and Western blotting with anti-TAP antibody (Open Biosystems).
TABLE 3.
Genotypes of strains used
Identification of SUP3
SUP3 was confirmed to be MTR4 by genetic linkage analysis. Y190 (ura3-52, gcn2-101, trm6-504, sup +) was mated to Y364 (ura3-52, gcn2-101, trm6-504, sup3, ubx∷URA3). The resulting diploids were identified by the inability to mate with either of the mating testers bearing Matα or a genotype. The diploid identified was subjected to sporulation and tetrad analysis. Tetrad analysis on 20 tetrads was done to look for cosegregation of the Ts+, 3-ATs, and uracil phenotypes.
Mtr4p complex (TRAMP) purification
MTR4 TAP-tagged strains were grown in 2.0 L of YPD to OD600 2.0–3.0 and harvested by centrifugation. Cells were resuspended in buffer (10 mM K-HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylmethysulfonyl fluoride (PMSF), 2.0 mM benzamidine, 1.0 μM leupepein, 2.0 μM pepstatin A, 4.0 μM chymostatin, 2.6 μM aprotinin). Cell disruption was conducted by using a French press. The extract was cleared by centrifugation (25,000g, 30 min), and the supernatant was subjected to further centrifugation (100,000g, 1 h). The clarified protein lysate was bound to IgG Sepharose beads (Amersham Biosciences) for 2 h at 4°C, after which the beads were washed three times with 10 mL of IPP 150 [10 mM Tris-Cl at pH 8.0, 150 mM NaCl, 0.1% Nonidet (NP-40)] (Puig et al. 2001). Proteins were recovered by adding TEV protease in TEV cleavage buffer (IPP 150 adjusted to 0.5 mM EDTA and 1.0 mM DTT) and incubating the beads for 2 h at 16°C. The presence of Mtr4p and Trf4p was confirmed by Western blotting using polyclonal antibody against Mtr4p or Trf4p (obtained from Patrick Linder, Université de Genève, Geneva and Michael Christman, Boston University Medical Center, MA, respectively).
RNA isolation, Northern blotting, and purification of poly(A)+ RNA
Total RNA was isolated as described (Kohrer and Domdey 1991). RNAs were separated on a 6% polyacrylamide, 8.0 M urea gel in 1× TBE buffer. The separated RNA was transferred to a HyBond N+ membrane (Pall Biosciences) in the cold at 14 V for 3 h in 0.5× TBE. Blots were probed with radiolabeled deoxyoligonucletides as described previously (Kadaba et al. 2006). The sequences of the oligonucleotides used in this study can be found in Table 4. Poly(A)+ RNA was purified using the protocol described previously (Kadaba et al. 2004).
In vitro polyadenylation assay
Yeast tRNAi Met transcripts were produced by in vitro transcription using HG300 as a template (Senger et al. 1992). Transcripts were treated with calf intestinal phosphatase (CIAP) (Promega) and 5′-end-labeled using polynucleotide kinase and [γ-32P]ATP. The radiolabeled tRNA transcripts were gel-purified on 6% polyacrylamide gels. Polyadenylation assays were performed in a buffer containing 20 mM Tris (pH 7.6), 5.0 mM MgCl2, 50 mM KCl, 2.0 mM DTT, 100 μg/mL BSA, 0.8 U/μl RNasin, and 1.0 mM ATP. Equal amounts of WT and mutant TRAMP complexes judged from the Mtr4p level by immunoblot were used in a reaction. In a standard RNA polyadenylation reaction, ∼80 nM Mtr4p and 100 nM labeled tRNA were incubated together. Reactions were stopped by adding formamide/EDTA gel loading buffer. Products of in vitro polyadenylation were analyzed by electrophoresis on 6% polyacrylamide gels and visualized by autoradiography.
Expression and purification of recombinant Mtr4p
B324, B325, and B327 were grown in 2.0 L of LB medium to A 600 = 0.4 at 37°C. The expression of Mtr4p was induced by addition of 1.0 mM isopropyl-1-thio-β-galactopyranoside (IPTG) (GBiosciences) for 3 h at 30°C. The cells were harvested and disrupted by sonication in equilibration/wash buffer (50 mM sodium phosphate at pH 7.4, 500 mM NaCl, 10 mM β-ME, 10% glycerol, 0.5 mM PMSF). The lysate was clarified by centrifugation at 12,000 rpm for 20 min at 4°C;. 4.0 mL of pre-equilibrated TALON metal affinity resin (Clotech) were incubated with the cell lysate for 2 h at 4°C. The resin was first washed by 10-column volumes of equilibration/wash buffer and then further washed by 10-column volumes of low salt buffer (50 mM NaCl, 50 mM sodium phosphate at pH 7.4, 10 mM β-ME, 10% glycerol); 2.0 mg Mtr4p was eluted in 10 mL of low salt buffer containing 200 mM imidazole. The volume of the Mtr4p sample was reduced to 2.0 mL before being loaded onto a Mono Q anion exchange column (Amersham Biosciences) using a centricon (Millipore). The bound proteins were washed with 5-column volumes of starting buffer (40 mM sodium phosphate at pH 7.4, 50 mM NaCl, 10 mM β-ME, and 10% glycerol), then eluted by development of a salt (NaCl) gradient from 100 to 500 mM in a buffer containing 40 mM sodium phosphate (pH 7.4), 10 mM β-ME, and 10% glycerol. Fractions were analyzed by SDS-PAGE and silver staining. Mtr4p was eluted at ∼150 mM NaCl.
ATPase assay
The reactions were carried out in a buffer containing 20 mM Tris (pH 7.6), 5.0 mM MgCl2, 50 mM KCl, 2.0 mM DTT, 100 μg/mL BSA, 0.8 U/μL RNasin, 5.0 μM ATP, 1.0 μCi of [α-32P]ATP (3000 Ci/mmol), and 500 nM WT or mutant purified recombinant Mtr4p. Then 2.5 or 5.0 μg of E. coli tRNA was added to stimulate ATPase activity. Reactions were incubated at 30°C and stopped by adding 0.1% SDS, 5.0 mM EDTA. In a control reaction, 0.025 U of CIAP was incubated with 1.0 μCi of [α-32P]ATP for 15 min. Aliquots of each reaction were spotted onto a PEI cellulose thin layer chromatography plate (20 cm × 20 cm; Sigma-Aldrich). The plates were developed in a buffer of 0.75 M KH2PO4 (pH 4.2). The plates were dried, and the radiolabeled ATP, ADP, AMP, and Pi were visualized by autoradiography. The amount of ADP was quantified with a Storm 840 PhosphorImager (Molecular Dynamics) and ImageQuant software.
tRNA binding assays
tRNA binding assays were accomplished by incubating 500 nM enzyme with 2.0 nM 32P-end-labeled E. coli tRNA in a total volume of 20 μL in buffer containing 20 mM Tris (pH 7.6), 5.0 mM MgCl2, 50 mM KCl, 2.0 mM DTT, 100 μg/mL BSA, and 0.8 U/μL RNasin. Reactions were incubated for 30 min at 30°C with or without 500 nM AMP-PNP (Sigma-Aldrich) and then put on ice for 10 min. tRNA bound to Mtr4p was collected on a GN-6 metrical membrane disc filter (Pall Biosciences) using a vacuum manifold. Filters were washed with cold buffer, dried, and counted by liquid scintillation.
RNA unwinding reactions
The RNA substrates used in the reactions were prepared as described (Yang and Jankowsky 2005). Substrates were formed as follows (duplex regions are underlined):
S[19]: T19+B44 (5′-AGCACCGUAAAGACGCAGC-3′+5′-GCUGCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA-3′);
S[16]: T16+B41 (5′-AGCACCGUAAAGACGC-3′+5′-GCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA-3′); and
F[16]: T16+B41 (5′-GCGUCUUUACGGUGCU-3′+5′-AAAACAAAACAAAACAAAACAAAAUAGCACCGUAAAGACGC-3′).
Reaction mixtures (30 μL) containing 40 mM NaH2PO4 (pH 6.0), 50 mM NaCl, 0.5 mM MgCl2, 2 mM DTT, 0.8 U/μL RNasin, 0.01% NP-40, and 0.5 nM 32P-labeled RNA substrate were incubated with 800 nM Mtr4p for 5 min at 30°C. At the time indicated, aliquots (3 μL) were removed, and the reaction was stopped with a buffer containing 1% SDS, 50 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue, and 20% glycerol as previously described (Yang and Jankowsky 2005). Aliquots were applied to a 15% nondenaturing polyacrylamide gel, and duplex and single-stranded RNAs were separated at room temperature at 100 V/cm for 1 h. Subsequently, gels were dried, and the radiolabeled RNAs were visualized and quantified with a Molecular Dynamics PhosphorImager and the ImageQuant software (Molecular Dynamics).
ACKNOWLEDGMENTS
We thank Quansheng Yang (Case Western Reserve University) for help with the initial RNA unwinding studies and Sarah Ozanick (Department of Biological Sciences, Marquette University, WI) for reading the manuscript. This work was supported by grants from the NIH to J.T.A. (GM069949) and E.J. (GM067700).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.808608.
REFERENCES
- Anderson, J., Phan, L., Cuesta, R., Carlson, B.A., Pak, M., Asano, K., Bjork, G.R., Tamame, M., Hinnebusch, A.G. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., Hinnebusch, A.G. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz, J., Kressler, D., Tollervey, D., Linder, P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae . EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu, D.E., Henras, A.K., Chanfreau, G.F. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., Sugino, A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base-pair restriction sites. Genes. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K., Phizicky, E.M. tRNA transfers to the limelight. Genes & Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Houseley, J., Tollervey, D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost, I., Dreyfus, M., Linder, P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;18:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukada, Y., Murata, K., Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky, E., Fairman, M.E. RNA helicases—One fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Jankowsky, E., Gross, C.H., Shuman, S., Pyle, A.M. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae . Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Wang, X., Anderson, J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Chen, S., Hitomi, M., Jacobs, E., Kumagai, C., Liang, S., Schneiter, R., Singleton, D., Wisniewska, J., Tartakoff, A.M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer, K., Domdey, H. Preparation of high molecular weight RNA. In: Guthrie C., Fink G.R., editors. Methods in enzymology: Guide to yeast genetics and molecular biology. Academic Press; San Diego, CA: 1991. pp. 398–405. [DOI] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Liang, S., Hitomi, M., Hu, Y.H., Liu, Y., Tartakoff, A.M. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol. Cell. Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. Purification of yeast exosome. Methods Enzymol. 2001;342:356–364. doi: 10.1016/s0076-6879(01)42558-4. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Tollervey, D. An NMD pathway in yeast involving accelerated deadenylation and exosome- mediated 3′ → 5′ degradation. Mol. Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., Tollervey, D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Pause, A., Sonenberg, N. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., Seraphin, B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., Séraphin, B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rocak, S., Linder, P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- Senger, B., Despons, L., Walter, P., Fasiolo, F. The anticodon triplet is not sufficient to confer methionine acceptance to a transfer RNA. Proc. Natl. Acad. Sci. 1992;89:10768–10771. doi: 10.1073/pnas.89.22.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth, R.B., Sun, L., Chen, Z.J. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., Lawrence, C.W. Methods of yeast genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1974. [Google Scholar]
- Tanner, N.K., Linder, P. DExD/H Box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:986–997. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Lennertz, P., Parker, R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, S.L., Matera, A.G. The trials and travels of tRNA. Genes & Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- Wyers, F., Rougemaille, M., Badis, G., Rousselle, J.-C., Dufour, M.-E., Boulay, J., Rgnault, B., Devaux, F., Namane, A., Sraphin, B. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Jankowsky, E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Jankowsky, E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]