Abstract
The replication-dependent histone mRNAs in metazoa are not polyadenylated, in contrast to the bulk of mRNA. Instead, they contain an RNA stem–loop (SL) structure close to the 3′ end of the mature RNA, and this 3′ end is generated by cleavage using a machinery involving the U7 snRNP and protein factors such as the stem–loop binding protein (SLBP). This machinery of 3′ end processing is related to that of polyadenylation as protein components are shared between the systems. It is commonly believed that histone 3′ end processing is restricted to metazoa and green algae. In contrast, polyadenylation is ubiquitous in Eukarya. However, using computational approaches, we have now identified components of histone 3′ end processing in a number of protozoa. Thus, the histone mRNA stem–loop structure as well as the SLBP protein are present in many different protozoa, including Dictyostelium, alveolates, Trypanosoma, and Trichomonas. These results show that the histone 3′ end processing machinery is more ancient than previously anticipated and can be traced to the root of the eukaryotic phylogenetic tree. We also identified histone mRNAs from both metazoa and protozoa that are polyadenylated but also contain the signals characteristic of histone 3′ end processing. These results provide further evidence that some histone genes are regulated at the level of 3′ end processing to produce either polyadenylated RNAs or RNAs with the 3′ end characteristic of replication-dependent histone mRNAs.
Keywords: histone, stem–loop, U7 RNA, U7 snRNP, SLBP, stem–loop binding protein, protozoa, RNA, evolution
INTRODUCTION
Eukaryotic mRNAs typically contain poly(A) tails at their 3′ ends, mediating important functions in export, stability, and translation. Formation of the poly(A) tail occurs in a two-step reaction in which first the pre-mRNA is cleaved at a site defined by two signals, a highly conserved upstream AAUAAA sequence and a downstream G/U-rich sequence. In a second step, the poly(A) tail is added to the 3′ end of the RNA. A complex machinery is involved in these reactions including the cleavage/polyadenylation specificity factor (CPSF) and the cleavage stimulation factor (CstF). A subunit of CPSF, CPSF-73, has been shown to be the enzyme responsible for the cleavage reaction (Dominski et al. 2005).
The metazoan replication-dependent histone mRNAs are unusual as they are the only eukaryotic mRNAs that lack poly(A) tails. These RNAs are produced mainly in the S-phase of somatic cells and produce histones to package newly replicated DNA into chromatin. Typically the replication-dependent histone mRNAs lack introns, and their genes are arranged in clusters. A small number of histone proteins like H3.3, H2a.Z, H1°, H3-cid, and macroH2a are encoded by polyadenylated mRNAs. This class of mRNAs encodes the variant histones used for chromatin remodeling and repair (Ausio 2006). They are expressed throughout the cell cycle (Wu and Bonner 1981), and many of their genes contain introns and are not arranged in clusters.
The 3′ end processing of replication-dependent histone mRNAs takes place with a mechanism distinct from that of polyadenylation. Processed histone mRNAs contain a stem–loop structure at their 3′ ends. This end is formed by specific machinery that recognizes features of the histone pre-mRNA, the stem–loop as well a downstream purine-rich element (HDE) 15–20 nucleotides (nt) downstream from the stem–loop (Marzluff and Duronio 2002). The stem–loop sequence is highly conserved in metazoa and consists of a stem with 6 base pairs (bp) and a 4-nt loop. Cleavage occurs between these two elements, after the fourth or fifth nucleotide, which is typically an adenosine.
A number of transacting factors are also involved in histone mRNA processing (Dominski and Marzluff 2007). One of these factors is the U7 snRNP that interacts with the HDE element where a 5′ terminal portion of the U7 RNA forms base-pairing interactions with the HDE. The 3′ part of the U7 RNA forms a helical structure, and the central part of U7 RNA contains an Sm site, a site where the Sm protein core assembles. The U7 snRNP contains a number of subunits shared with spliceosomal snRNPs, referred to as B, D3, G, E, and F. However, it also contains the subunits Lsm10 and Lsm11 specific to the U7 snRNP and that replace D1 and D2 in the spliceosomal snRNP (Pillai et al. 2001; Pillai et al. 2003).
The histone mRNA stem–loop is a binding site for the stem–loop binding protein, SLBP (Wang et al. 1996). This protein has a centrally located RNA-binding domain. The SLBP probably stabilizes binding of U7 snRNP to the HDE (Melin et al. 1992; Streit et al. 1993; Spycher et al. 1994; Dominski et al. 1999). Yet another protein interacting with SL is the 3′hExo protein (Dominski et al. 2003). The zinc finger protein ZFP-100 bridges between the SLBP and the U7-specific protein Lsm11 (Dominski et al. 2002). Another factor essential in histone 3′ end processing is symplekin (Kolev and Steitz 2005), also involved in polyadenylation (Takagaki and Manley 2000; Barnard et al. 2004; Xing et al. 2004). It occurs in a complex with subunits of CPSF and CstF. The actual histone mRNA cleavage reaction is performed by the CPSF-73 subunit (Dominski et al. 2005), the same protein responsible for cleavage in the context of polyadenylation. This means that at least two components are shared between the polyadenylation and histone 3′ end processing machineries, CPSF-73 and symplekin. Another feature shared by polyadenylation and histone 3′ end processing is their tight association with transcription termination (Chodchoy et al. 1991; Buratowski 2005).
Whereas normally the replication-dependent histone genes are not polyadenylated, the RNA transcribed from a few histone genes may be processed either by polyadenylation or by the mechanism characteristic of histone mRNA. Thus, transcripts have been found that are polyadenylated and that also contained the SL and HDE motifs (Challoner et al. 1989; Cheng et al. 1989; Kirsh et al. 1989; Mannironi et al. 1989; Collart et al. 1992; Moss et al. 1994; Wang et al. 1997; Lanzotti et al. 2002). Thus, it would seem that under certain conditions the histone mRNA processing signals are disregarded and instead the RNA is polyadenylated.
As histone mRNA specific processing is mechanistically related to polyadenylation, an interesting question is how these processing events are evolutionarily related. It is commonly believed that histone 3′ end processing is restricted to metazoa and green algae (Dominski and Marzluff 2007). This assumption is mainly based on the observation that histone mRNAs of protozoa, plants, and yeast are polyadenylated (Chaubet et al. 1988; Liu and Gorovsky 1993; Aslund et al. 1994; Sanchez et al. 1994). In contrast, polyadenylation is present in all eukaryotes. It would therefore seem that the polyadenylation machinery is more ancestral than that of histone 3′ end processing. However, we have now examined computationally the occurrence of the histone stem–loop structure in a variety of organisms and have shown that it is present in histone mRNAs in a number of additional species, including protozoa and a few species of Streptophyta. In addition, most of these species have an SLBP homolog. These findings have important implications as to the evolution of histone 3′ end processing and the relationship of this machinery to that of polyadenylation.
RESULTS AND DISCUSSION
A 3′ end stem–loop is present in a subset of nonmetazoan eukaryotic histone genes
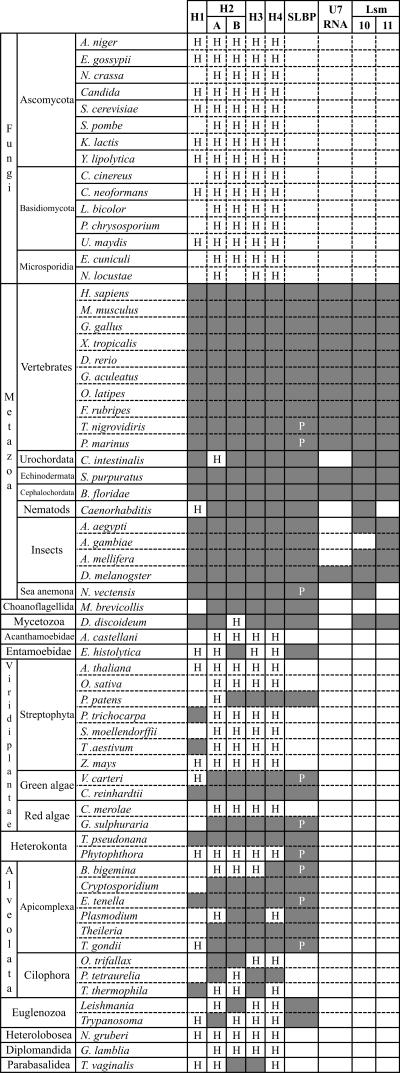
Histone mRNA stem–loops, hereafter referred to as SL motifs, have been reported previously in metazoa and green algae, but searches for these motifs have not been carried out in a systematic fashion. We therefore developed a procedure in order to mine genomic sequence databases for SL motifs as well as to examine whether they are present in other eukaryotes than metazoa and green algae. First, a collection of 8302 sequences downstream from histone coding sequences was assembled from the organisms listed in Figure 1 (for more details, see Supplemental Material Document 2). This was carried out as described under Materials and Methods with BLAST searches using as queries a collection of previously known histone protein (H1, H2A, H2B, H3, and H4) sequences. As some of the histone variants (Ausio 2006) are very similar to the replication-dependent histones, we were not able to distinguish these two categories for most of the organisms considered here. As a result, a fraction of sequences in our data set corresponds to the histone variants.
FIGURE 1.
Phylogenetic distribution of components involved in histone 3′ end formation. The distribution of identified stem–loop motifs in regions downstream from histone coding sequences are shown as well as SLBP, U7 RNA, Lsm10, and Lsm11. In the case of histones, “H” indicates that a histone downstream sequence was identified but no stem–loop motif. Shaded boxes represent cases in which a histone downstream sequence including a stem–loop motif was identified, and empty boxes are cases in which no histone or histone downstream sequence at all could be identified. For the SLBP, a “P” indicates that the available protein sequence covers only the RNA-binding domain. In instances where a genus name only is indicated, multiple species were analyzed: C. elegans and Caenorhabditis briggsae; Phytophthora infestans and P. sojae; C. hominis and C. parvum; Plasmodium berghei, Plasmodium chabaudi, P. falciparum, and Plasmodium yoelii; T. annulata and T. parva; Leishmania infantum and Leishmania major; Trypanosoma brucei and T. cruzi; Candida albicans, Candida glabrata, and Candida tropicalis.
SL motifs in the histone downstream regions were predicted by applying a combination of pattern searches and the Infernal software (Eddy 2002) together with the SL motif covariance model of Rfam (Griffiths-Jones et al. 2003) or models created from multiple sequence alignments when the existing Rfam model was not adequate. We identified an SL in 4208 of the 8302 sequences referred to above (Supplemental Material Documents 3 and 4).
In some of the organisms, we failed to identify a particular histone gene or a histone gene with an accompanying SL. However, some of the genomes are not fully sequenced, and we cannot exclude that the histone and an accompanying SL will be found once the genome is complete. In a number of protozoa, for instance, in Cryptosporidium, Theileria, and Oxytricha, we are missing H1 histone genes, perhaps because these particular genes are present in a lower copy number and for this reason are less likely to be identified when the genome sequence is not complete. It is also possible that the H1 sequences escape detection because they are strongly divergent in these organisms.
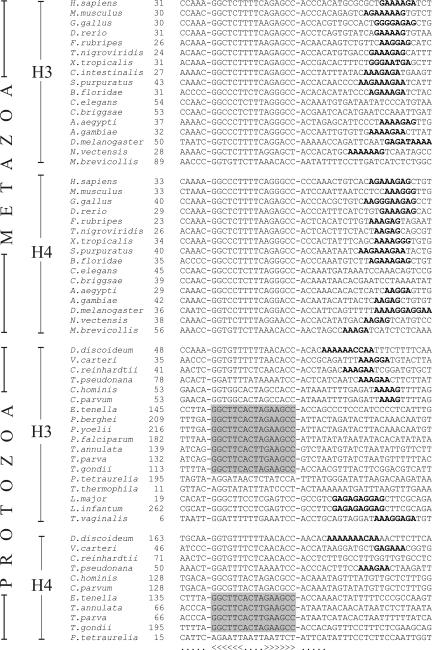
An overview of the occurrence of the SL motif is shown in Figure 1, and selected H3 and H4 sequences containing this motif as well as the downstream purine-rich HDE element are shown in Figure 2 (complete collection in Supplemental Material Document 3). With few exceptions, an SL is identified in all metazoa for all the histone types (Fig. 1). We do not know what characterizes the histone genes without an SL, but in metazoa many of these should correspond to the histone variants.
FIGURE 2.
Selected stem–loop motifs of metazoa and protozoa. Selected stem–loop motifs of H3 and H4 histone genes were manually aligned. A more comprehensive collection of sequences are in the Supplemental Material Document 3. Potential HDE regions are shown in boldface, and the number following the species name is the distance between the stop codon and the first nucleotide of the conserved stem–loop motif. Regions in stem–loop motifs that are identical in a group of alveolates are shown with a shaded background. Full species names are in Figure 1.
Not only were SL motifs identified in metazoa, but also in many protozoa (Fig. 2; Supplemental Material Documents 3 and 4). Out of 2417 protozoan histone downstream sequences, 318 contained an SL motif. Examples for H3 and H4 histones are shown in Figure 2, but genes for all the other histone classes were found to have SL motifs (Fig. 1). We regard these as very strong predictions because they scored well with the covariance model and the presence of SL got strong support from comparative genomics. For example, in the group of the alveolates Eimeria, Plasmodium, Theileria, and Toxoplasma, the SL sequence is strongly conserved compared to the flanking sequences as indicated in Figure 2 (for more details, see Supplemental Material Document 3). The reason that all the novel SL motifs previously escaped detection (and are lacking in Rfam) is mainly that the primary sequences strongly diverge from the vertebrate SL motifs.
A comparison between metazoa and protozoa shows that the SL motifs are less frequent in the protozoa. SL motifs are, for instance, very rare among histone genes in Oxytricha, Phytophthora, and Trypanosoma. It therefore seems reasonable to assume that the majority of histone transcripts in these organisms are polyadenylated, consistent with previous reports (Chaubet et al. 1988; Liu and Gorovsky 1993; Aslund et al. 1994; Sanchez et al. 1994). Another difference is that in metazoa, the distance between the stop codon and SL is fairly well conserved, whereas this distance in protozoa is much more variable (Fig. 2).
In addition to the SL of the green algae Volvox and Chlamydomonas that were previously identified (Osley 1991; Fabry et al. 1995), we identified an SL in H1 genes from Triticum aestivum and Populus trichocarpa. We also identified an SL candidate in H2B, H3, and H4 genes of the moss Physcomitrella patens but not in any other plant species. The significance of these results is not clear but could suggest that there are remnants from the histone mRNA processing machinery in these species of the Streptophyta group.
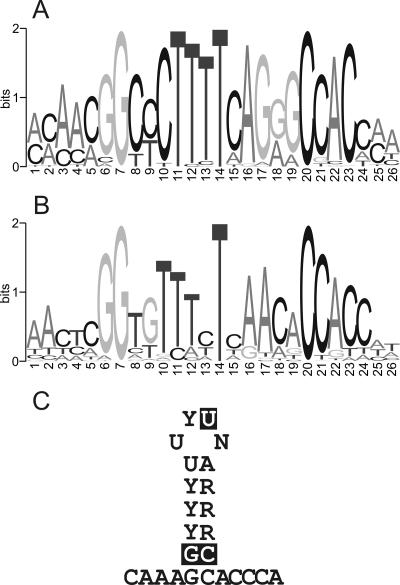
No SL motif at all could be identified in Giardia lamblia, Naegleria gruberi, Acanthamoeba castellani, and Cyanidioschyzon merolae. This could be because SL motifs are even more strongly divergent in these organisms and escape detection using our method. However, we find it more likely that such motifs are absent in these species, which is also consistent with the absence of an SLBP homolog as discussed below. It would therefore seem that these organisms are lacking a machinery for histone mRNA processing. In addition, an SL motif could not be identified in any of the fungal histone genes that were analyzed (Fig. 1). Conserved features of metazoan and protozoan SL motifs are summarized in Figure 3. Typically, the loop of SL has 4 nt. An exception is Dictyostelium discoideum, where the loop of H1 has 5 nt. The strongly conserved G-C pair at the base of the stem is occasionally changed to an A-U pair, as in Tetrahymena and in insects.
FIGURE 3.
Conserved features of histone stem–loop motifs. Sequence logos of histone SL motifs in (A) metazoa and in (B) protozoa. (C) Conserved bases in the stem–loop motif are shown in the context of the secondary structure; the most highly conserved positions are shown with a black background.
A purine-rich histone downstream element (HDE) has been proposed to pair with a region of the U7 RNA. With the exception of Caenorhabditis, such an HDE region was identified in all histone downstream regions of metazoa with an SL element (Fig. 2; Supplemental Material Document 3). In the protozoa, such a purine-rich region is less common, with exceptions of heterokonta, Volvox, and Trypanosoma. The absence of a metazoan type of HDE indicates that the corresponding protozoan U7 RNA, if such an RNA is at all present, is different from its metazoan homologs.
The SL motif is specific to histone mRNAs
To address the question if the SL motif is restricted to histone mRNAs, we also searched non-histone mRNAs for SL motifs. For this study, we analyzed all sequences annotated as 3′ UTRs in the EMBL database with a pattern-based search combined with Infernal searches. SL motifs were not identified in any non-histone mRNAs, providing strong evidence that the SL motif is very specific to histone mRNA. This result is consistent with the observation that SLBP only binds to histone mRNAs (Townley-Tilson et al. 2006).
EST sequences provide evidence that some histone genes could be subject to either histone-specific processing or to polyadenylation
There is evidence that some histone genes in vertebrates may be either processed as determined by the SL and HDE elements, or processed using the polyadenylation machinery (Challoner et al. 1989; Cheng et al. 1989; Kirsh et al. 1989; Mannironi et al. 1989; Collart et al. 1992; Moss et al. 1994; Wang et al. 1997; Lanzotti et al. 2002). We examined publicly available EST sequences in order to find more examples of such cases and to study whether RNAs with poly(A) tails as well as SL/HDE signals are present also in protozoa. ESTs coding for histones were identified using BLAST (Altschul et al. 1990), and the presence of poly(A) tails (as defined here by a sequence of 20 or more consecutive As at the 3′ end), polyadenylation signals (Beaudoing et al. 2000), and SL motifs was examined. We identified ESTs with a poly(A) tail as well as SL/HDE signals from Mus musculus, Danio rerio, Xenopus tropicalis, Strongylocentrotus purpuratus, Drosophila melanogaster, D. discoideum, Plasmodium falciparum, and Theileria annulata as listed in Table 1. Table 1 also shows previously described polyadenylated histone mRNAs with SL motifs. In addition, we analyzed a histone mRNA sequence from Entamoeba histolytica reported by Sanchez et al. (1994) to be polyadenylated and found that it also has an SL motif. Because relatively few ESTs/mRNAs of this category have been identified (Table 1), the co-occurrence of an SL and a poly(A) tail is probably a rare event. Nevertheless, these results show that a few histone genes may be alternatively processed.
TABLE 1.
Histone mRNAs with both a stem–loop motif and a poly(A) tail
Homologs to the metazoan SLBP protein are identified in protozoa
Homologs of SLBP have been identified in human, mouse, frog, fruit fly, and Caenorhabditis elegans (Wang et al. 1996; Michel et al. 2000; Sullivan et al. 2001). These SLBP homologs are known to contain a functionally important and centrally located RNA-binding domain, i.e., a domain interacting with the stem–loop structure. This domain is also the most strongly conserved part of the protein. For instance, C. elegans SLBP shares a high identity to mouse SLBP only at the RBD (Kodama et al. 2002). It has also been shown that the minimal version of the human protein that efficiently supports cleavage of histone pre-mRNA and interacts with ZFP-100 consists of 93 amino acids containing the 73 amino acid RNA binding domain and 20 amino acids downstream (Dominski et al. 1999, 2002).
We noted that BLAST searches using metazoan SLBP as queries identified possible protozoan homologs and used PSI-BLAST (Altschul et al. 1997; Altschul and Koonin 1998) as described under Materials and Methods to more efficiently identify SLBP homologs. The species in which SLBP homologs were identified are shown in Figure 1 (SLBP protein sequences are in Supplemental Material Document 5). The RDB portion of SLBP is the region conserved, and a multiple sequence alignment of this domain of selected metazoa and protozoa is shown in Figure 4.
FIGURE 4.
Conserved RNA-binding domain of SLBP homologs. Multiple alignment of stem–loop binding protein (SLBP) homologs was produced with T-Coffee (Notredame et al. 2000) and visualized with Jalview (Clamp et al. 2004). Full species names are in Figure 1.
In a number of protozoa, D. discoideum, Volvox carteri, Chlamydomonas reinhardtii, Thalassiosira pseudonana, five different Apicomplexa, Leishmania, Trypanosoma, and E. histolytica, we were able to identify both SL and SLBP, providing strong evidence that these organisms have a 3′ end processing machinery related to that in metazoa. Conversely, no SL motif or SLBP protein could be identified in G. lamblia, N. gruberi, A. castellani, and C. merolae, suggesting that an SL dependent processing is absent in these organisms. Ambiguous cases are Phytophthora, Oxytricha, Paramecium, and Trichomonas, where we identified SL motifs but no SLBP. However, in these cases, we could be missing the protein because the genome sequences for these organisms are not yet complete. In addition, we cannot exclude the possibility that for any organisms where we have failed to identify an SLBP homolog, another protein with a similar structure could be functionally replacing SLBP.
Novel U7 snRNA sequences and potential pairing to histone mRNA
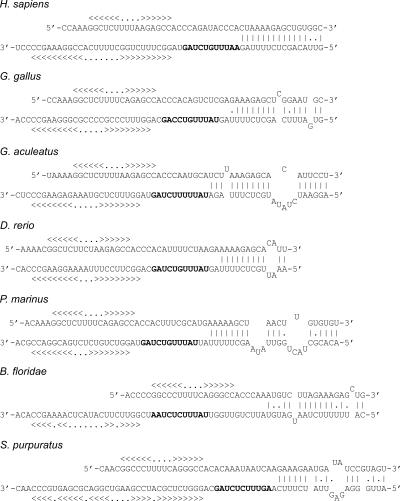
The U7 snRNP takes part in histone 3′ end processing, and a region of the U7 RNA pairs with the histone mRNA HDE sequence (Fig. 5). Previously homologs to U7 RNA were identified in mammals, sand urchin, and insects (Dominski and Marzluff 1999). We used BLAST or FASTA to identify potential U7 RNA candidates in a number of metazoan genomes and examined these using a covariance model of U7 RNA. This procedure resulted in novel homologs, including those from a number of teleosts and Petromyzon marinus (sea lamprey), S. purpuratus, and Branchiostoma floridae (Fig. 1; Supplemental Material Document 7). The resulting RNAs are reliable predictions because they show strong primary sequence similarity to previously known U7 RNAs, and they have the structural properties of previously known U7 RNAs, i.e., an Sm site, a hairpin loop, and a region pairing with the HDE (Fig. 5). In many organisms, more than one U7 RNA gene candidate was found. The prediction of U7 RNA in protozoa was very unreliable, and we were not able to reach a conclusion as to the presence of a U7 RNA in these phylogenetic groups.
FIGURE 5.
Proposed secondary structure of U7 RNA orthologs and possible interaction with histone mRNA. Upper sequences are histone mRNA sequences, and lower sequences are U7 RNAs. The U7 RNA sequence from Homo sapiens (Mowry and Steitz 1987) is shown, as well as U7 sequences not previously reported from Gallus gallus, D. rerio, G. aculeatus, P. marinus, B. floridae, and S. purpuratus. Helices in the SL motif as well as in U7 RNA are shown with a bracket notation. Potential Sm sites in U7 RNA are shown in boldface. See Supplemental Figure 7 for more detailed information on the origin of U7 RNA sequences.
Potential pairing of some of the different U7 RNAs to a histone HDE region is shown in Figure 5. In these examples, the mRNA with the best match to the HDE region was selected. These results give support to an evolutionarily conserved pairing between U7 RNA and HDE, although the pairing is not as extensive as in human.
Identification of Lsm10 and Lsm11 orthologs
In human the Lsm10 and Lsm11 proteins are known to be specific to the U7 snRNP. Using profile-based searches, we identified these proteins in metazoa but not in protozoa, with the exception of Dictyostelium (Fig. 1). On the basis of these findings it is interesting to note that the Lsm proteins and U7 RNA seem to have a similar phylogenetic distribution (Fig. 1), but it is premature to conclude that a U7 snRNP is missing in the protozoa. If there is a U7 RNA in the protozoa, it probably has properties different from the metazoan U7 RNA as, with few exceptions, the histone mRNA does not seem to have a HDE region characteristic of metazoa.
Evolution of histone 3′ end processing
In conclusion, we have demonstrated that important components of histone 3′ end processing are present in many different protozoa. Thus, both the SL motif and SLBP are present in a set of protozoa, including the very deeply branching Trichomonas and Euglenozoa, i.e., organisms that are close to the root of the eukaryotic tree (Baldauf 2003; Steenkamp et al. 2006). It therefore seems highly likely that these elements of histone 3′ end processing developed very early in eukaryotic evolution.
There are mechanistic links between histone 3′ end processing and polyadenylation as protein components are shared between the two systems. At the same time they are conceptually different in the sense that an snRNP is involved in histone mRNA processing. As both polyadenylation and histone mRNA processing now may be traced to the root of the eukaryotic tree, we are not able to reach a conclusion as to which of the two mechanisms developed first. Specifically, our results do not offer support to a model of evolution in which histone 3′ end processing was developed from the polyadenylation machinery (Dominski and Marzluff 2007), although such a mode of evolution cannot be excluded.
We have also observed that the SL motifs and SLBP are missing in most plants, in fungi, and in some protozoa. Furthermore, in the protozoa with SL motifs, these motifs are less frequent. Therefore, we favor a model of evolution where the histone 3′ end processing developed very early, but where this machinery was partially or completely lost in the development of protozoa, plants, and fungi.
MATERIALS AND METHODS
Sources of genomic and protein sequences
Genomic sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov/entrez/; ftp.ncbi.nih.gov/genomes), EMBL (http://www.ebi.ac.uk), ENSEMBL (http://www.ensembl.org), and TraceDB (ftp.ncbi.nlm.nih.gov/pub/TraceDB). In the case of Gasterosteus aculeatus, D. rerio, and Oryzias latipes, we used the ENSEMBL versions v46.1d, v46.7, and v46.1c, respectively. Sequences of Fugu rubripes (v4.0), B. floridae (v1.0), X. tropicalis (v4.0), and Nematostella vectensis were from the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). P. marinus (v3.0) data were from the WU Genome Sequencing Center (http://genome.wustl.edu/). In addition, we made use of genome sequence data of Plasmodium (PlasmoDB at http://www.plasmodb.org), Trypanosoma cruzi, E. histolytica, Theileria parva, Toxoplasma gondii, and Trichomonas vaginalis (TIGR), Cryptosporidium hominis, and Cryptosporidium parvum (http://www.cryptodb.org/cryptodb/), Leishmania species (http://www.sanger.ac.uk), C. reinhardtii (http://genome.jgi-psf.org/chlre2/chlre2.home.html), and G. lamblia (http://www.jbpc.mbl.edu/Giardia-HTML/index2.html). Expressed sequence tags (ESTs) were downloaded from the dbEST at NCBI (http://www.ncbi.nlm.nih.gov/dbEST/), and protein sequences were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/blast/db/).
Identification of the stem–loop motif downstream from histone coding sequences
A set of the histone protein (H1, H2A, H2B, H3, and H4) sequences was assembled (Supplemental Material Document 1) and used as queries with TBLASTN (Altschul et al. 1990) searches against genomic sequences to locate histone genes and to classify them as H1, H2A, H2B, H3, or H4. A script was developed to identify the exact position of the stop codon in the histone coding sequence. A portion 12 nt upstream and 300 nt downstream of the stop codon was extracted from the significant hits of the TBLASTN searches. To identify SL motifs, the covariance model of the metazoan SL from Rfam (RF00032; http://www.sanger.ac.uk/Software/Rfam/) was used with cmsearch of the Infernal package (Eddy 2002; Griffiths-Jones et al. 2003). Typically, a filtering step was first performed with pattern searches using rnabob (http://www.genetics.wustl.edu/eddy/softare/#rnabob). A subset of protozoan SL motifs could only be found using a new covariance model that was constructed from stem–loop motifs of protozoa, initially found with the metazoan model. Multiple alignments of sequences downstream from histone coding sequences were created using ClustalW 1.83 (Thompson et al. 1994) or T-Coffee (Notredame et al. 2000). Conserved elements were identified with MEME (Bailey et al. 2006).
Analysis of histone ESTs
Histone sequences in the EST database were identified with TBLASTN (Altschul et al. 1990) searches with the set of histone proteins referred to above as queries. An E-value of 1e-5 was used as the threshold. The presence of poly(A) tails was examined in addition to downstream polyadenylation signals (Beaudoing et al. 2000). The SL motifs were identified as described above.
Identification of protein homologs
PSI-BLAST (Altschul et al. 1997; Altschul and Koonin 1998) was used to identify homologs to the SLBP, Lsm10, and Lsm11 proteins. The default E-value of 0.001 was used as the threshold for inclusion in PSI-BLAST iterations. The database used was the NCBI GenBank set of proteins (Benson et al. 2006). Most of the SLBP homologs from protozoa were not in the protein sequence databases but were identified by TBLASTN (Altschul et al. 1990) searches of genome sequences. Multiple alignments were created using T-Coffee (Notredame et al. 2000) and visualized with JalView (Clamp et al. 2004).
Identification of U7 snRNA
Novel U7 RNA homologs were identified using a two-step procedure in which, first, potential candidates were detected using high sensitivity with the sequence homology-based methods BLAST, FASTA (Pearson 2000), profile matching using hmmsearch (HMMER package), or pattern-based matching using rnabob. In a second step, the candidates were tested using cmsearch of the Infernal package. The resulting U7 RNA predictions were also checked for conserved primary sequence motifs and the ability to fold into a secondary structure typical for U7 snRNA. Secondary structure predictions were carried out by MFOLD (Zuker 1989). In order to examine possible pairing between U7 RNA and the histone mRNA HDE region, these two sequences were concatenated and folded with MFOLD using appropriate constraints.
SUPPLEMENTAL DATA
Supplemental Materials can be found at http://bio.lundberg.gu.se/sl/.
ACKNOWLEDGMENTS
M.D.L. was supported by a grant from CONACYT, The National Council for Science and Technology, Mexico.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.782308.
REFERENCES
- Altschul, S.F., Koonin, E.V. Iterated profile searches with PSI-BLAST—A tool for discovery in protein databases. Trends Biochem. Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslund, L., Carlsson, L., Henriksson, J., Rydaker, M., Toro, G.C., Galanti, N., Pettersson, U. A gene family encoding heterogeneous histone H1 proteins in Trypanosoma cruzi . Mol. Biochem. Parasitol. 1994;65:317–330. doi: 10.1016/0166-6851(94)90082-5. [DOI] [PubMed] [Google Scholar]
- Ausio, J. Histone variants—The structure behind the function. Brief. Funct. Genomic. Proteomic. 2006;5:228–243. doi: 10.1093/bfgp/ell020. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L., Williams, N., Misleh, C., Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf, S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Barnard, D.C., Ryan, K., Manley, J.L., Richter, J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Beaudoing, E., Freier, S., Wyatt, J.R., Claverie, J.M., Gautheret, D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Wheeler, D.L. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski, S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Challoner, P.B., Moss, S.B., Groudine, M. Expression of replication-dependent histone genes in avian spermatids involves an alternate pathway of mRNA 3′ end formation. Mol. Cell. Biol. 1989;9:902–913. doi: 10.1128/mcb.9.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubet, N., Chaboute, M.E., Clement, B., Ehling, M., Philipps, G., Gigot, C. The histone H3 and H4 mRNAs are polyadenylated in maize. Nucleic Acids Res. 1988;16:1295–1304. doi: 10.1093/nar/16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, G.H., Nandi, A., Clerk, S., Skoultchi, A.I. Different 3′ end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc. Natl. Acad. Sci. 1989;86:7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodchoy, N., Pandey, N.B., Marzluff, W.F. An intact histone 3′-processing site is required for transcription termination in a mouse histone H2a gene. Mol. Cell. Biol. 1991;11:497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp, M., Cuff, J., Searle, S.M., Barton, G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Collart, D., Romain, P.L., Huebner, K., Pockwinse, S., Pilapil, S., Cannizzaro, L.A., Lian, J.B., Croce, C.M., Stein, J.L., Stein, G.S. A human histone H2B.1 variant gene, located on chromosome 1, utilizes alternative 3′ end processing. J. Cell. Biochem. 1992;50:374–385. doi: 10.1002/jcb.240500406. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Marzluff, W.F. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Marzluff, W.F. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Zheng, L.X., Sanchez, R., Marzluff, W.F. Stem–loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Erkmann, J.A., Yang, X., Sanchez, R., Marzluff, W.F. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem–loop binding protein in the histone pre-mRNP to stimulate 3′ end processing. Genes & Dev. 2002;16:58–71. doi: 10.1101/gad.932302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.C., Kaygun, H., Dadlez, M., Marzluff, W.F. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.C., Marzluff, W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics. 2002;3:18–33. doi: 10.1186/1471-2105-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry, S., Muller, K., Lindauer, A., Park, P.B., Cornelius, T., Schmitt, R. The organization structure and regulatory elements of Chlamydomonas histone genes reveal features linking plant and animal genes. Curr. Genet. 1995;28:333–345. doi: 10.1007/BF00326431. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A., Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsh, A.L., Groudine, M., Challoner, P.B. Polyadenylation and U7 snRNP-mediated cleavage: Alternative modes of RNA 3′ processing in two avian histone H1 genes. Genes & Dev. 1989;3:2172–2179. doi: 10.1101/gad.3.12b.2172. [DOI] [PubMed] [Google Scholar]
- Kodama, Y., Rothman, J.H., Sugimoto, A., Yamamoto, M. The stem–loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans . Development. 2002;129:187–196. doi: 10.1242/dev.129.1.187. [DOI] [PubMed] [Google Scholar]
- Kolev, N.G., Steitz, J.A. Symplekin and multiple other polyadenylation factors participate in 3′ end maturation of histone mRNAs. Genes & Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti, D.J., Kaygun, H., Yang, X., Duronio, R.J., Marzluff, W.F. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol. Cell. Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Gorovsky, M.A. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE) Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannironi, C., Bonner, W.M., Hatch, C.L. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3′ processing signals. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W.F., Duronio, R.J. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- Melin, L., Soldati, D., Mital, R., Streit, A., Schumperli, D. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F., Schumperli, D., Muller, B. Specificities of Caenorhabditis elegans and human hairpin binding proteins for the first nucleotide in the histone mRNA hairpin loop. RNA. 2000;6:1539–1550. doi: 10.1017/s135583820000056x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, S.B., Ferry, R.A., Groudine, M. An alternative pathway of histone mRNA 3′ end formation in mouse round spermatids. Nucleic Acids Res. 1994;22:3160–3166. doi: 10.1093/nar/22.15.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry, K.L., Steitz, J.A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Osley, M.A. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Will, C.L., Luhrmann, R., Schumperli, D., Muller, B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Grimmler, M., Meister, G., Will, C.L., Luhrmann, R., Fischer, U., Schumperli, D. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes & Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, L.B., Enea, V., Eichinger, D. Increased levels of polyadenylated histone H2B mRNA accumulate during Entamoeba invadens cyst formation. Mol. Biochem. Parasitol. 1994;67:137–146. doi: 10.1016/0166-6851(94)90103-1. [DOI] [PubMed] [Google Scholar]
- Spycher, C., Streit, A., Stefanovic, B., Albrecht, D., Koning, T.H., Schumperli, D. 3′ end processing of mouse histone pre-mRNA: Evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp, E.T., Wright, J., Baldauf, S.L. The protistan origins of animals and fungi. Mol. Biol. Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Streit, A., Koning, T.W., Soldati, D., Melin, L., Schumperli, D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993;21:1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, E., Santiago, C., Parker, E.D., Dominski, Z., Yang, X., Lanzotti, D.J., Ingledue, T.C., Marzluff, W.F., Duronio, R.J. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes & Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki, Y., Manley, J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley-Tilson, W.H., Pendergrass, S.A., Marzluff, W.F., Whitfield, M.L. Genome-wide analysis of mRNAs bound to the histone stem–loop binding protein. RNA. 2006;12:1853–1867. doi: 10.1261/rna.76006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.F., Whitfield, M.L., Ingledue T.C., III, Dominski, Z., Marzluff, W.F. The protein that binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Wang, Z.F., Sirotkin, A.M., Buchold, G.M., Skoultchi, A.I., Marzluff, W.F. The mouse histone H1 genes: Gene organization and differential regulation. J. Mol. Biol. 1997;271:124–138. doi: 10.1006/jmbi.1997.1166. [DOI] [PubMed] [Google Scholar]
- Wu, R.S., Bonner, W.M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981;27:321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Xing, H., Mayhew, C.N., Cullen, K.E., Park-Sarge, O.K., Sarge, K.D. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J. Biol. Chem. 2004;279:10551–10555. doi: 10.1074/jbc.M311719200. [DOI] [PubMed] [Google Scholar]
- Zuker, M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]